Unsupervised Deep Learning Approach for Characterizing Fractality in Dried Drop Patterns of Differently Mixed Viscum album Preparations
Abstract
:1. Introduction
2. Materials and Methods
- 1
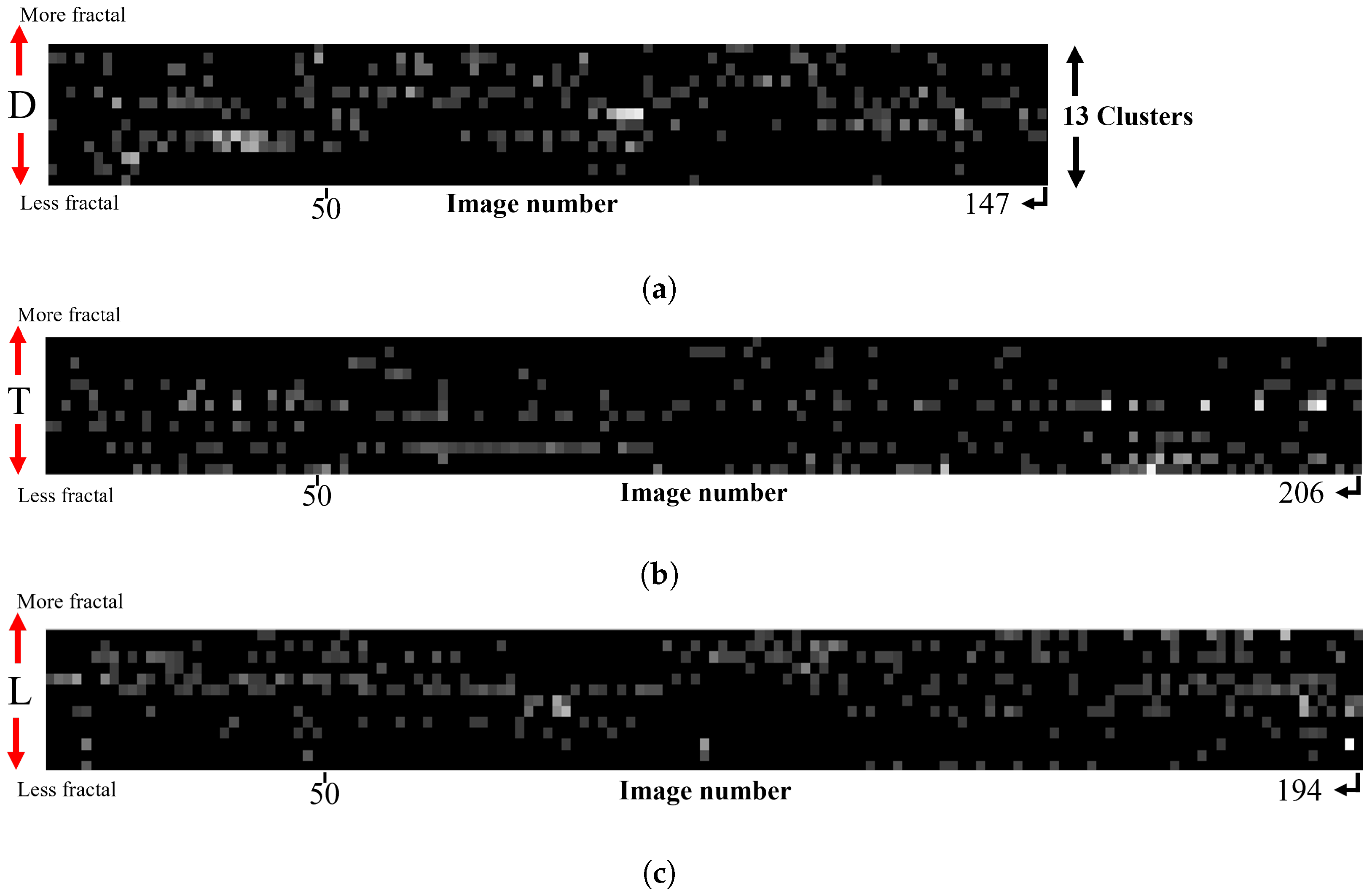
- Turbulent Flow (Variant T): The mixing process involves using a machine to apply vertical strokes. This set comprises a total of 206 images.
- 2
- Laminar Flow (Variant L): The mixing process is carried out manually, inducing a sequence of vortex-like flows. This set comprises a total of 196 images.
- 3
- Diffusion Control (Variant D): This set, consisting of 204 images, represents the control variant where the mixture remains as undisturbed as possible, relying on diffusion processes for dilution.
