Enhancing Retinal Blood Vessel Segmentation through Self-Supervised Pre-Training †
Abstract
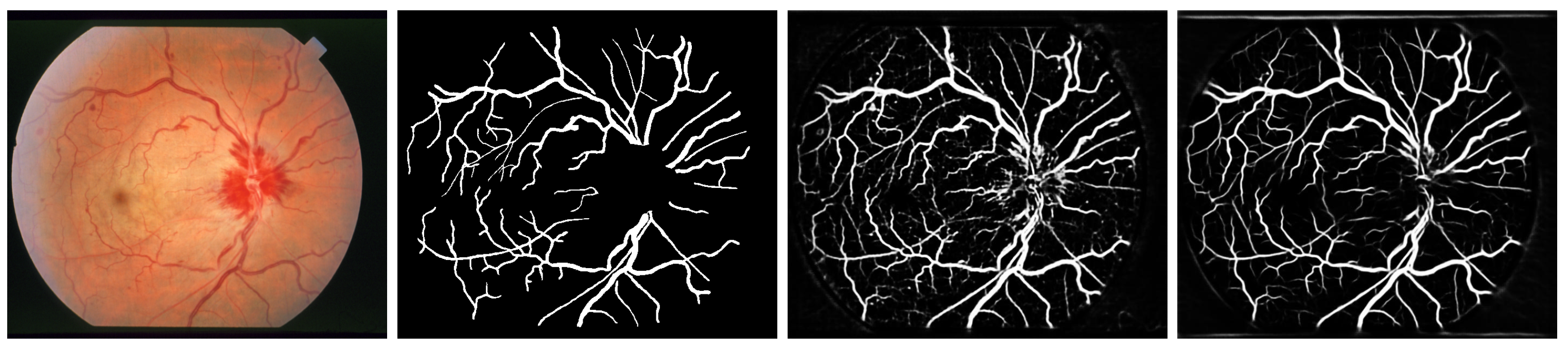
:1. Introduction
2. Methodology
3. Results and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hervella, Á.S.; Rouco, J.; Novo, J.; Ortega, M. Retinal Image Understanding Emerges from Self-Supervised Multimodal Reconstruction. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI, Granada, Spain, 16–20 September 2018; Volume 11070, pp. 321–328. [Google Scholar]
- Álvaro, S.; Hervella.; Rouco, J.; Novo, J.; Ortega, M. Self-supervised multimodal reconstruction of retinal images over paired datasets. Expert Syst. Appl. 2020, 161, 113674. [Google Scholar]
- Hervella, Á.S.; Rouco, J.; Novo, J.; Ortega, M. Learning the retinal anatomy from scarce annotated data using self-supervised multimodal reconstruction. Appl. Soft Comput. 2020, 91, 106210. [Google Scholar] [CrossRef]
- Morano, J.; Hervella, Á.S.; Barreira, N.; Novo, J.; Rouco, J. Multimodal Transfer Learning-based Approaches for Retinal Vascular Segmentation. In Proceedings of the European Conference on Artificial Intelligence (ECAI), Santiago de Compostela, Spain, 29 August–5 September 2020. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; 9 October 2015; Volume 9351, pp. 234–241. [Google Scholar]
- Jégou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017; pp. 1175–1183. [Google Scholar] [CrossRef]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. ENet: A Deep Neural Network Architecture for Real-Time Semantic Segmentation. arXiv 2016, arXiv:1606.02147. [Google Scholar]
- Canziani, A.; Culurciello, E.; Paszke, A. Evaluation of neural network architectures for embedded systems. In Proceedings of the 2017 IEEE International Symposium on Circuits and Systems (ISCAS), Baltimore, MD, USA, 28–31 May 2017; pp. 1–4. [Google Scholar] [CrossRef]
- Kashefpur, M.; Kafieh, R.; Jorjandi, S.; Golmohammadi, H.; Khodabande, Z.; Abbasi, M.; Fakharzadeh, A.A.; Kashefpoor, M.; Rabbani, H. Isfahan MISP Dataset. J. Med. Signals Sens. 2016, 7, 43–48. [Google Scholar]
- Hervella, Á.S.; Rouco, J.; Novo, J.; Ortega, M. Multimodal registration of retinal images using domain-specific landmarks and vessel enhancement. Procedia Comput. Sci. 2018, 126, 97–104. [Google Scholar] [CrossRef]
- Staal, J.; Abramoff, M.D.; Niemeijer, M.; Viergever, M.A.; van Ginneken, B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Hoover, A.D.; Kouznetsova, V.; Goldbaum, M. Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response. IEEE Trans. Med. Imaging 2000, 19, 203–210. [Google Scholar] [CrossRef] [PubMed]
| U-Net | FC-DenseNet | ENet | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SSMP | FS | SSMP | FS | SSMP | FS | ||||||
| ROC | PR | ROC | PR | ROC | PR | ROC | PR | ROC | PR | ROC | PR |
| 0.9834 | 0.9051 | 0.9728 | 0.8590 | 0.9794 | 0.8924 | 0.9699 | 0.8468 | 0.9694 | 0.8434 | 0.8349 | 0.4472 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morano, J.; Hervella, Á.S.; Barreira, N.; Novo, J.; Rouco, J. Enhancing Retinal Blood Vessel Segmentation through Self-Supervised Pre-Training. Proceedings 2020, 54, 44. https://doi.org/10.3390/proceedings2020054044
Morano J, Hervella ÁS, Barreira N, Novo J, Rouco J. Enhancing Retinal Blood Vessel Segmentation through Self-Supervised Pre-Training. Proceedings. 2020; 54(1):44. https://doi.org/10.3390/proceedings2020054044
Chicago/Turabian StyleMorano, José, Álvaro S. Hervella, Noelia Barreira, Jorge Novo, and José Rouco. 2020. "Enhancing Retinal Blood Vessel Segmentation through Self-Supervised Pre-Training" Proceedings 54, no. 1: 44. https://doi.org/10.3390/proceedings2020054044
APA StyleMorano, J., Hervella, Á. S., Barreira, N., Novo, J., & Rouco, J. (2020). Enhancing Retinal Blood Vessel Segmentation through Self-Supervised Pre-Training. Proceedings, 54(1), 44. https://doi.org/10.3390/proceedings2020054044






