Margin-Based Training of HDC Classifiers
Abstract
:1. Introduction
- Establishment of criteria and decision rules to trigger the refinement of HDC classifiers, based on the confidence levels of individual sample classifications.
- Development of HDC classifiers trained using either a multiplicative or additive margin applied to confidence scores, which operate on quantized class prototypes, and a comparative analysis of these methods.
- Configuration and enhancement of margin-based HDC classifiers for scalability, enabling effective operation on larger and more diverse datasets, exemplified by experimentation on 121 UCI datasets.
- Comprehensive experimental evaluation across 121 UCI datasets, demonstrating improved mean accuracy of the proposed HDC classifiers compared to other HDC models.
- Assessment of the impact of design choices on the classification performance of the proposed HDC classifiers.
2. Background on HDC and Basic HDC Classifiers
2.1. Formation of HDC Representations
2.2. HDC Operations
2.3. Hypervector Representation of Scalars and Numerical Vectors
- Quantizing the numerical values of each feature to the range of values such that .
- Representing the quantized feature values as their HVs using linear mapping.
- Assigning a random HV to each of the d features .
- Binding each feature value HV with its corresponding feature HV using the component-wise XOR, i.e., .
- Finally, obtaining the HV by the addition of all d bound feature–value pairs followed by a majority vote:
2.4. Basic HDC Classifiers
- For the current training sample i, predict its class using the classifier.
- If the prediction is incorrect, then update both the correct class prototype and the incorrect class prototype.
- If the prediction is correct, leave all class prototypes unchanged.
3. Related Work
3.1. Prototype Classifiers
3.2. Prototype Refinement
3.3. Classification Scores and Confidence in Classification Results
3.4. Other HDC Classifiers
4. Methods
4.1. HDC Classifiers with Additive Margin
4.2. HDC Classifiers with Multiplicative Margin
4.3. Design Choices
4.3.1. Input Data Preprocessing/Standardization
4.3.2. Initialization of Class Prototypes
4.3.3. Quantization of Class Prototypes
4.3.4. Similarity Measures Between Hypervectors and Prototypes
4.3.5. The Training Procedure
4.3.6. The Choice of Design Setup for Experiments
- Input data standardization: Per-feature z-score standardization. In preliminary evaluations, we investigated more intensive approaches for managing outliers; however, these yielded inconsistent effects across datasets. Consequently, we adopt the standard z-score approach for consistency.
- Input data quantization: Uniform quantization into the [0, Q] range. We use .
- Input data transformation to hypervectors (Section 2.3): Here, instead of flipping bits per quantization level as in our previous work [48], we adopt a scheme that flips bits to generate HVs for scalar values. This effectively halves the dimensionality of the hypervectors.
- Hypervector standardization: We maintain the binary format of generated HVs by avoiding further standardization.
- Prototypes initialization: Before starting error-driven iterative training to refine class prototypes, we initialize the two sets of prototypes (integer and quantized) using the initial “incremental” prototypes from Prot-HDCL.
- The similarity measures between HVs and class prototypes: For AM-HDCL, we use the Hamming similarity normalized by the HV dimensionality, and for MM-HDCL, we apply dot-product similarity.
- Selection of values: The hyperparameter is evaluated over the range of [0.00, 0.02, 0.04, 0.06, 0.08, 0.10, 0.12, 0.15, 0.2, 0.25, 0.3, 0.4, 0.5, 0.7] for both AM-HDCL and MM-HDCL.
- Integer prototype update rule: According to rule (7).
- Binary prototype update rule: Binarization of integer prototypes is applied after each epoch. For AM-HDCL, binarization is achieved by thresholding based on class counter values, while for MM-HDCL, the thresholding is performed with zero values.
- Training stopping: In contrast to [48], we significantly reduce the number of training epochs from 2500 to 100, making training feasible for the large collection of 121 UCI datasets. Additionally, we halt training early if the training accuracy reached 0.9999.
- Selection of class prototypes for deployment: Upon completion of the training, we do not use the final class prototypes for testing. Nor do we apply any procedure or validate some hyperparameters to select some intermediate set of prototypes. Instead, we select the prototypes obtained at the epoch with the highest training accuracy, as in our previous work [48]. While this method may not always be optimal (we observed that maximum testing accuracy often occurs in earlier epochs, before potential “overtraining”), it remains reasonable since prototypes obtained after the training completion may lead to worse performance.