2.1. Automatic Full Texture Patch Selection
2.2. Deep Texture Representation Using Convolutional Neural Network
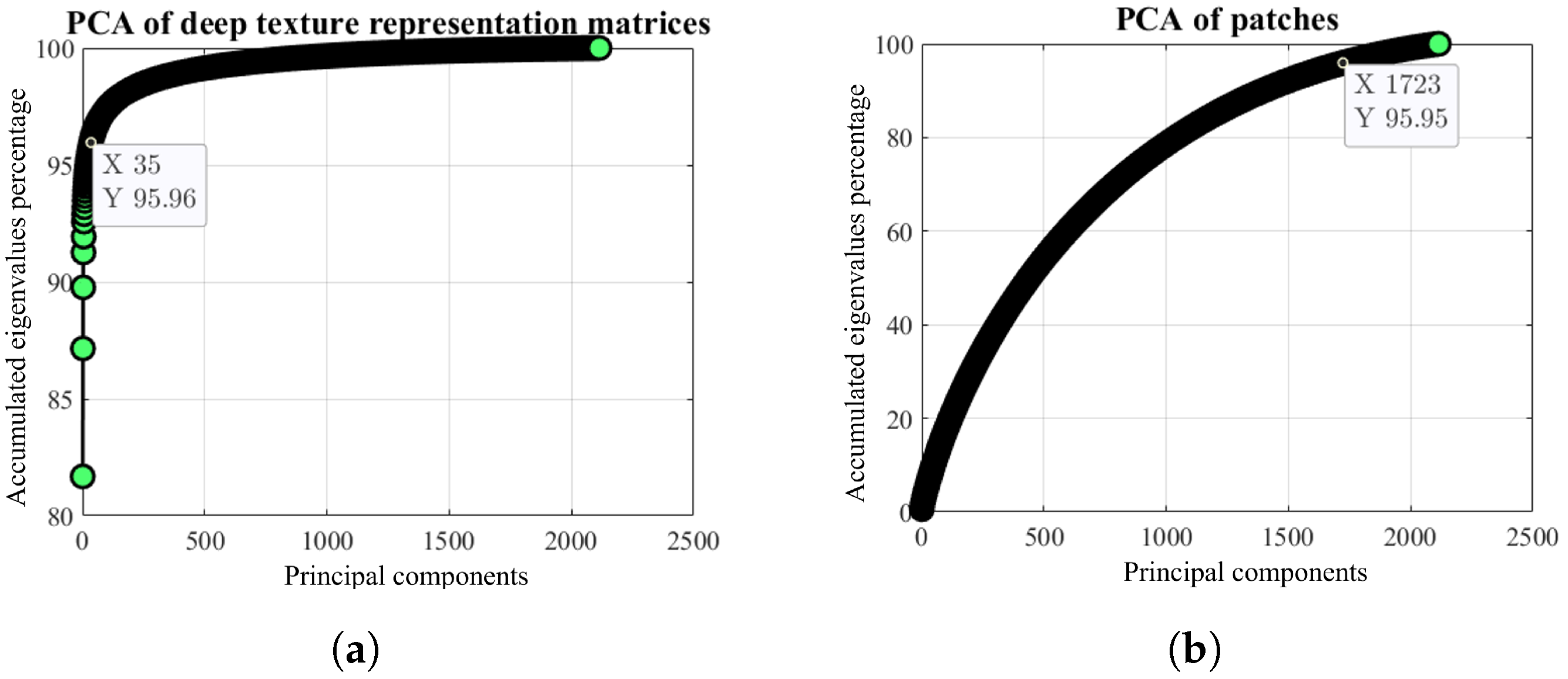
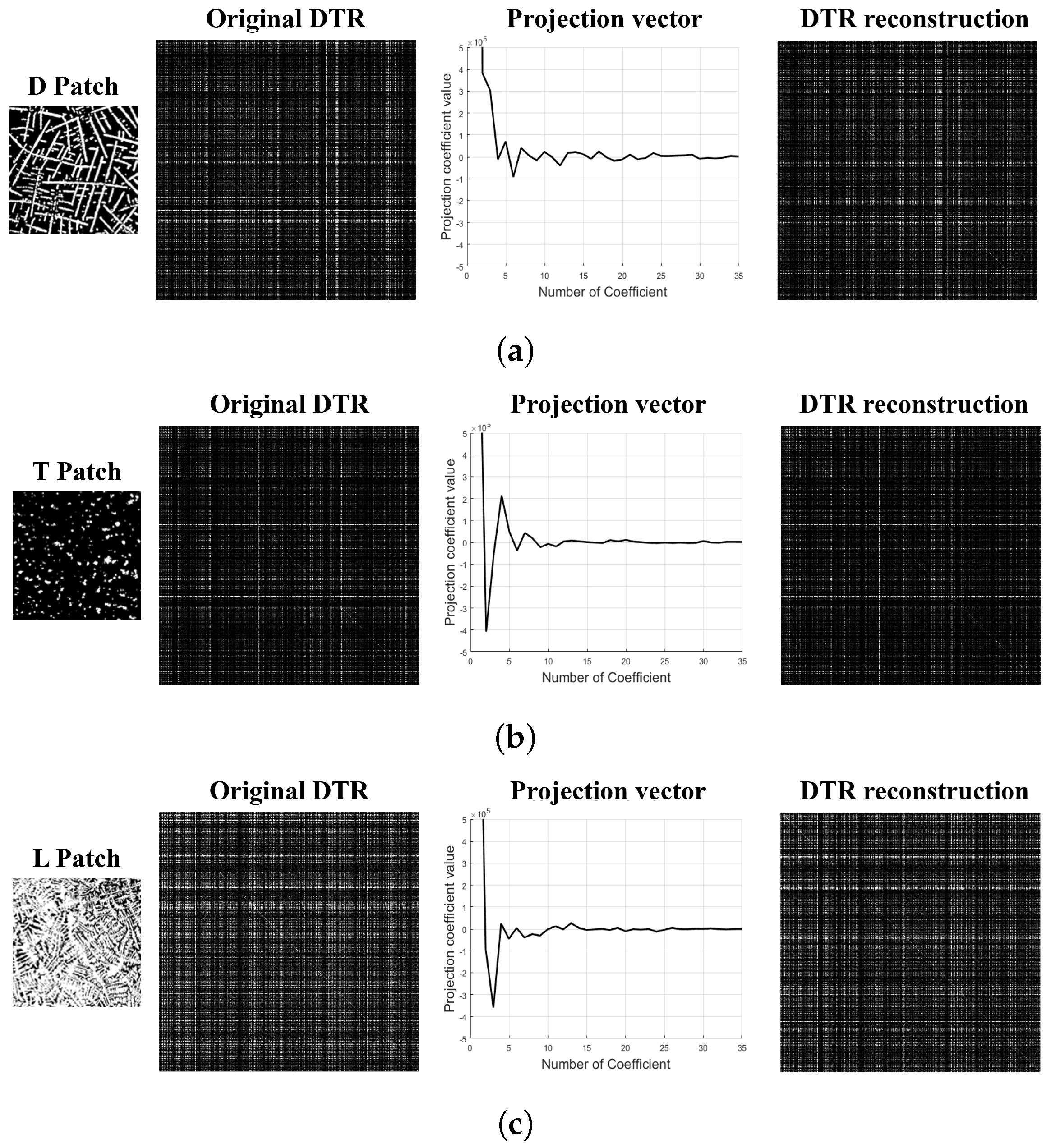
2.3. Dimensionality Reduction of the Deep Texture Representation
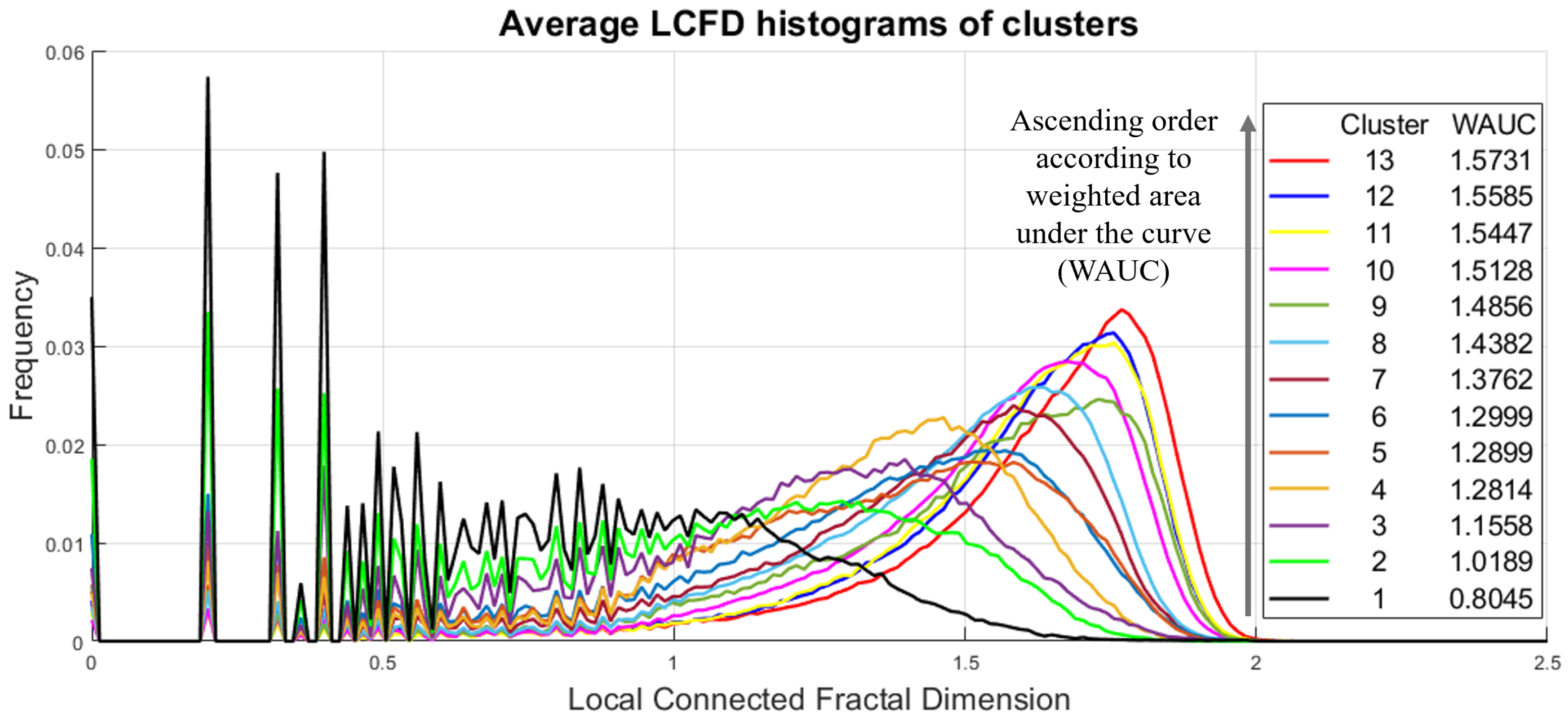
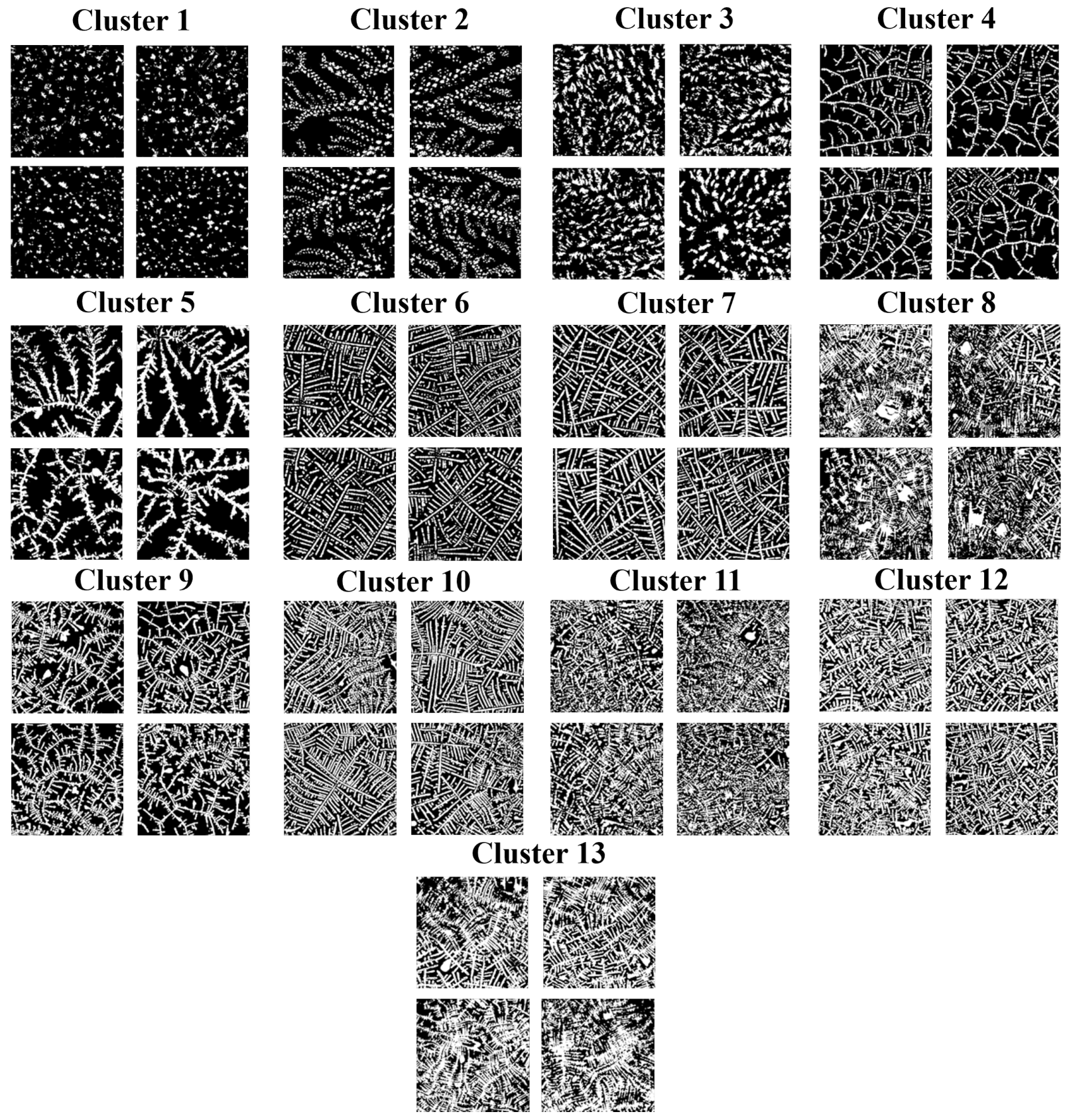
2.4. DTR Clusterization in the Reduced-Dimension Space
2.5. Clustering Refinement
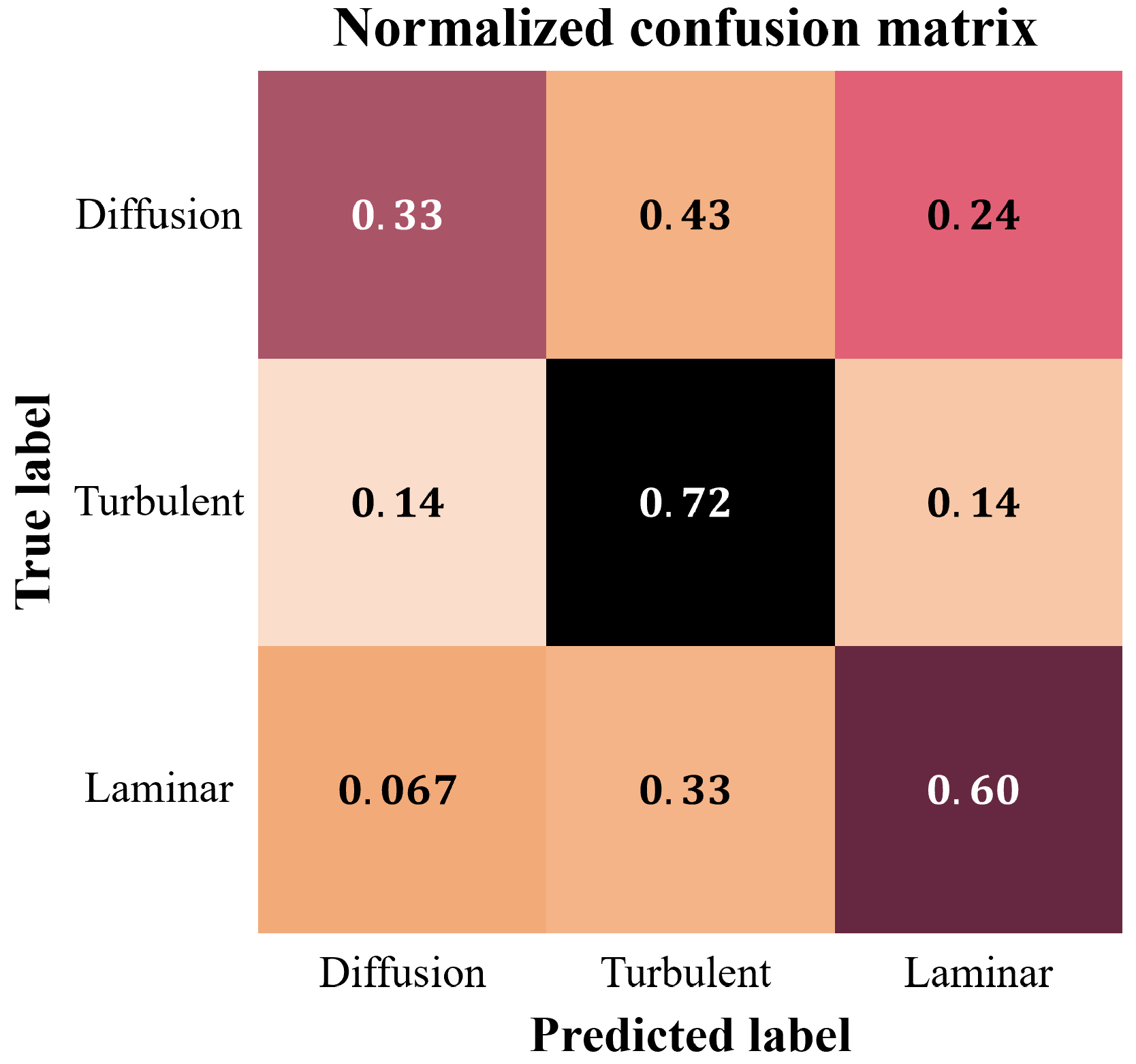
3. Results at the Image Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| VAQ | Viscum album Quercus |
| CNN | Convolutional Neural Network |
| DTR | Deep Texture Representation |
| DEM | Droplet Evaporation Method |
| PCA | Principal Component Analysis |
| DenseNet | Dense Convolutional Neural Network |
| WAUC | Weighted Area Under the Curve |
| LCFD | Local Connected Fractal Dimension |
| SVM | Support Vector Machine |
| GLCM | Gray Level Co-occurrence Matrix |
References
- Xing, F.; Xie, Y.; Su, H.; Liu, F.; Yang, L. Deep Learning in Microscopy Image Analysis: A Survey. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 4550–4568. [Google Scholar] [CrossRef] [PubMed]
- Razzak, M.I.; Naz, S.; Zaib, A. Deep Learning for Medical Image Processing: Overview, Challenges and Future. arXiv 2017, arXiv:1704.06825. [Google Scholar]
- Englbrecht, F.; Ruider, I.E.; Bausch, A.R. Automatic image annotation for fluorescent cell nuclei segmentation. PLoS ONE 2021, 16, e0250093. [Google Scholar] [CrossRef]
- Weeks, A.R. Morphological operations on color images. J. Electron. Imaging 2001, 10, 548. [Google Scholar] [CrossRef]
