Artificial Intelligence-Based Plant Disease Classification in Low-Light Environments
Abstract
1. Introduction
1.1. Using Handcrafted Features
1.2. Using Deep Features
1.2.1. Using Normal Illumination Images
1.2.2. Using Low-Light Noisy Images
- -
- To the best of our knowledge, this is the first approach that effectively performs plant disease classification from low-light noisy images, and we propose DPA-Net.
- -
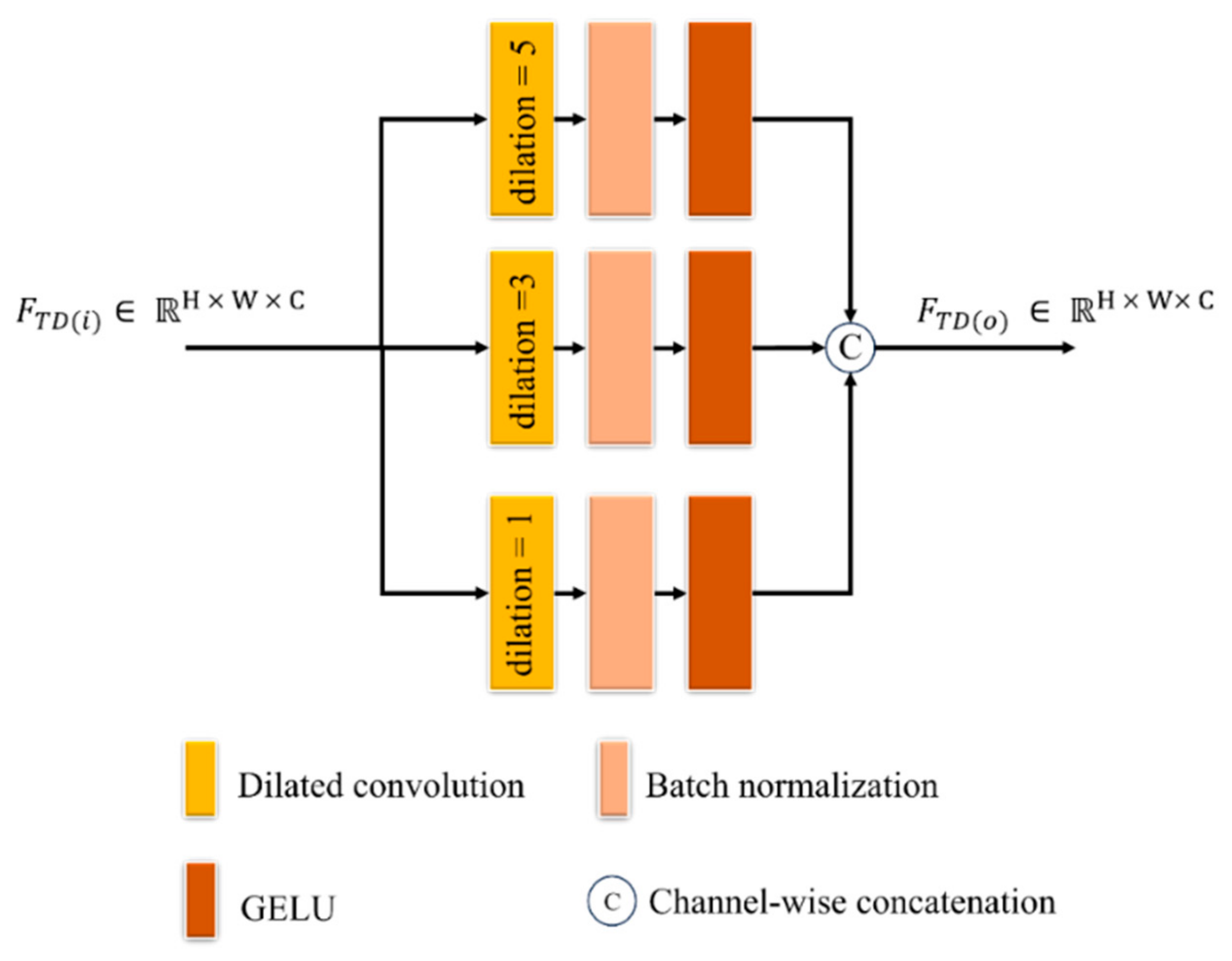
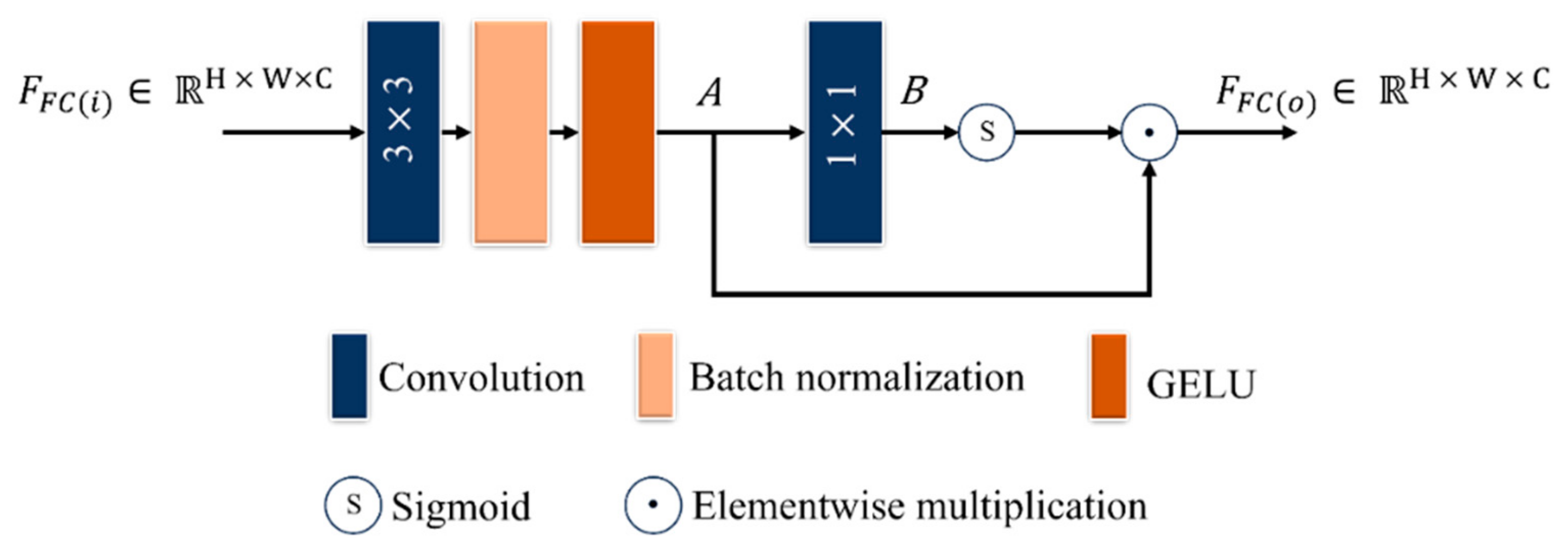
- A triple dilated convolution block (TDCB) is proposed to extract both global and local contextual information, focusing on disease patterns at various locations along the leaf edges and distinguishing relevant features from noisy low-illumination images with a wider receptive field by concentrating on signal consistency. A fused convolution block (FCB) is proposed to improve low contrast by accentuating differences in pixel intensities and highlighting subtle features to provide information about small, localized, disease-affected areas on leaves.
- -
- A multi-scale feature extraction block (MFEB) is proposed to extract deep features at different scales and aids the model in capturing fine-grained details with a wider spatial relationship of disease spread over the leaf simultaneously, which provides context-aware representation of features for noisy images with low contrast.
- -
- Moreover, to validate the classification results of our proposed DPA-Net and analyze structural irregularities, we performed fractal dimension estimation on diseased and healthy leaves. In addition, the real low-illumination dataset is constructed by capturing images at 0 lux using a smartphone at night.
