1. Introduction
SARS-CoV-2 is a strain of coronavirus responsible for the global COVID-19 pandemic of 2020. This virus causes a severe acute respiratory syndrome (SARS-CoV-2) and viral pneumonia that may leave lungs severely and irreparably damaged [
1,
2].
For the detection of this virus, sputum samples are analyzed. In these tests, using RT-qPCR or Reverse Transcription Quantitative Polymerase Chain Reaction, the RNA chains that are present in the obtained samples are transcribed into DNA. These tests use deoxyribonucleotides with fluorescent markers that, as soon as they successfully couple with the reference sequence, emit light. This way, by measuring the fluorescence emitted during each cycle, we can know if the reference gene has been found. The gene to be detected is called “E”, common to the family to which COVID-19 belongs to group 2 coronavirus.
2. Methodology
To study these signals, we will analyze different features, in the search for one that best separates positive from negative patients. In this case, we have analyzed the mean, standard deviation and the percentiles 25% and 75% of the PCR signals of the patient (as well as the positive and negative reference signals for the PCR batch of that given sample). To study their separability, we will use a kernel density estimation with a gaussian kernel over a trained support vector machine (SVM) with a lineal kernel.
3. Results
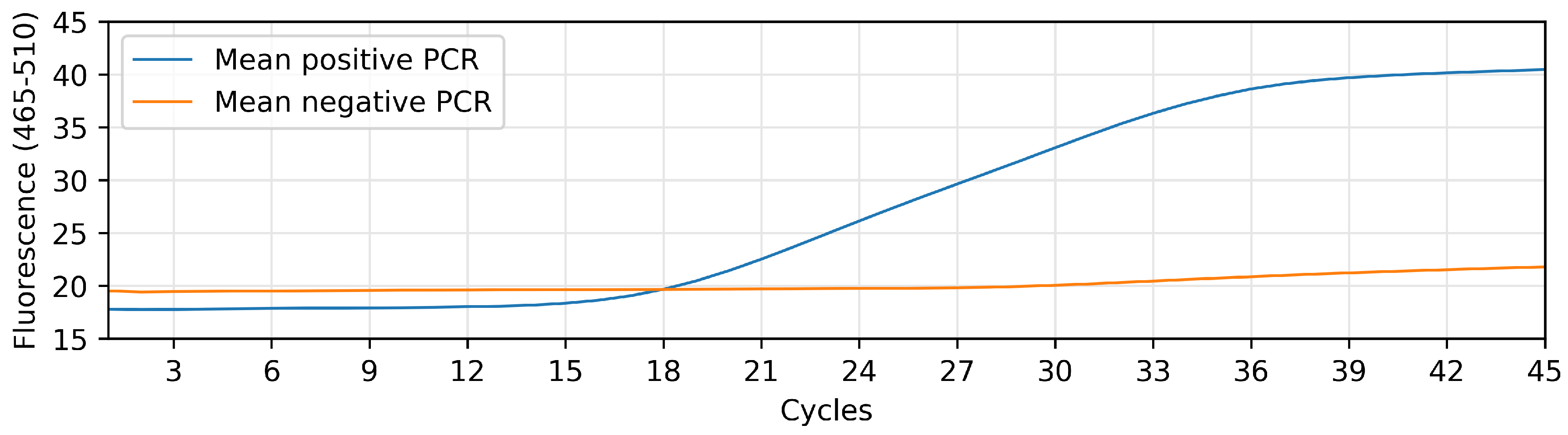
Our dataset is composed by 65 positive and 65 negative patients. These signals have been generated with a LightCycler 480 Real-Time PCR System from Roche, using fluorophores with wavelength of excitation of 465 nm and wavelength of detection of 510 nm over 45 PCR cycles. In
Figure 1, we can see the mean relative fluorescence returned by the positive PCRs and the negative PCRs. This figure clearly shows the general condition that separates a positive sample from a negative one: the excitation of the signal that is obtained after a certain number of PCR cycles. However, this slope and cycle threshold is highly dependent on the batch (thus the need for the reference signals).