- Sharma, N.; Ray, A.; Shukla, K.; Sharma, S.; Pradhan, S.; Srivastva, A.; Aggarwal, L. Automated medical image segmentation techniques. J. Med. Phys. 2010, 35, 3. [Google Scholar] [CrossRef]
- Salau, A.O.; Jain, S. Feature Extraction: A Survey of the Types, Techniques, Applications. In Proceedings of the 2019 International Conference on Signal Processing and Communication (ICSC), Noida, India, 7–9 March 2019. [Google Scholar] [CrossRef]
- Almeida-Prieto, S.; Blanco-Méndez, J.; Otero-Espinar, F.J. Microscopic image analysis techniques for the morphological characterization of pharmaceutical particles: Influence of process variables. J. Pharm. Sci. 2006, 95, 348–357. [Google Scholar] [CrossRef]
- Raje, C.; Rangole, J. Detection of Leukemia in microscopic images using image processing. In Proceedings of the 2014 International Conference on Communication and Signal Processing, Melmaruvathur, India, 3–5 April 2014. [Google Scholar] [CrossRef]
- Cruz, D.; Jennifer, C.; Valiente; Castor, L.C.; Mendoza, C.M.T.; Jay, B.A.; Jane, L.S.C.; Brian, P.T.B. Determination of blood components (WBCs, RBCs, and Platelets) count in microscopic images using image processing and analysis. In Proceedings of the 2017 IEEE 9th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment and Management (HNICEM), Manila, Philippines, 1–3 December 2017. [Google Scholar] [CrossRef]
- Liu, Z.; Jin, L.; Chen, J.; Fang, Q.; Ablameyko, S.; Yin, Z.; Xu, Y. A survey on applications of deep learning in microscopy image analysis. Comput. Biol. Med. 2021, 134, 104523. [Google Scholar] [CrossRef]