2. Materials and Methods
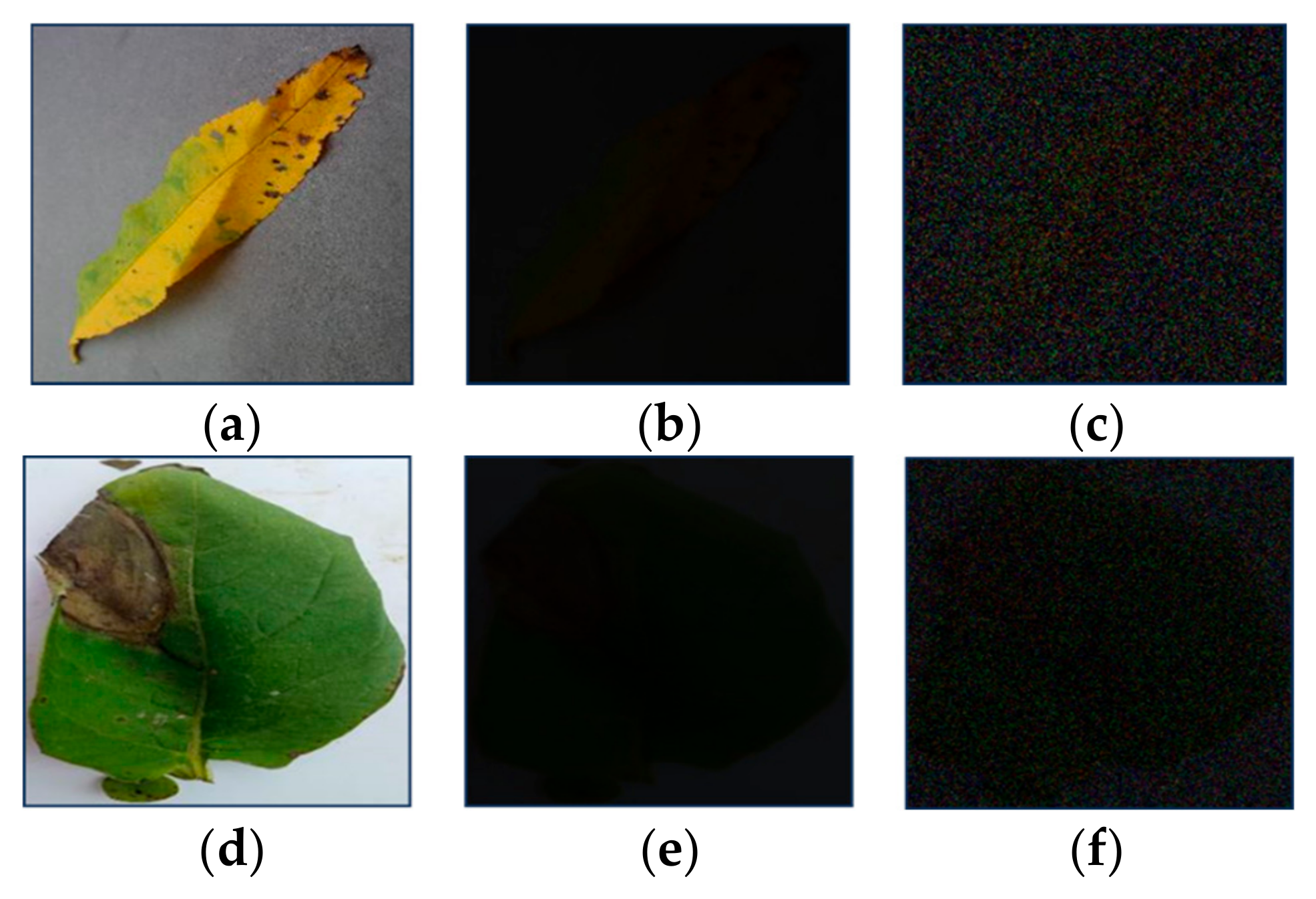
2.1. Experimental Setup
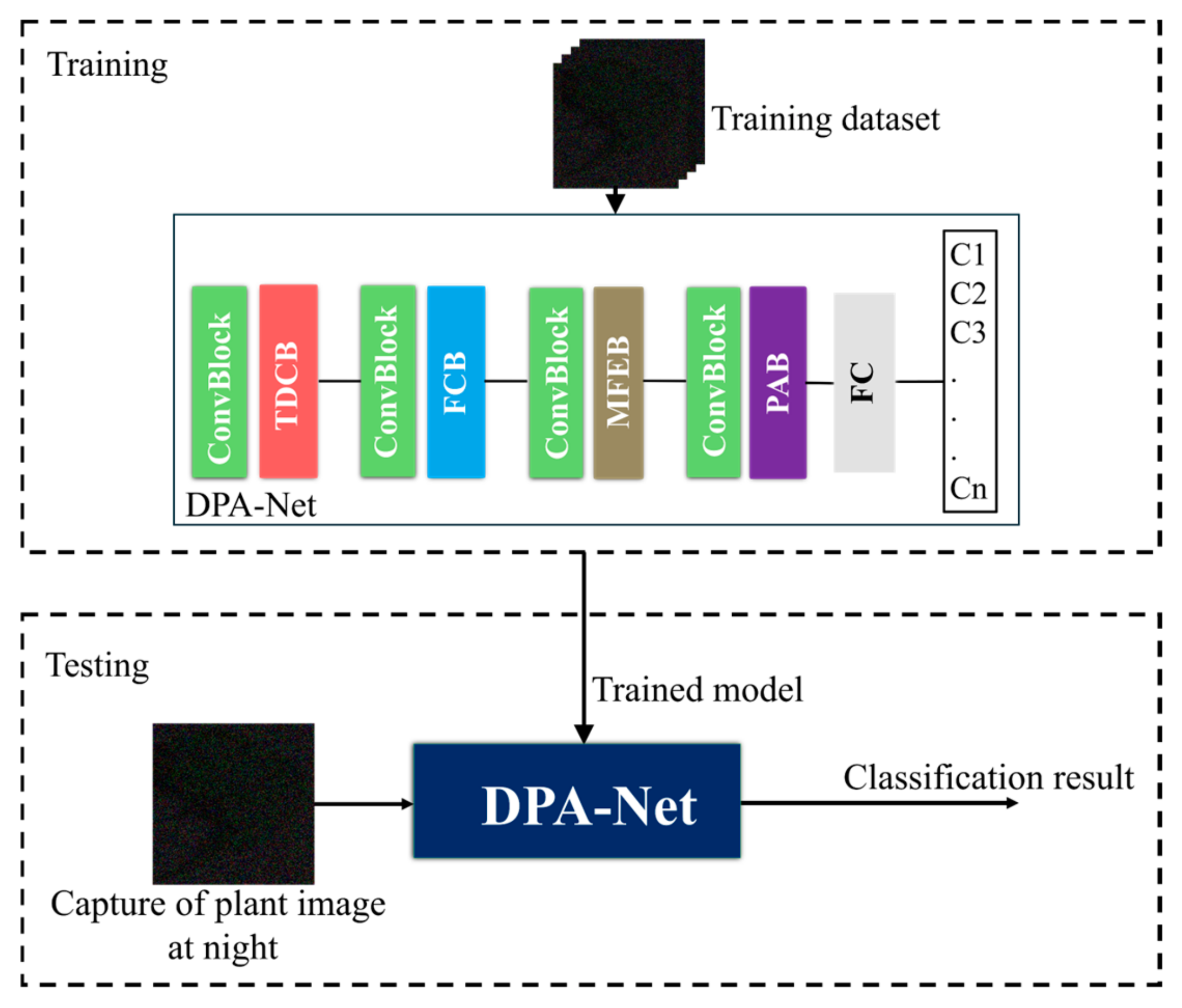
2.2. Overview of the Proposed Method
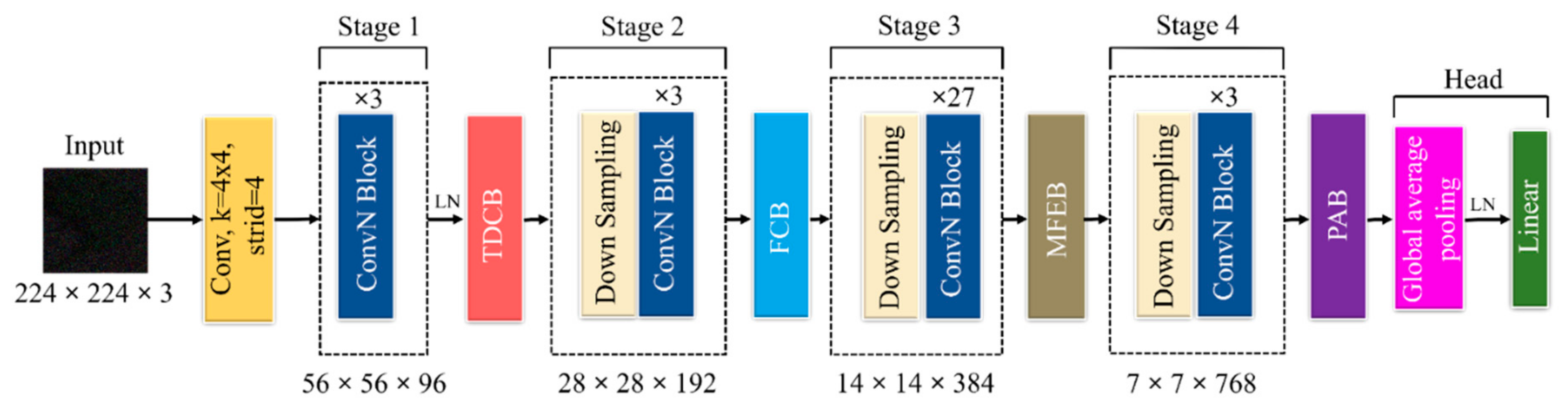
2.3. DPA-Net Structure
2.3.1. TDCB
2.3.2. FCB
2.3.3. MFEB
2.3.4. PAB
3. Experimental Results
3.1. Training Details
3.2. Evaluation Metrics
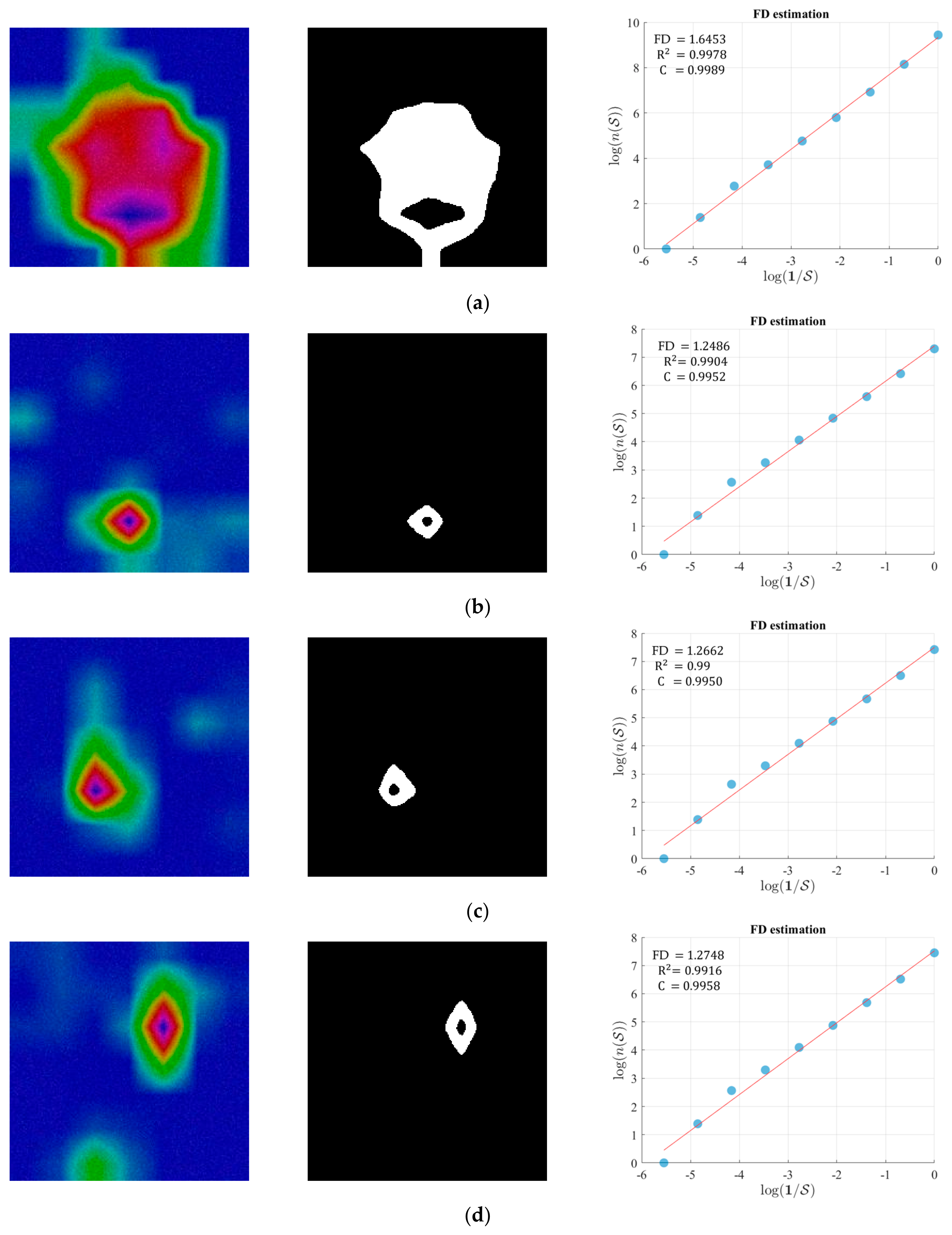
3.3. Fractal Dimension Estimation
| Algorithm 1: Procedure for estimating FD | |
| Input: I: Binary activated image derived from the proposed DPA-Net Output: : Fractal dimension | |
| Step 1: | Set the size of box to the largest dimensions aligned with the nearest power of 2. = 2 ^[log(max(size(I)))/log2] |
| Step 2: | Make the dimensions of I equal to the using padding if size(I) < size(): padding (I) = end |
| Step 3: | Assign the starting number of boxes k = zeros(1, +1) |
| Step 4: | Count the number of boxes K() containing at least one pixel of diseased area k(+1) = sum(I(:)) |
| Step 5: | while > 1: I. Reduce the size of box as = /2 II. Compute again K() end |
| Step 6: | Calculate log() and log(1/) for each value of |
| Step 7: | Determine the best-fit line for [log(), log(1/)] using least square regression. |
| Step 8: | The slop of fitted line is fractal dimension Return |
3.4. Ablation Study
3.5. Comparison of DPA-Net with State-of-the-Art (SOTA) Methods
3.6. Comparisons of Model Complexity
4. Discussion
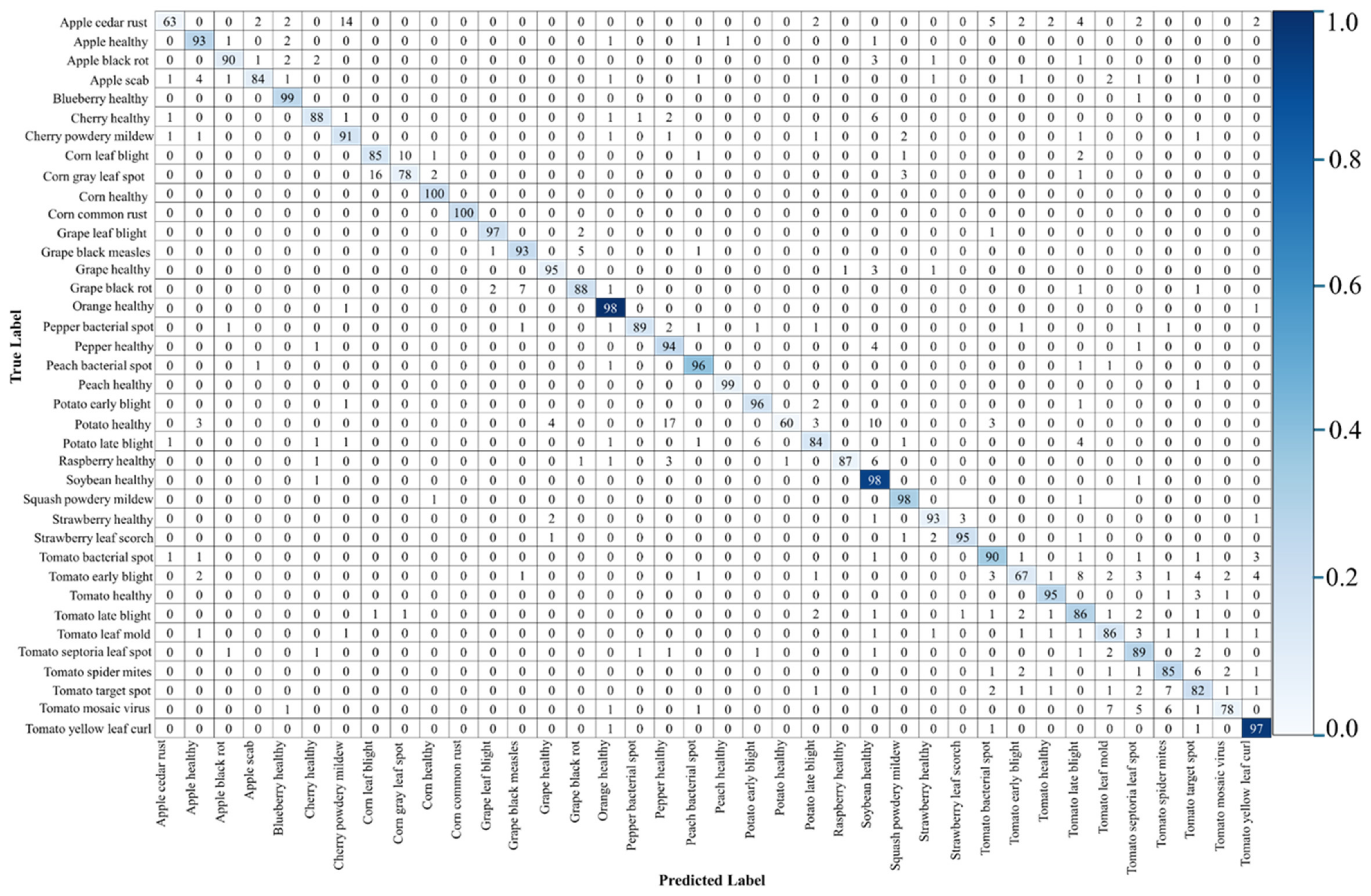
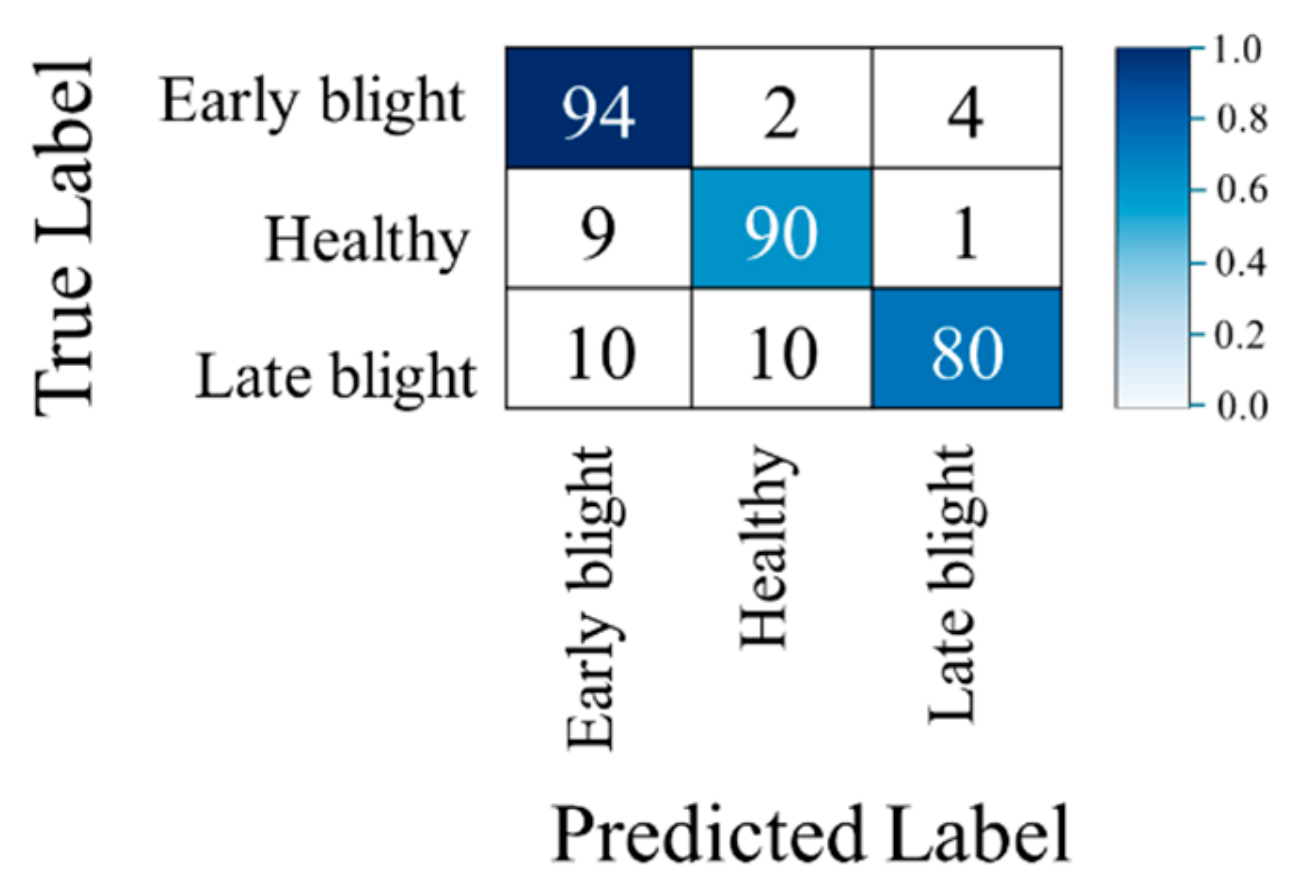
4.1. Confusion Matrices, Robustness to the Illumination and Noise Level, and Experiments with Real Low-Illumination Dataset
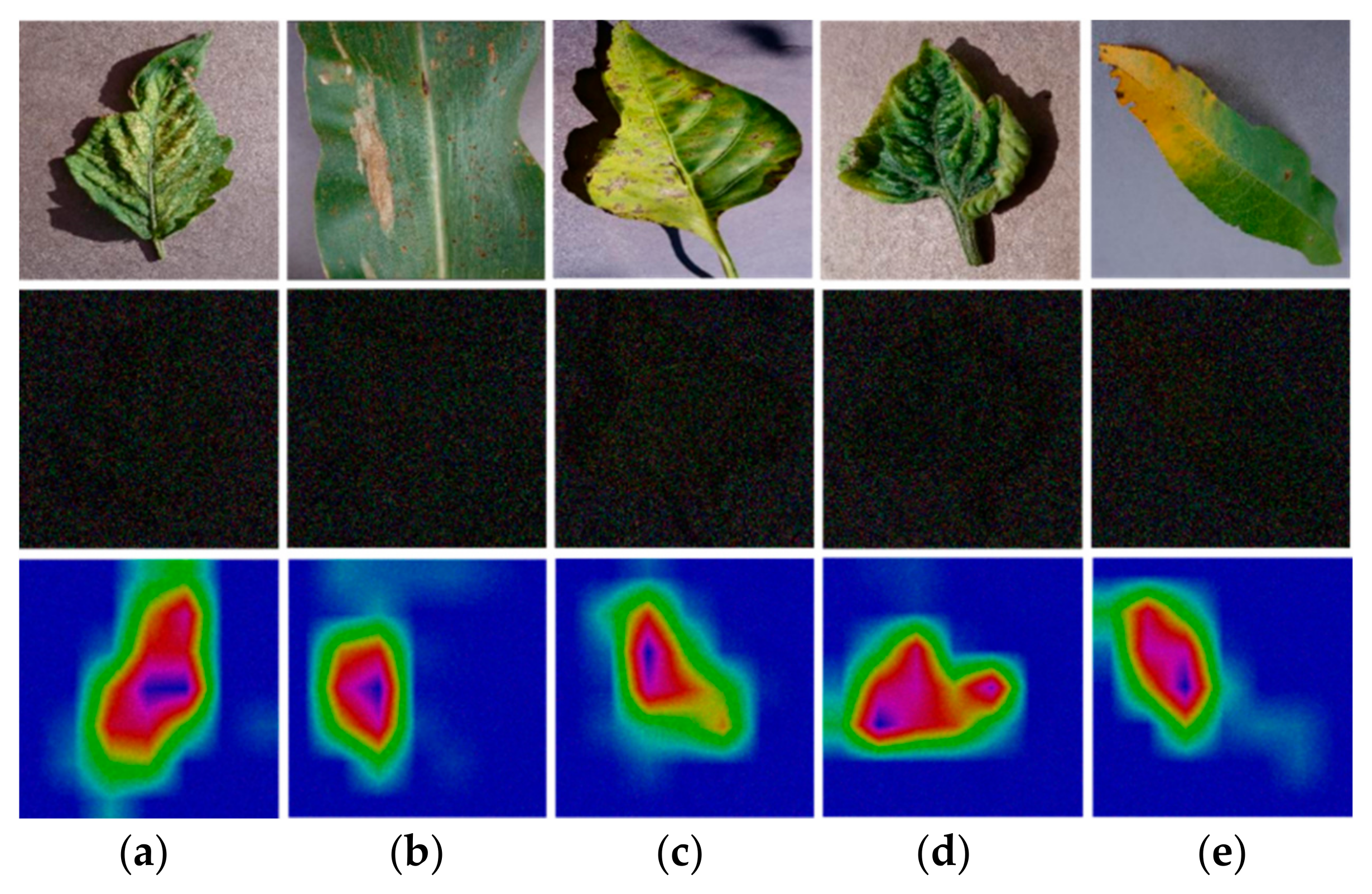
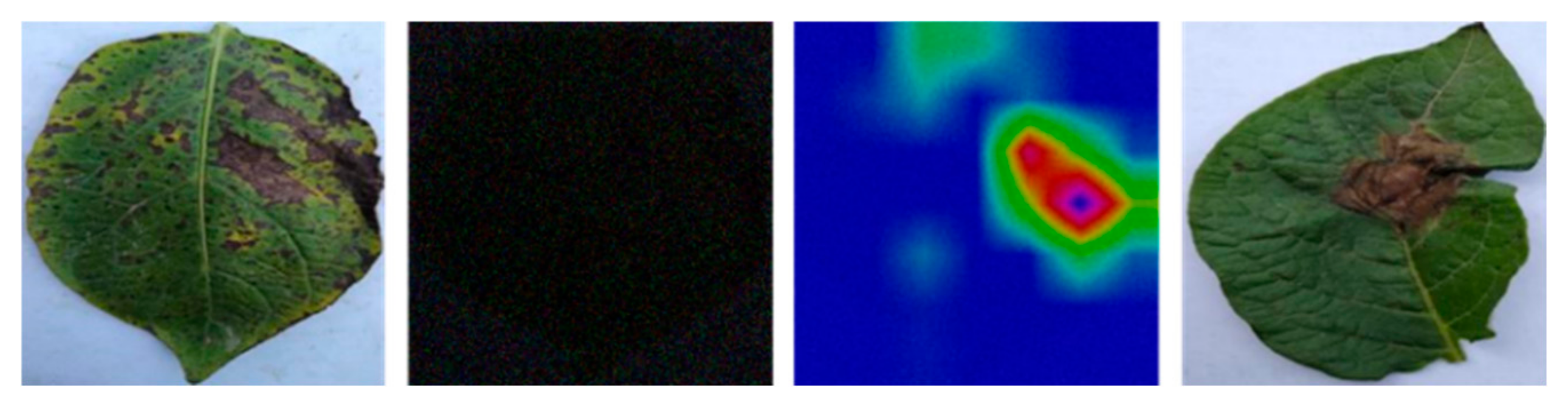
4.2. Statistical Analyses, and Grad-CAM
4.3. Performance Evaluation of DPA-Net by FD Estimation
4.4. Integration of FD in Classification Results
5. Limitations of the Proposed DPA-Net
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vásconez, J.P.; Vásconez, I.N.; Moya, V.; Calderón-Díaz, M.J.; Valenzuela, M.; Besoain, X.; Seeger, M.; Auat Cheein, F. Deep Learning-Based Classification of Visual Symptoms of Bacterial Wilt Disease Caused by Ralstonia Solanacearum in Tomato Plants. Comput. Electron. Agric. 2024, 227, 109617. [Google Scholar] [CrossRef]
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global Food Demand and the Sustainable Intensification of Agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- Habib, M.T.; Majumder, A.; Jakaria, A.Z.M.; Akter, M.; Uddin, M.S.; Ahmed, F. Machine Vision Based Papaya Disease Recognition. J. King Saud Univ.—Comput. Inf. Sci. 2020, 32, 300–309. [Google Scholar] [CrossRef]