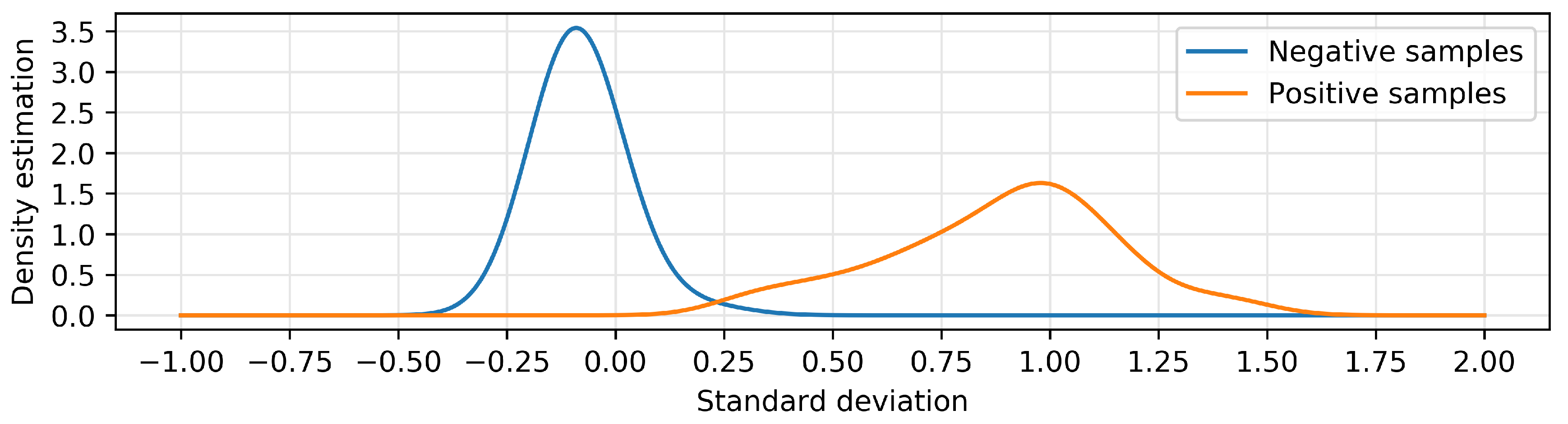
Nonetheless, the best results were obtained by using only the standard deviation of the main signal, with an F1 score of 0.94. As shown in
Figure 2, we can satisfactorily classify the majority of the patients without the necessity of the reference signals of the PCRs and with a minimal overlap between classes.
As future work, we plan a more in-depth study with a larger dataset. This will allow us to generate more complex features, as well as using modern machine learning and statistical strategies to develop a robust system that can, effectively, further speed up the process of diagnosing this disease.
Author Contributions
All the authors contributed equally to this work and have read and agreed to the published version of the manuscript.
Funding
Instituto de Salud Carlos III, Government of Spain, DTS18/00136 research project; Ministerio de Ciencia, Innovación y Universidades, Government of Spain, RTI2018-095894-B-I00 research project, Ayudas para la formación de profesorado universitario (FPU), grant ref. FPU18/02271; CITIC, Centro de Investigación de Galicia ref. ED431G 2019/01, receives financial support from Consellería de Educación, Universidade e Formación Profesional, Xunta de Galicia, through the ERDF (80%) and Secretaría Xeral de Universidades (20%).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- De Moura, J.; Novo, J.; Ortega, M. Fully automatic deep convolutional approaches for the analysis of Covid-19 using chest X-ray images. medRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- De Moura, J.; Ramos, L.; Vidal, P.L.; Cruz, M.; Abelairas, L.; Castro, E.; Novo, J.; Ortega, M. Deep convolutional approaches for the analysis of Covid-19 using chest X-Ray images from portable devices. medRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).