- Galata, D.L.; Mészáros, L.A.; Kállai-Szabó, N.; Szabó, E.; Pataki, H.; Marosi, G.; Nagy, Z.K. Applications of machine vision in pharmaceutical technology: A review. Eur. J. Pharm. Sci. 2021, 159, 105717. [Google Scholar] [CrossRef]
- Kokornaczyk, M.O.; Trebbi, G.; Dinelli, G.; Marotti, I.; Bregola, V.; Nani, D.; Borghini, F.; Betti, L. Droplet evaporation method as a new potential approach for highlighting the effectiveness of ultra high dilutions. Complement. Ther. Med. 2014, 22, 333–340. [Google Scholar] [CrossRef]
- Kokornaczyk, M.O.; Würtenberger, S.; Baumgartner, S. Impact of succussion on pharmaceutical preparations analyzed by means of patterns from evaporated droplets. Sci. Rep. 2020, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Kokornaczyk, M.O.; Kunz, C.; Baumgartner, S. Impact of vertical succussion strokes vs. vortex potentization on droplet evaporation patterns obtained from Iscador Quercus 3x potency. Int. J. High Dilution Res. 2021, 18, 3. [Google Scholar] [CrossRef]
- Abdul-Fattah, A.M.; Kalonia, D.S.; Pikal, M.J. The Challenge of Drying Method Selection for Protein Pharmaceuticals: Product Quality Implications. J. Pharm. Sci. 2007, 96, 1886–1916. [Google Scholar] [CrossRef]
- Scherr, C.; Bodrova, N.B.; Baumgartner, S.; Kokornaczyk, M. Phase-Transition-Induced Pattern Formation Applied to Basic Research on Homeopathy: A Systematic Review. Homeopathy 2018, 107, 181–188. [Google Scholar] [CrossRef]