- Sajitha, P.; Andrushia, A.D.; Anand, N.; Naser, M.Z. A Review on Machine Learning and Deep Learning Image-Based Plant Disease Classification for Industrial Farming Systems. J. Ind. Inf. Integr. 2024, 38, 100572. [Google Scholar] [CrossRef]
- Shirahatti, J.; Patil, R.; Akulwar, P. A Survey Paper on Plant Disease Identification Using Machine Learning Approach. In Proceedings of the 3rd IEEE International Conference on Communication and Electronics Systems, Coimbatore, India, 15–16 October 2018; pp. 1171–1174. [Google Scholar]
- Panigrahi, K.P.; Das, H.; Sahoo, A.K.; Moharana, S.C. Maize Leaf Disease Detection and Classification Using Machine Learning Algorithms. In Proceedings of the Springer Progress in Computing, Analytics and Networking, Singapore, 27 March 2020; pp. 659–669. [Google Scholar]
- Bhatia, A.; Chug, A.; Singh, A.P. Application of Extreme Learning Machine in Plant Disease Prediction for Highly Imbalanced Dataset. J. Stat. Manag. Syst. 2020, 23, 1059–1068. [Google Scholar] [CrossRef]
- Mathew, A.; Antony, A.; Mahadeshwar, Y.; Khan, T.; Kulkarni, A. Plant Disease Detection Using GLCM Feature Extractor and Voting Classification Approach. Mater. Today Proc. 2022, 58, 407–415. [Google Scholar] [CrossRef]
- Panchal, P.; Raman, V.C.; Mantri, S. Plant Diseases Detection and Classification Using Machine Learning Models. In Proceedings of the 4th IEE International Conference on Computational Systems and Information Technology for Sustainable Solution, Bengaluru, India, 20–21 December 2019; pp. 1–6. [Google Scholar]
- Thakur, P.S.; Khanna, P.; Sheorey, T.; Ojha, A. Trends in Vision-Based Machine Learning Techniques for Plant Disease Identification: A Systematic Review. Expert Syst. Appl. 2022, 208, 118117. [Google Scholar] [CrossRef]
- Wani, J.A.; Sharma, S.; Muzamil, M.; Ahmed, S.; Sharma, S.; Singh, S. Machine Learning and Deep Learning Based Computational Techniques in Automatic Agricultural Diseases Detection: Methodologies, Applications, and Challenges. Arch. Comput. Methods Eng. 2022, 29, 641–677. [Google Scholar] [CrossRef]
- Yu, M.; Ma, X.; Guan, H. Recognition Method of Soybean Leaf Diseases Using Residual Neural Network Based on Transfer Learning. Ecol. Inform. 2023, 76, 102096. [Google Scholar] [CrossRef]
- Reddy, S.R.G.; Varma, G.P.S.; Davuluri, R.L. Resnet-Based Modified Red Deer Optimization with DLCNN Classifier for Plant Disease Identification and Classification. Comput. Electr. Eng. 2023, 105, 108492. [Google Scholar] [CrossRef]