5. Experiments
6. Results and Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AM-HDCL | HDC classifier with additive margin |
| AM-HDCL = 0 | AM-HDCL with = 0 |
| HDC | Hyperdimensional computing |
| HV | Hypervector |
| MM-HDCL | HDC classifier with multiplicative margin |
| MM-HDCL = 0 | MM-HDCL with = 0 |
| Prot-HDCL | Prototype HDC classifier |
| Ref-HDCL | HDC classifier with prototype refinement |
Appendix A. Details on Datasets
| ID | Dataset | No. Samples | No. Features | No. Classes | Ratio Feat/Samples | Avg Samples per Class |
|---|---|---|---|---|---|---|
| 1 | abalone | 4177 | 8 | 3 | 0.0019 | 1392 |
| 2 | acute-inflammation | 120 | 6 | 2 | 0.0500 | 60 |
| 3 | acute-nephritis | 120 | 6 | 2 | 0.0500 | 60 |
| 4 | adult | 48,842 | 14 | 2 | 0.0003 | 24,421 |
| 5 | annealing | 898 | 31 | 5 | 0.0345 | 180 |
| 6 | arrhythmia | 452 | 262 | 13 | 0.5796 | 35 |
| 7 | audiology-std | 196 | 59 | 18 | 0.3010 | 11 |
| 8 | balance-scale | 625 | 4 | 3 | 0.0064 | 208 |
| 9 | balloons | 16 | 4 | 2 | 0.2500 | 8 |
| 10 | bank | 4521 | 16 | 2 | 0.0035 | 2260 |
| 11 | blood | 748 | 4 | 2 | 0.0053 | 374 |
| 12 | breast-cancer | 286 | 9 | 2 | 0.0315 | 143 |
| 13 | breast-cancer-wisc | 699 | 9 | 2 | 0.0129 | 350 |
| 14 | breast-cancer-wisc-diag | 569 | 30 | 2 | 0.0527 | 284 |
| 15 | breast-cancer-wisc-prog | 198 | 33 | 2 | 0.1667 | 99 |
| 16 | breast-tissue | 106 | 9 | 6 | 0.0849 | 18 |
| 17 | car | 1728 | 6 | 4 | 0.0035 | 432 |
| 18 | cardiotocography-10clases | 2126 | 21 | 10 | 0.0099 | 213 |
| 19 | cardiotocography-3clases | 2126 | 21 | 3 | 0.0099 | 709 |
| 20 | chess-krvk | 28,056 | 6 | 18 | 0.0002 | 1559 |
| 21 | chess-krvkp | 3196 | 36 | 2 | 0.0113 | 1598 |
| 22 | congressional-voting | 435 | 16 | 2 | 0.0368 | 218 |
| 23 | conn-bench-sonar-mines-rocks | 208 | 60 | 2 | 0.2885 | 104 |
| 24 | conn-bench-vowel-deterding | 990 | 11 | 11 | 0.0111 | 90 |
| 25 | connect-4 | 67,557 | 42 | 2 | 0.0006 | 33,778 |
| 26 | contrac | 1473 | 9 | 3 | 0.0061 | 491 |
| 27 | credit-approval | 690 | 15 | 2 | 0.0217 | 345 |
| 28 | cylinder-bands | 512 | 35 | 2 | 0.0684 | 256 |
| 29 | dermatology | 366 | 34 | 6 | 0.0929 | 61 |
| 30 | echocardiogram | 131 | 10 | 2 | 0.0763 | 66 |
| 31 | ecoli | 336 | 7 | 8 | 0.0208 | 42 |
| 32 | energy-y1 | 768 | 8 | 3 | 0.0104 | 256 |
| 33 | energy-y2 | 768 | 8 | 3 | 0.0104 | 256 |
| 34 | fertility | 100 | 9 | 2 | 0.0900 | 50 |
| 35 | flags | 194 | 28 | 8 | 0.1443 | 24 |
| 36 | glass | 214 | 9 | 6 | 0.0421 | 36 |
| 37 | haberman-survival | 306 | 3 | 2 | 0.0098 | 153 |
| 38 | hayes-roth | 160 | 3 | 3 | 0.0188 | 53 |
| 39 | heart-cleveland | 303 | 13 | 5 | 0.0429 | 61 |
| 40 | heart-hungarian | 294 | 12 | 2 | 0.0408 | 147 |
| 41 | heart-switzerland | 123 | 12 | 5 | 0.0976 | 25 |
| 42 | heart-va | 200 | 12 | 5 | 0.0600 | 40 |
| 43 | hepatitis | 155 | 19 | 2 | 0.1226 | 78 |
| 44 | hill-valley | 1212 | 100 | 2 | 0.0825 | 606 |
| 45 | horse-colic | 368 | 25 | 2 | 0.0679 | 184 |
| 46 | ilpd-indian-liver | 583 | 9 | 2 | 0.0154 | 292 |
| 47 | image-segmentation | 2310 | 18 | 7 | 0.0078 | 330 |
| 48 | ionosphere | 351 | 33 | 2 | 0.0940 | 176 |
| 49 | iris | 150 | 4 | 3 | 0.0267 | 50 |
| 50 | led-display | 1000 | 7 | 10 | 0.0070 | 100 |