- Crivoi, A.; Duan, F. Evaporation-induced formation of fractal-like structures from nanofluids. Phys. Chem. Chem. Phys. 2012, 14, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Akhtar, M.N.; Navascués, M.A. Fractal Dimension of Fractal Functions on the Real Projective Plane. Fractal Fract. 2023, 7, 510. [Google Scholar] [CrossRef]
- Pantic, I.; Valjarevic, S.; Cumic, J.; Paunkovic, I.; Terzic, T.; Corridon, P.R. Gray Level Co-Occurrence Matrix, Fractal and Wavelet Analyses of Discrete Changes in Cell Nuclear Structure following Osmotic Stress: Focus on Machine Learning Methods. Fractal Fract. 2023, 7, 272. [Google Scholar] [CrossRef]
- Urgilez-Clavijo, A.; Rivas-Tabares, D.A.; Martín-Sotoca, J.J.; Alfonso, A.M.T. Local Fractal Connections to Characterize the Spatial Processes of Deforestation in the Ecuadorian Amazon. Entropy 2021, 23, 748. [Google Scholar] [CrossRef]
- Carreón, Y.J.P.; Díaz-Hernández, O.; Santos, G.J.E.; Cipriano-Urbano, I.; Solorio-Ordaz, F.J.; González-Gutiérrez, J.; Zenit, R. Texture Analysis of Dried Droplets for the Quality Control of Medicines. Sensors 2021, 21, 4048. [Google Scholar] [CrossRef]
- Zuñiga, A.G.; Florindo, J.B.; Bruno, O.M. Gabor wavelets combined with volumetric fractal dimension applied to texture analysis. Pattern Recognit. Lett. 2014, 36, 135–143. [Google Scholar] [CrossRef]
- Scoutaris, N.; Ross, S.; Douroumis, D. Current Trends on Medical and Pharmaceutical Applications of Inkjet Printing Technology. Pharm. Res. 2016, 33, 1799–1816. [Google Scholar] [CrossRef] [PubMed]
- Sachar, S.; Kumar, A. Deep ensemble learning for automatic medicinal leaf identification. Int. J. Inf. Technol. 2022, 14, 3089–3097. [Google Scholar] [CrossRef] [PubMed]