- Kaya, Y.; Gürsoy, E. A Novel Multi-Head CNN Design to Identify Plant Diseases Using the Fusion of RGB Images. Ecol. Inform. 2023, 75, 101998. [Google Scholar] [CrossRef]
- Malik, A.; Vaidya, G.; Jagota, V.; Eswaran, S.; Sirohi, A.; Batra, I.; Rakhra, M.; Asenso, E. Design and Evaluation of a Hybrid Technique for Detecting Sunflower Leaf Disease Using Deep Learning Approach. J. Food Qual. 2022, 2022, 9211700. [Google Scholar] [CrossRef]
- Guo, M.-H.; Xu, T.-X.; Liu, J.-J.; Liu, Z.-N.; Jiang, P.-T.; Mu, T.-J.; Zhang, S.-H.; Martin, R.R.; Cheng, M.-M.; Hu, S.-M. Attention Mechanisms in Computer Vision: A Survey. Comput. Vis. Media 2022, 8, 331–368. [Google Scholar] [CrossRef]
- Yang, W.; Yuan, Y.; Zhang, D.; Zheng, L.; Nie, F. An Effective Image Classification Method for Plant Diseases with Improved Channel Attention Mechanism aECAnet Based on Deep Learning. Symmetry 2024, 16, 451. [Google Scholar] [CrossRef]
- Dai, G.; Tian, Z.; Fan, J.; Sunil, C.K.; Dewi, C. DFN-PSAN: Multi-Level Deep Information Feature Fusion Extraction Network for Interpretable Plant Disease Classification. Comput. Electron. Agric. 2024, 216, 108481. [Google Scholar] [CrossRef]
- Yang, L.; Yu, X.; Zhang, S.; Long, H.; Zhang, H.; Xu, S.; Liao, Y. GoogLeNet Based on Residual Network and Attention Mechanism Identification of Rice Leaf Diseases. Comput. Electron. Agric. 2023, 204, 107543. [Google Scholar] [CrossRef]
- Xu, L.; Cao, B.; Zhao, F.; Ning, S.; Xu, P.; Zhang, W.; Hou, X. Wheat Leaf Disease Identification Based on Deep Learning Algorithms. Physiol. Mol. Plant Pathol. 2023, 123, 101940. [Google Scholar] [CrossRef]
- Tang, L.; Yi, J.; Li, X. Improved Multi-Scale Inverse Bottleneck Residual Network Based on Triplet Parallel Attention for Apple Leaf Disease Identification. J. Integr. Agric. 2024, 23, 901–922. [Google Scholar] [CrossRef]
- Bi, C.; Xu, S.; Hu, N.; Zhang, S.; Zhu, Z.; Yu, H. Identification Method of Corn Leaf Disease Based on Improved Mobilenetv3 Model. Agronomy 2023, 13, 300. [Google Scholar] [CrossRef]
- Nagasubramanian, G.; Sakthivel, R.K.; Patan, R.; Sankayya, M.; Daneshmand, M.; Gandomi, A.H. Ensemble Classification and IoT-Based Pattern Recognition for Crop Disease Monitoring System. IEEE Internet Things J. 2021, 8, 12847–12854. [Google Scholar] [CrossRef]
- Garg, G.; Gupta, S.; Mishra, P.; Vidyarthi, A.; Singh, A.; Ali, A. CROPCARE: An Intelligent Real-Time Sustainable IoT System for Crop Disease Detection Using Mobile Vision. IEEE Internet Things J. 2023, 10, 2840–2851. [Google Scholar] [CrossRef]
- DPA-Net. Available online: https://github.com/gondalalihamza/DPA-Net (accessed on 4 June 2025).
- Hughes, D.P.; Salathé, M. An Open Access Repository of Images on Plant Health to Enable the Development of Mobile Disease Diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Rashid, J.; Khan, I.; Ali, G.; Almotiri, S.H.; AlGhamdi, M.A.; Masood, K. Multi-Level Deep Learning Model for Potato Leaf Disease Recognition. Electronics 2021, 10, 2064. [Google Scholar] [CrossRef]
- GeForce 10 Series Graphics Cards. Available online: https://www.nvidia.com/en-in/geforce/10-series/ (accessed on 16 May 2024).
- Shen, L.; Yue, Z.; Feng, F.; Chen, Q.; Liu, S.; Ma, J. MSR-Net: Low-Light Image Enhancement Using Deep Convolutional Network. arXiv 2017, arXiv:1711.02488. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, Louisiana, USA, 19–24 June 2022. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Ho, Y.; Wookey, S. The Real-World-Weight Cross-Entropy Loss Function: Modeling the Costs of Mislabeling. IEEE Access 2020, 8, 4806–4813. [Google Scholar] [CrossRef]
- Akram, R.; Kim, J.S.; Jeong, M.S.; Gondal, H.A.H.; Tariq, M.H.; Irfan, M.; Park, K.R. Attention-Driven and Hierarchical Feature Fusion Network for Crop and Weed Segmentation with Fractal Dimension Estimation. Fractal Fract. 2025, 9, 592. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, Q.; Huang, X. An Algorithm for Crack Detection, Segmentation, and Fractal Dimension Estimation in Low Light Environments by Fusing FFT and Convolutional Neural Network. Fractal Fract. 2023, 7, 820. [Google Scholar] [CrossRef]
- Arsalan, M.; Haider, A.; Hong, J.S.; Kim, J.S.; Park, K.R. Deep Learning-Based Detection of Human Blastocyst Compartments with Fractal Dimension Estimation. Fractal Fract. 2024, 8, 267. [Google Scholar] [CrossRef]
- González-Sabbagh, S.P.; Robles-Kelly, A. A Survey on Underwater Computer Vision. ACM Comput. Surv. 2023, 55, 1–39. [Google Scholar] [CrossRef]
- Akram, R.; Hong, J.S.; Kim, S.G.; Sultan, H.; Usman, M.; Gondal, H.A.H.; Tariq, M.H.; Ullah, N.; Park, K.R. Crop and Weed Segmentation and Fractal Dimension Estimation Using Small Training Data in Heterogeneous Data Environment. Fractal Fract. 2024, 8, 285. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, X.; Yin, Y. Duality Revelation and Operator-Based Method in Viscoelastic Problems. Fractal Fract. 2025, 9, 274. [Google Scholar] [CrossRef]
- Sultan, H.; Ullah, N.; Hong, J.S.; Kim, S.G.; Lee, D.C.; Jung, S.Y.; Park, K.R. Estimation of Fractal Dimension and Segmentation of Brain Tumor with Parallel Features Aggregation Network. Fractal Fract. 2024, 8, 357. [Google Scholar] [CrossRef]
- Kim, S.G.; Hong, J.S.; Kim, J.S.; Park, K.R. Estimation of Fractal Dimension and Detection of Fake Finger-Vein Images for Finger-Vein Recognition. Fractal Fract. 2024, 8, 646. [Google Scholar] [CrossRef]
- Tariq, M.H.; Sultan, H.; Akram, R.; Kim, S.G.; Kim, J.S.; Usman, M.; Gondal, H.A.H.; Seo, J.; Lee, Y.H.; Park, K.R. Estimation of Fractal Dimensions and Classification of Plant Disease with Complex Backgrounds. Fractal Fract. 2025, 9, 315. [Google Scholar] [CrossRef]
- Brouty, X.; Garcin, M. Fractal Properties, Information Theory, and Market Efficiency. Chaos Solitons Fractals 2024, 180, 114543. [Google Scholar] [CrossRef]
- Yang, B.; Li, M.; Li, F.; Wang, Y.; Liang, Q.; Zhao, R.; Li, C.; Wang, J. A Novel Plant Type, Leaf Disease and Severity Identification Framework Using CNN and Transformer with Multi-Label Method. Sci. Rep. 2024, 14, 11664. [Google Scholar] [CrossRef]
- Akuthota, U.C.; Abhishek; Bhargava, L. A Lightweight Low-Power Model for the Detection of Plant Leaf Diseases. SN Comput. Sci. 2024, 5, 327. [Google Scholar] [CrossRef]
- Chilakalapudi, M.; Jayachandran, S. Optimized Deep Learning Network for Plant Leaf Disease Segmentation and Multi-Classification Using Leaf Images. Netw. Comput. Neural Syst. 2025, 36, 615–648. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.S.; Catal, C.; Yalic, H.Y.; Temucin, H.; Tekinerdogan, B. Analysis of Transfer Learning for Deep Neural Network Based Plant Classification Models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Altabaji, W.I.A.E.; Umair, M.; Tan, W.-H.; Foo, Y.-L.; Ooi, C.-P. Comparative Analysis of Transfer Learning, LeafNet, and Modified LeafNet Models for Accurate Rice Leaf Diseases Classification. IEEE Access 2024, 12, 36622–36635. [Google Scholar] [CrossRef]
- Desanamukula, V.S.; Dharma Teja, T.; Rajitha, P. An In-Depth Exploration of ResNet-50 and Transfer Learning in Plant Disease Diagnosis. In Proceedings of the 2024 IEEE International Conference on Inventive Computation Technologies, Lalitpur, Nepal, 24–26 April 2024; pp. 614–621. [Google Scholar]
- Li, E.; Wang, L.; Xie, Q.; Gao, R.; Su, Z.; Li, Y. A Novel Deep Learning Method for Maize Disease Identification Based on Small Sample-Size and Complex Background Datasets. Ecol. Inform. 2023, 75, 102011. [Google Scholar] [CrossRef]
- Mohameth, F.; Bingcai, C.; Sada, K.A. Plant Disease Detection with Deep Learning and Feature Extraction Using Plant Village. J. Comput. Commun. 2020, 8, 10–22. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017, 2917536. [Google Scholar] [CrossRef] [PubMed]
- Nandhini, S.; Ashokkumar, K. An Automatic Plant Leaf Disease Identification Using DenseNet-121 Architecture with a Mutation-Based Henry Gas Solubility Optimization Algorithm. Neural Comput. Appl. 2022, 34, 5513–5534. [Google Scholar] [CrossRef]
- Wu, Q.; Ma, X.; Liu, H.; Bi, C.; Yu, H.; Liang, M.; Zhang, J.; Li, Q.; Tang, Y.; Ye, G. A Classification Method for Soybean Leaf Diseases Based on an Improved ConvNeXt Model. Sci. Rep. 2023, 13, 19141. [Google Scholar] [CrossRef]
- Wang, F.; Rao, Y.; Luo, Q.; Jin, X.; Jiang, Z.; Zhang, W.; Li, S. Practical Cucumber Leaf Disease Recognition Using Improved Swin Transformer and Small Sample Size. Comput. Electron. Agric. 2022, 199, 107163. [Google Scholar] [CrossRef]
- Parashar, N.; Johri, P. ModularSqueezeNet: A Modified Lightweight Deep Learning Model for Plant Disease Detection. In Proceedings of the 2nd IEEE International Conference on Disruptive Technologies, Greater Noida, India, 15–16 March 2024; pp. 1331–1334. [Google Scholar]
- Muiz Fayyaz, A.; A. Al-Dhlan, K.; Ur Rehman, S.; Raza, M.; Mehmood, W.; Shafiq, M.; Choi, J.-G. Leaf Blights Detection and Classification in Large Scale Applications. Intell. Autom. Soft Comput. 2022, 31, 507–522. [Google Scholar] [CrossRef]
- Arya, S.; Singh, R. A Comparative Study of CNN and AlexNet for Detection of Disease in Potato and Mango Leaf. In Proceedings of the 2019 IEEE International Conference on Issues and Challenges in Intelligent Computing Techniques, Ghaziabad, India, 27–28 September 2019; pp. 1–6. [Google Scholar]
- Shinde, N.; Ambhaikar, A. Fine-Tuned Xception Model for Potato Leaf Disease Classification. In Proceedings of the 5th Springer International Conference on Computing, Communications, and Cyber-Security, Jammu, India, 29 February–1 March 2024; pp. 663–676. [Google Scholar]
- Ashikuzzaman, M.; Roy, K.; Lamon, A.; Abedin, S. Potato Leaf Disease Detection By Deep Learning: A Comparative Study. In Proceedings of the 6th IEEE International Conference on Electrical Engineering and Information & Communication Technology, Dhaka, Bangladesh, 2–4 May 2024; pp. 278–283. [Google Scholar]
- Indira, K.; Mallika, H. Classification of Plant Leaf Disease Using Deep Learning. J. Inst. Eng. India Ser. B 2024, 105, 609–620. [Google Scholar] [CrossRef]
- Sholihati, R.A.; Sulistijono, I.A.; Risnumawan, A.; Kusumawati, E. Potato Leaf Disease Classification Using Deep Learning Approach. In Proceedings of the 2020 IEEE International Electronics Symposium, Surabaya, Indonesia, 29–30 September 2020; pp. 392–397. [Google Scholar]
- Chugh, G.; Sharma, A.; Choudhary, P.; Khanna, R. Potato Leaf Disease Detection Using Inception V3. Int. Res. J. Eng. Technol. 2020, 7, 1363–1366. [Google Scholar]
- Mahum, R.; Munir, H.; Mughal, Z.-U.-N.; Awais, M.; Sher Khan, F.; Saqlain, M.; Mahamad, S.; Tlili, I. A Novel Framework for Potato Leaf Disease Detection Using an Efficient Deep Learning Model. Hum. Ecol. Risk Assess. Int. J. 2023, 29, 303–326. [Google Scholar] [CrossRef]
- Nazir, T.; Iqbal, M.M.; Jabbar, S.; Hussain, A.; Albathan, M. EfficientPNet—An Optimized and Efficient Deep Learning Approach for Classifying Disease of Potato Plant Leaves. Fractal Fract. 2023, 13, 841. [Google Scholar] [CrossRef]
- Luong, H.H. Improving Potato Diseases Classification Based on Custom ConvNeXtSmall and Combine with the Explanation Model. Int. J. Adv. Comput. Sci. Appl. 2024, 15, 1206–1219. [Google Scholar] [CrossRef]
- Jetson TX2 Module. Available online: https://developer.nvidia.com/embedded/jetson-tx2 (accessed on 23 August 2024).