- Acuña, C.; y Terán, A.M.; Kokornaczyk, M.O.; Baumgartner, S.; Castelán, M. Deep learning applied to analyze patterns from evaporated droplets of Viscum album extracts. Sci. Rep. 2022, 12, 15332. [Google Scholar] [CrossRef]
- Monograph, T. European Pharmacopoeia. In European Directorate for the Quality of Medicine & Health Care of the Council of 376 Europe (EDQM), 9th ed.; Communications and Events Division: Strasbourg, France, 2017; Volume 9, pp. 3104–3105. [Google Scholar]
- Malhi, U.S.; Zhou, J.; Yan, C.; Rasool, A.; Siddeeq, S.; Du, M. Unsupervised Deep Embedded Clustering for High-Dimensional Visual Features of Fashion Images. Appl. Sci. 2023, 13, 2828. [Google Scholar] [CrossRef]
- Fard, M.M.; Thonet, T.; Gaussier, E. Deep k-Means: Jointly clustering with k-Means and learning representations. Pattern Recognit. Lett. 2020, 138, 185–192. [Google Scholar] [CrossRef]
- Ren, Y.; Pu, J.; Yang, Z.; Xu, J.; Li, G.; Pu, X.; Yu, P.S.; He, L. Deep Clustering: A Comprehensive Survey. arXiv 2022, arXiv:2210.04142. [Google Scholar]
- Stauder, G.M.; Matthes, H.; Friedel, W.E.; Bock, P.R. Use of fermented mistletoe (Viscum album L.) extract from oak tree (Quercus) as supportive treatment for patients with pancreatic cancer. J. Clin. Oncol. 2009, 27, e15656. [Google Scholar] [CrossRef]
- Tröger, W.; Galun, D.; Reif, M.; Schumann, A.; Stanković, N.; Milićević, M. Quality of Life of Patients with Advanced Pancreatic Cancer During Treatment with Mistletoe. Dtsch. Ärzteblatt Int. 2014, 111, 493. [Google Scholar] [CrossRef]