- Meyer-Baese, A.; Schmid, V. Foundations of Neural Networks. In Pattern Recognition and Signal Analysis in Medical Imaging; Elsevier: Amsterdam, The Netherlands, 2014; pp. 197–243. ISBN 978-0-12-409545-8. [Google Scholar]
- SL-C3510ND. Available online: https://www.samsung.com/sec/support/model/SL-C3510ND/ (accessed on 9 June 2025).
- Galaxy A23 Self-Pay. Available online: https://www.samsung.com/sec/support/model/SM-A235NLBOKOO/ (accessed on 9 June 2025).
- LM-81LX Mini Light Meter. Available online: https://thelabk.com/goods/view?no=3247 (accessed on 9 June 2025).
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of Student’s t-Test, Analysis of Variance, and Covariance. Ann. Card. Anaesth. 2019, 22, 407. [Google Scholar] [CrossRef]
- Cohen, J. A Power Primer. Psychol. Bull. 1992, 112, 155–159. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
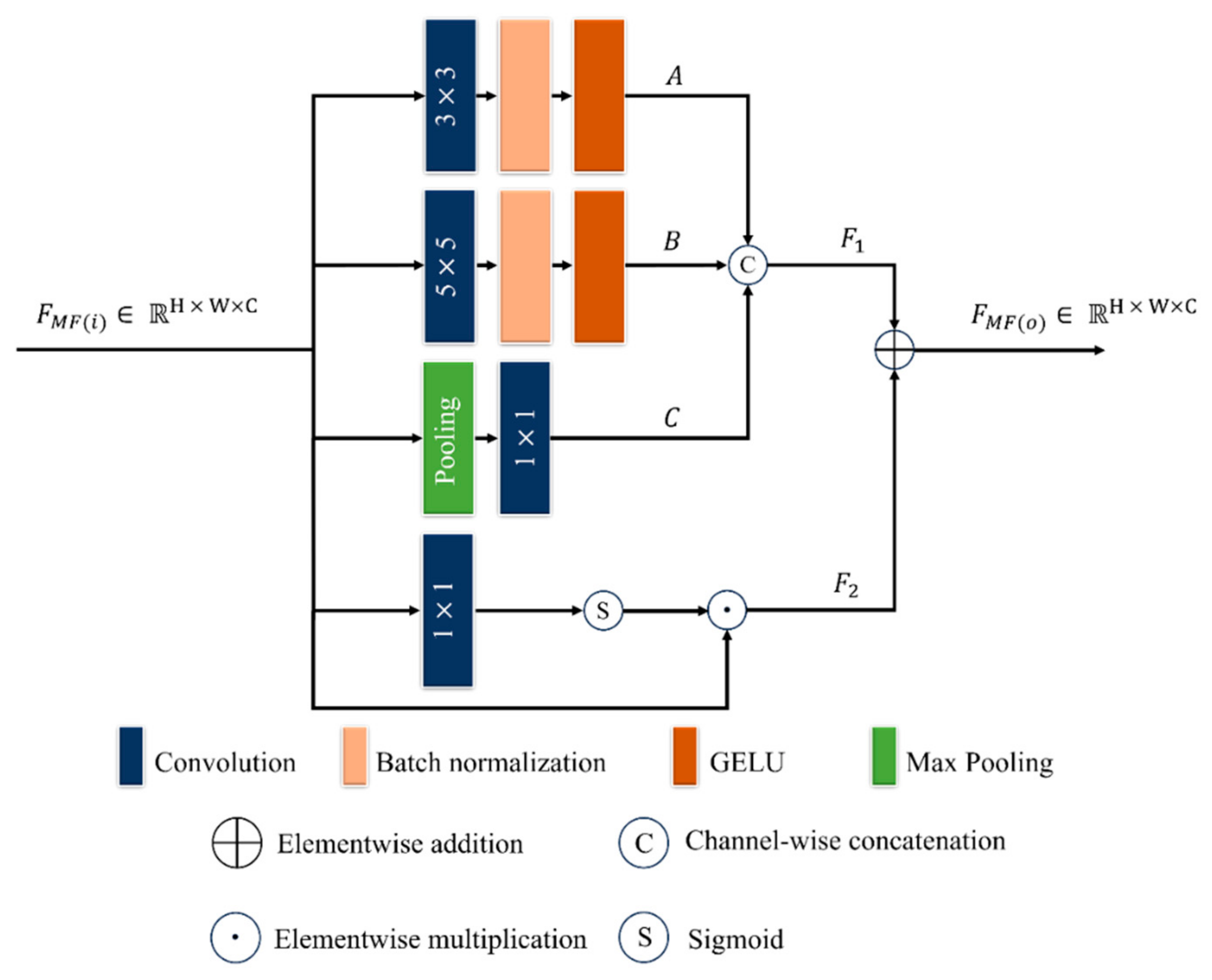
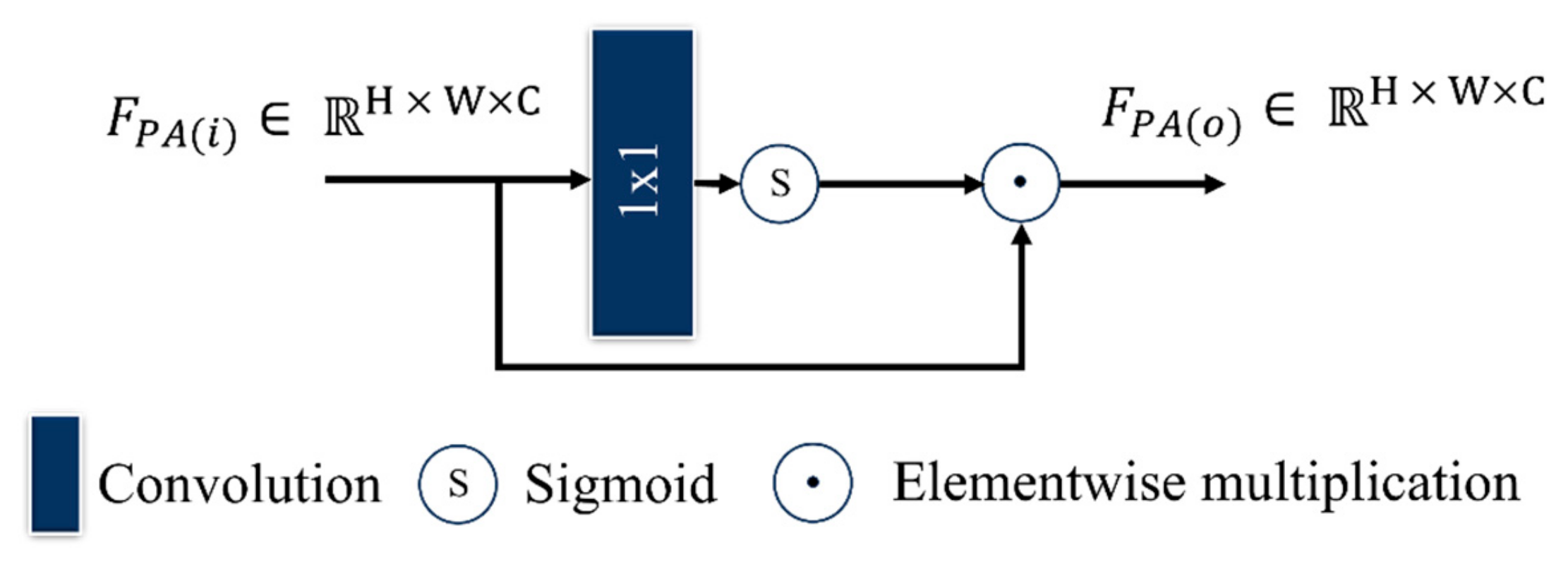




| Category | Method | Dataset | Classes | Accuracy (%) | Strengths | Limitation | |
|---|---|---|---|---|---|---|---|
| Using handcrafted features | EfficientNetB7 [6] | PlantVillage | 4 | 79.23 | Early detection with less memory requirements | Low accuracy on untrained datasets | |
| ELM [7] | TPMD | 2 | 88.57 | Efficient for imbalanced dataset |
| ||
| Voting Classifier [8] | PlantVillage | 3 | 92.60 | Improved accuracy with respect to SVM | Limited to two classes of early and late blight diseases only | ||
| HSV + GLCM+ RF [9] | Self-collected | 4 | 98 | High accuracy with less computational complexity | Lack of accuracy comparison | ||
| Using deep features | Using normal illumination images | ResNet-18 with transfer learning [12] | Self-collected | 4 | 99.53 | High accuracy with fast processing speed |
|
| ResNet-50 + MRDOA [13] | PlantVillage | 18 | 99.72 |
| Require extensive preprocessing | ||
| Rice Plant dataset | 3 | 99.68 | |||||
| DenseNet + RGB Fusion [14] | PlantVillage | 38 | 98.17 |
| More training time and lack of hyperparameter optimization | ||
| MobileNet + VGG-16 with transfer learning [15] | Self-collected | 5 | 89.2 | Outperform due to ensemble of DL models |
| ||
| ResNet-34+ aECAnet [17] | Peanut | 3 | 97.7 |
|
| ||
| PlantVillage | 39 | 98.5 | |||||
| YOLO5 + PSA [18] | Katra-twelve | 12 | 98.25 |
|
| ||
| BARI-sunflower | 4 | 94.47 | |||||
| FGVC8 | 12 | 93.55 | |||||
| GoogleNet + ECA [19] | Self-collected | 8 | 99.58 |
| High complexity of model | ||
| CNN + RCAB + FB [20] | Self-collected | 5 | 99.95 |
|
| ||
| ResNet-50 + ResNext blocks [21] | New PlantVillage | 7 | 98.73 |
|
| ||
| MobileNet-V3 + ECA [22] | Images from PlantVillage and PlantDoc | 4 | 98.23 |
|
| ||
| Using low-light noisy images | DPA-Net (proposed) | PlantVillage | 38 | 92.11 | First study on plant disease classification of low-light noisy images | Complex background is not considered | |
| Potato Leaf Disease | 3 | 88.92 | |||||
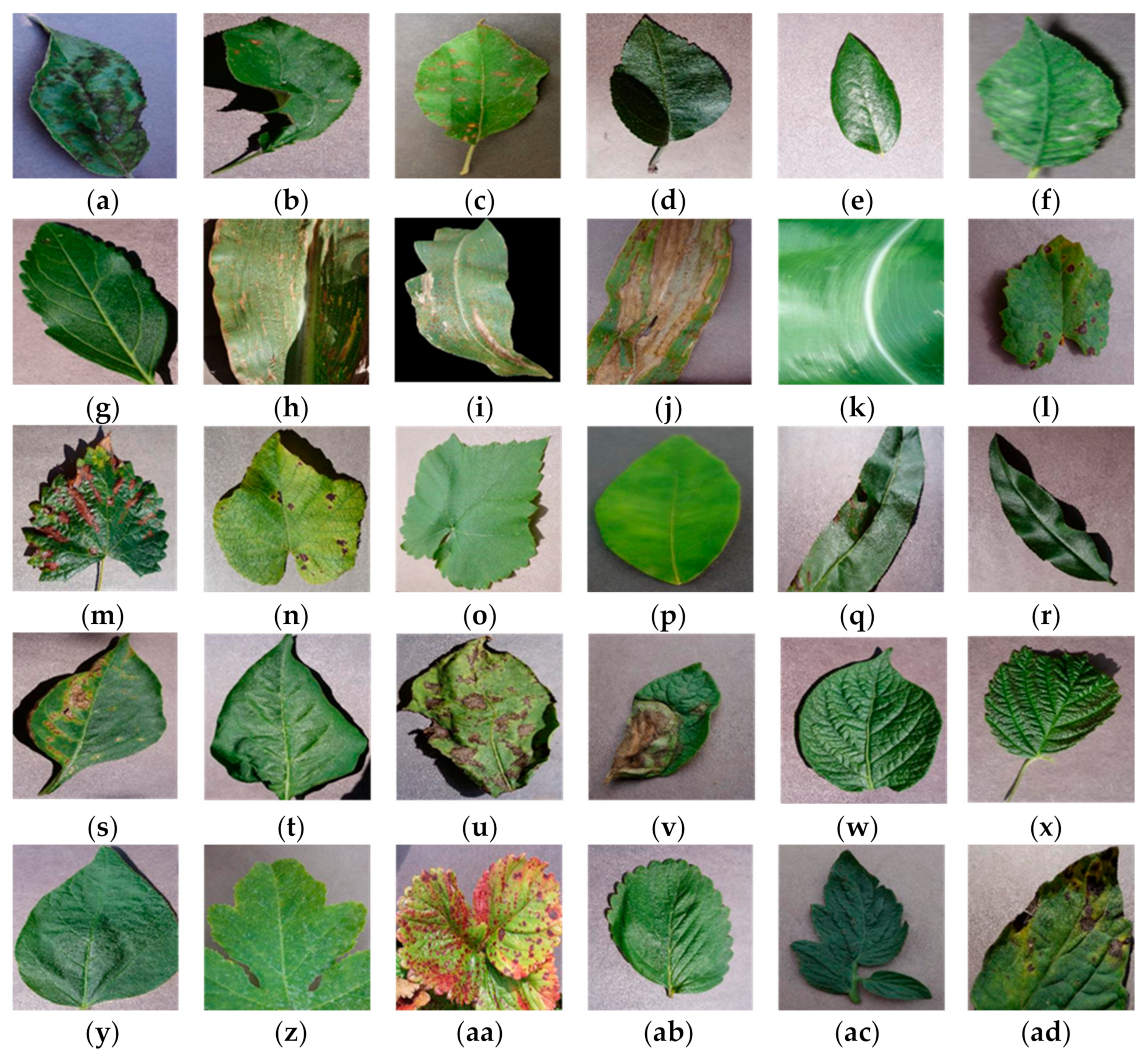
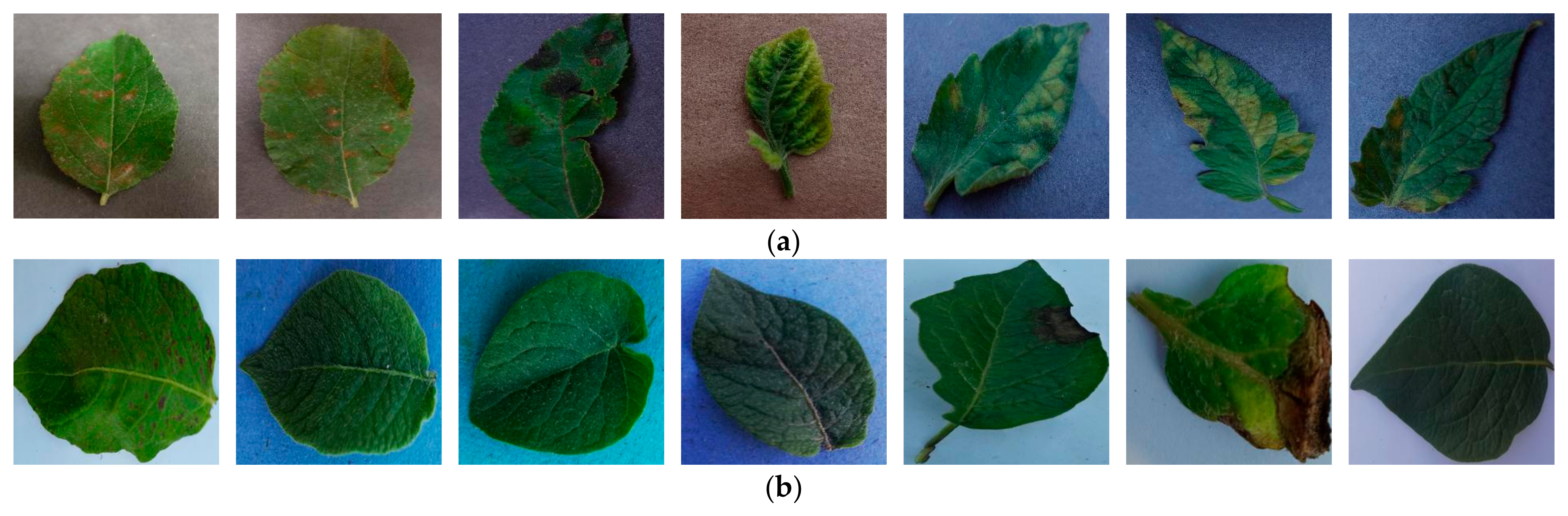
| Plant Name | Class Name | Sample | Total Number of Samples | |
|---|---|---|---|---|
| Apple | Disease | Scab | 630 | 3171 |
| Black rot | 621 | |||
| Cedar apple rust | 275 | |||
| Healthy | 1645 | |||
| Blueberry | Healthy | 1502 | 1502 | |
| Cherry | Disease | Powdery mildew | 1052 | 1906 |
| Healthy | 854 | |||
| Corn | Disease | Gray leaf spot | 513 | 3852 |
| Common rust | 1192 | |||
| Northern leaf blight | 985 | |||
| Healthy | 1162 | |||
| Grape | Disease | Black rot | 1180 | 4062 |
| Black measles | 1383 | |||
| Leaf blight | 1076 | |||
| Healthy | 423 | |||
| Orange | Healthy | 5507 | 5507 | |
| Peach | Disease | Bacterial spot | 2297 | 2657 |
| Healthy | 360 | |||
| Pepper | Disease | Bacterial spot | 997 | 2475 |
| Healthy | 1478 | |||
| Potato | Disease | Early blight | 1000 | 2152 |
| Late blight | 1000 | |||
| Healthy | 152 | |||
| Raspberry | Healthy | 371 | 371 | |
| Soybean | Healthy | 5090 | 5090 | |
| Squash | Disease | Powdery mildew | 1835 | 1835 |
| Strawberry | Disease | Leaf scorch | 1109 | 1565 |
| Healthy | 456 | |||
| Tomato | Disease | Bacterial spot | 2127 | 18,160 |
| Early blight | 1000 | |||
| Late blight | 1909 | |||
| Leaf mold | 952 | |||
| Septoria leaf spot | 1771 | |||
| Spider mites | 1676 | |||
| Target spot | 1404 | |||
| Mosaic virus | 373 | |||
| Yellow leaf curl virus | 5357 | |||
| Healthy | 1591 | |||
| Total number of images | 54,305 | |||
| Plant Name | Class Name | Total Number of Samples | |
|---|---|---|---|
| Potato | Disease | Early blight | 1628 |
| Late blight | 1424 | ||
| Healthy | 1020 | ||
| Total number of images | 4072 | ||
| Case | DCB | FCB | MFEB | PAB | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|---|---|---|
| 1 | 89.84 | 86.97 | 85.54 | 86.25 | ||||
| 2 | ✓ | 90.79 | 88.06 | 86.94 | 87.49 | |||
| 3 | ✓ | 90.89 | 88.26 | 87.02 | 87.63 | |||
| 4 | ✓ | 90.22 | 87.25 | 86.32 | 86.78 | |||
| 5 | ✓ | 90.36 | 87.61 | 86.36 | 86.98 | |||
| 6 | ✓ | ✓ | 91.17 | 88.64 | 87.18 | 87.90 | ||
| 7 | ✓ | ✓ | ✓ | 91.54 | 89.15 | 87.88 | 88.51 | |
| Proposed (DPA-Net) | ✓ | ✓ | ✓ | ✓ | 92.11 | 89.73 | 88.49 | 89.11 |
| Dilation Rate | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| 1, 2, 4 | 91.51 | 88.94 | 87.89 | 88.41 |
| 1, 4, 6 | 91.54 | 89.22 | 87.84 | 88.52 |
| 1, 3, 5 (proposed) | 92.11 | 89.73 | 88.49 | 89.11 |
| Dilation Layers | Dilate Rate | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| 2 | 3, 5 | 91.67 | 89.23 | 88.21 | 88.72 |
| 3 (proposed) | 1, 3, 5 | 92.11 | 89.73 | 88.49 | 89.11 |
| 4 | 1, 3, 5, 7 | 91.84 | 89.53 | 88.27 | 88.89 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| With 3 × 3 convolution layer only | 91.88 | 89.23 | 88.32 | 88.77 |
| With 1 × 1 convolution layer only | 91.66 | 89.03 | 87.98 | 88.50 |
| Without attention | 92.04 | 89.73 | 88.47 | 89.10 |
| With attention (proposed) | 92.11 | 89.73 | 88.49 | 89.11 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Without A | 91.91 | 89.20 | 88.33 | 88.76 |
| Without B | 91.94 | 89.49 | 88.36 | 88.92 |
| Without C | 91.99 | 89.87 | 88.33 | 89.09 |
| Without F2 | 91.54 | 88.93 | 87.97 | 88.38 |
| With A, B, C, and F2 (proposed) | 92.11 | 89.73 | 88.49 | 89.11 |
| Case | Learning Rate | Batch Size | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|---|
| 1 | 0.00005 | 8 | 68.10 | 67.65 | 67.39 | 67.52 |
| 2 | 0.0005 | 8 | 63.88 | 67.31 | 63.10 | 65.14 |
| 3 | 0.0001 | 16 | 76.31 | 76.05 | 75.84 | 75.94 |
| 4 | 0.0001 | 4 | 79.01 | 78.63 | 78.61 | 78.62 |
| Proposed | 0.0001 | 8 | 88.92 | 88.88 | 88.32 | 88.60 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Swin-T [43] | 69.13 | 60.77 | 59.89 | 60.32 |
| SqueezeNet [44] | 76.72 | 72.34 | 67.80 | 70.00 |
| ShuffleNet [45] | 76.96 | 70.96 | 66.77 | 68.79 |
| AlexNet [46] | 81.14 | 74.79 | 75.08 | 74.93 |
| XceptionNet [47] | 88.00 | 83.90 | 83.41 | 83.65 |
| Resnet-50 [48] | 88.12 | 84.07 | 83.94 | 84.01 |
| MobileNet-V2 [49] | 88.61 | 84.94 | 84.21 | 84.57 |
| VGG-16 [50] | 88.95 | 85.58 | 84.58 | 85.08 |
| InceptionNet [51] | 89.40 | 85.89 | 85.70 | 85.80 |
| DenseNet-121 [52] | 89.63 | 86.07 | 85.68 | 85.87 |
| ConvNext-small [53] | 89.84 | 86.97 | 85.54 | 86.25 |
| DPA-Net (proposed) | 92.11 | 89.73 | 88.49 | 89.11 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Swin-T [54] | 39.94 | 13.11 | 33.33 | 19.03 |
| SqueezeNet [55] | 54.07 | 55.94 | 49.15 | 51.60 |
| ShuffleNet [56] | 66.55 | 66.47 | 65.26 | 65.85 |
| AlexNet [57] | 43.08 | 25.72 | 36.75 | 28.18 |
| XceptionNet [58] | 71.30 | 71.09 | 70.52 | 70.80 |
| ResNet-50 [59] | 78.13 | 78.36 | 77.36 | 77.85 |
| MobileNet-V2 [60] | 73.53 | 73.58 | 72.49 | 73.02 |
| VGG-16 [61] | 75.64 | 75.69 | 75.21 | 75.44 |
| InceptionNet [62] | 75.70 | 74.80 | 75.05 | 74.93 |
| DenseNet-121 [63] | 77.59 | 77.18 | 77.10 | 77.14 |
| EfficientNetV2 [64] | 78.87 | 78.56 | 77.80 | 78.18 |