- Thronicke, A.; Schad, F.; Debus, M.; Grabowski, J.; Soldner, G. Viscum album L. Ther. Oncol. Update Curr. Evidence. Complement. Med. Res. 2022, 29, 362–368. [Google Scholar] [CrossRef]
- Kelley, J.B.; Paschal, B.M. Fluorescence-based quantification of nucleocytoplasmic transport. Methods 2019, 157, 106–114. [Google Scholar] [CrossRef]
- Gatys, L.A.; Ecker, A.S.; Bethge, M. Texture Synthesis Using Convolutional Neural Networks. arXiv 2015, arXiv:1505.07376. [Google Scholar]
- Nielsen, F. Hierarchical Clustering. In Introduction to HPC with MPI for Data Science; Springer International Publishing: Basel, Swizterland, 2016; pp. 195–211. [Google Scholar] [CrossRef]
- Huang, Q.; Gao, R.; Akhavan, H. An ensemble hierarchical clustering algorithm based on merits at cluster and partition levels. Pattern Recognit. 2023, 136, 109255. [Google Scholar] [CrossRef]
- Murtagh, F.; Contreras, P. Algorithms for hierarchical clustering: An overview. WIREs Data Min. Knowl. Discov. 2011, 2, 86–97. [Google Scholar] [CrossRef]
- Zambelli, A.E. A data-driven approach to estimating the number of clusters in hierarchical clustering. F1000Research 2016, 5, 2809. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Karypis, G.; Fayyad, U. Hierarchical Clustering Algorithms for Document Datasets. Data Min. Knowl. Discov. 2005, 10, 141–168. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2016, arXiv:1608.06993. [Google Scholar]
- de Mello, D.; Assunção, R.; Murai, F. Top-Down Deep Clustering with Multi-generator GANs. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual, 22 February–1 March 2022; AAAI Press: Palo Alto, CA, USA, 2021. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, H.; Zheng, Z.; Chen, J.; Li, Z.; Bu, J.; Wu, J.; Wang, X.; Zhu, W.; Ester, M. A Comprehensive Survey on Deep Clustering: Taxonomy, Challenges, and Future Directions. arXiv 2022, arXiv:2206.07579. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acuña, C.; Kokornaczyk, M.O.; Baumgartner, S.; Castelán, M. Unsupervised Deep Learning Approach for Characterizing Fractality in Dried Drop Patterns of Differently Mixed Viscum album Preparations. Fractal Fract. 2023, 7, 733. https://doi.org/10.3390/fractalfract7100733
Acuña C, Kokornaczyk MO, Baumgartner S, Castelán M. Unsupervised Deep Learning Approach for Characterizing Fractality in Dried Drop Patterns of Differently Mixed Viscum album Preparations. Fractal and Fractional. 2023; 7(10):733. https://doi.org/10.3390/fractalfract7100733
Chicago/Turabian StyleAcuña, Carlos, Maria Olga Kokornaczyk, Stephan Baumgartner, and Mario Castelán. 2023. "Unsupervised Deep Learning Approach for Characterizing Fractality in Dried Drop Patterns of Differently Mixed Viscum album Preparations" Fractal and Fractional 7, no. 10: 733. https://doi.org/10.3390/fractalfract7100733
APA StyleAcuña, C., Kokornaczyk, M. O., Baumgartner, S., & Castelán, M. (2023). Unsupervised Deep Learning Approach for Characterizing Fractality in Dried Drop Patterns of Differently Mixed Viscum album Preparations. Fractal and Fractional, 7(10), 733. https://doi.org/10.3390/fractalfract7100733