| ConvNext-small [65] | 82.60 | 82.63 | 81.86 | 82.23 |
| DPA-Net (proposed) | 88.92 | 88.88 | 88.32 | 88.60 |
| Method | #Param (M) | FLOPs (G) | Memory Usage (MB) |
|---|---|---|---|
| Swin-T [43] | 18.89 | 2.98 | 72.03 |
| SqueezeNet [44] | 0.75 | 0.74 | 2.88 |
| ShuffleNet [45] | 0.38 | 0.04 | 1.45 |
| AlexNet [46] | 57.16 | 0.71 | 218.05 |
| XceptionNet [47] | 20.89 | 4.60 | 79.67 |
| Resnet-50 [48] | 23.59 | 4.13 | 89.97 |
| MobileNet-V2 [49] | 2.27 | 0.33 | 8.67 |
| VGG-16 [50] | 134.42 | 15.52 | 512.79 |
| InceptionNet [51] | 25.19 | 5.75 | 96.09 |
| DenseNet-121 [52] | 6.99 | 2.90 | 26.68 |
| ConvNext-small [53] | 49.44 | 8.68 | 188.61 |
| DPA-Net (proposed) | 52.35 | 9.63 | 199.72 |
| Method | #Param (M) | FLOPs (G) | Memory Usage (MB) |
|---|---|---|---|
| Swin-T [54] | 18.85 | 2.98 | 71.92 |
| SqueezeNet [55] | 0.74 | 0.73 | 2.81 |
| ShuffleNet [56] | 0.34 | 0.04 | 1.32 |
| AlexNet [57] | 57.02 | 0.71 | 217.50 |
| XceptionNet [58] | 20.81 | 4.60 | 79.40 |
| ResNet-50 [59] | 23.51 | 4.13 | 89.70 |
| MobileNet-V2 [60] | 2.23 | 0.33 | 8.50 |
| VGG-16 [61] | 134.28 | 15.52 | 512.24 |
| InceptionNet [62] | 25.11 | 5.75 | 95.82 |
| DenseNet-121 [63] | 6.96 | 2.90 | 26.54 |
| EfficientNetV2 [64] | 20.18 | 8.37 | 76.99 |
| ConvNext-small [65] | 49.41 | 8.68 | 188.50 |
| DPA-Net (Proposed) | 52.33 | 9.63 | 199.61 |
| Method | Desktop Computer | Jetson TX2 |
|---|---|---|
| Swin-T [43] | 5.69 | 606.65 |
| SqueezeNet [44] | 4.26 | 11.28 |
| ShuffleNet [45] | 4.96 | 10.87 |
| AlexNet [46] | 4.15 | 8.58 |
| XceptionNet [47] | 5.06 | 29.92 |
| Resnet-50 [48] | 5.08 | 31.26 |
| MobileNet-V2 [49] | 4.73 | 11.82 |
| VGG-16 [50] | 8.2 | 84.61 |
| InceptionNet [51] | 7.7 | 55.51 |
| DenseNet-12 [52] | 6.67 | 29.61 |
| ConvNext-small [53] | 9.63 | 89.55 |
| DPA-Net (proposed) | 10.26 | 96.29 |
| Accuracy | Precision | Recall | F1-Score | |||
|---|---|---|---|---|---|---|
| 1.5 | 0.5 | 24 | 88.40 | 88.49 | 88.01 | 88.25 |
| 1.5 | 0.5 | 26 | 88.47 | 88.20 | 88.48 | 88.34 |
| 1.4 | 0.5 | 25 | 87.98 | 87.80 | 87.55 | 87.67 |
| 1.6 | 0.5 | 25 | 89.38 | 89.36 | 88.87 | 89.11 |
| 1.5 | 0.5 | 25 | 88.92 | 88.88 | 88.32 | 88.60 |
| 1.5 | 0.6 | 25 | 89.51 | 89.80 | 89.17 | 89.48 |
| Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|
| 85.00 | 86.26 | 85.00 | 85.63 |
| Results | Healthy Case | Disease Cases | ||
|---|---|---|---|---|
| Black Measles | Yellow Leaf Curl | Bacterial Spot | ||
| FD | 1.6453 | 1.2486 | 1.2662 | 1.2748 |
| R2 | 0.9978 | 0.9904 | 0.9900 | 0.9916 |
| C | 0.9989 | 0.9952 | 0.9950 | 0.9958 |
| Method | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| DPA-Net (proposed) | 88.92 | 88.88 | 88.32 | 88.60 |
| DPA-Net (proposed) with FD analysis | 93.68 | 96.75 | 95.07 | 95.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gondal, H.A.H.; Jeong, S.I.; Jang, W.H.; Kim, J.S.; Akram, R.; Irfan, M.; Tariq, M.H.; Park, K.R. Artificial Intelligence-Based Plant Disease Classification in Low-Light Environments. Fractal Fract. 2025, 9, 691. https://doi.org/10.3390/fractalfract9110691
Gondal HAH, Jeong SI, Jang WH, Kim JS, Akram R, Irfan M, Tariq MH, Park KR. Artificial Intelligence-Based Plant Disease Classification in Low-Light Environments. Fractal and Fractional. 2025; 9(11):691. https://doi.org/10.3390/fractalfract9110691
Chicago/Turabian StyleGondal, Hafiz Ali Hamza, Seong In Jeong, Won Ho Jang, Jun Seo Kim, Rehan Akram, Muhammad Irfan, Muhammad Hamza Tariq, and Kang Ryoung Park. 2025. "Artificial Intelligence-Based Plant Disease Classification in Low-Light Environments" Fractal and Fractional 9, no. 11: 691. https://doi.org/10.3390/fractalfract9110691
APA StyleGondal, H. A. H., Jeong, S. I., Jang, W. H., Kim, J. S., Akram, R., Irfan, M., Tariq, M. H., & Park, K. R. (2025). Artificial Intelligence-Based Plant Disease Classification in Low-Light Environments. Fractal and Fractional, 9(11), 691. https://doi.org/10.3390/fractalfract9110691