| 51 | lenses | 24 | 4 | 3 | 0.1667 | 8 |
| 52 | letter | 20,000 | 16 | 26 | 0.0008 | 769 |
| 53 | libras | 360 | 90 | 15 | 0.2500 | 24 |
| 54 | low-res-spect | 531 | 100 | 9 | 0.1883 | 59 |
| 55 | lung-cancer | 32 | 56 | 3 | 1.7500 | 11 |
| 56 | lymphography | 148 | 18 | 4 | 0.1216 | 37 |
| 57 | magic | 19,020 | 10 | 2 | 0.0005 | 9510 |
| 58 | mammographic | 961 | 5 | 2 | 0.0052 | 480 |
| 59 | miniboone | 130,064 | 50 | 2 | 0.0004 | 65,032 |
| 60 | molec-biol-promoter | 106 | 57 | 2 | 0.5377 | 53 |
| 61 | molec-biol-splice | 3190 | 60 | 3 | 0.0188 | 1063 |
| 62 | monks-1 | 556 | 6 | 2 | 0.0108 | 278 |
| 63 | monks-2 | 601 | 6 | 2 | 0.0100 | 300 |
| 64 | monks-3 | 554 | 6 | 2 | 0.0108 | 277 |
| 65 | mushroom | 8124 | 21 | 2 | 0.0026 | 4062 |
| 66 | musk-1 | 476 | 166 | 2 | 0.3487 | 238 |
| 67 | musk-2 | 6598 | 166 | 2 | 0.0252 | 3299 |
| 68 | nursery | 12,960 | 8 | 5 | 0.0006 | 2592 |
| 69 | oocytes_merluccius_nucleus_4d | 1022 | 41 | 2 | 0.0401 | 511 |
| 70 | oocytes_merluccius_states_2f | 1022 | 25 | 3 | 0.0245 | 341 |
| 71 | oocytes_trisopterus_nucleus_2f | 912 | 25 | 2 | 0.0274 | 456 |
| 72 | oocytes_trisopterus_states_5b | 912 | 32 | 3 | 0.0351 | 304 |
| 73 | optical | 5620 | 62 | 10 | 0.0110 | 562 |
| 74 | ozone | 2536 | 72 | 2 | 0.0284 | 1268 |
| 75 | page-blocks | 5473 | 10 | 5 | 0.0018 | 1095 |
| 76 | parkinsons | 195 | 22 | 2 | 0.1128 | 98 |
| 77 | pendigits | 10,992 | 16 | 10 | 0.0015 | 1099 |
| 78 | pima | 768 | 8 | 2 | 0.0104 | 384 |
| 79 | pittsburg-bridges-MATERIAL | 106 | 7 | 3 | 0.0660 | 35 |
| 80 | pittsburg-bridges-REL-L | 103 | 7 | 3 | 0.0680 | 34 |
| 81 | pittsburg-bridges-SPAN | 92 | 7 | 3 | 0.0761 | 31 |
| 82 | pittsburg-bridges-T-OR-D | 102 | 7 | 2 | 0.0686 | 51 |
| 83 | pittsburg-bridges-TYPE | 105 | 7 | 6 | 0.0667 | 18 |
| 84 | planning | 182 | 12 | 2 | 0.0659 | 91 |
| 85 | plant-margin | 1600 | 64 | 100 | 0.0400 | 16 |
| 86 | plant-shape | 1600 | 64 | 100 | 0.0400 | 16 |
| 87 | plant-texture | 1599 | 64 | 100 | 0.0400 | 16 |
| 88 | post-operative | 90 | 8 | 3 | 0.0889 | 30 |
| 89 | primary-tumor | 330 | 17 | 15 | 0.0515 | 22 |
| 90 | ringnorm | 7400 | 20 | 2 | 0.0027 | 3700 |
| 91 | seeds | 210 | 7 | 3 | 0.0333 | 70 |
| 92 | semeion | 1593 | 256 | 10 | 0.1607 | 159 |
| 93 | soybean | 683 | 35 | 18 | 0.0512 | 38 |
| 94 | spambase | 4601 | 57 | 2 | 0.0124 | 2300 |
| 95 | spect | 265 | 22 | 2 | 0.0830 | 132 |
| 96 | spectf | 267 | 44 | 2 | 0.1648 | 134 |
| 97 | statlog-australian-credit | 690 | 14 | 2 | 0.0203 | 345 |
| 98 | statlog-german-credit | 1000 | 24 | 2 | 0.0240 | 500 |
| 99 | statlog-heart | 270 | 13 | 2 | 0.0481 | 135 |
| 100 | statlog-image | 2310 | 18 | 7 | 0.0078 | 330 |
| 101 | statlog-landsat | 6435 | 36 | 6 | 0.0056 | 1072 |
| 102 | statlog-shuttle | 58,000 | 9 | 7 | 0.0002 | 8286 |
| 103 | statlog-vehicle | 846 | 18 | 4 | 0.0213 | 212 |
| 104 | steel-plates | 1941 | 27 | 7 | 0.0139 | 277 |
| 105 | synthetic-control | 600 | 60 | 6 | 0.1000 | 100 |
| 106 | teaching | 151 | 5 | 3 | 0.0331 | 50 |
| 107 | thyroid | 7200 | 21 | 3 | 0.0029 | 2400 |
| 108 | tic-tac-toe | 958 | 9 | 2 | 0.0094 | 479 |
| 109 | titanic | 2201 | 3 | 2 | 0.0014 | 1100 |
| 110 | trains | 10 | 29 | 2 | 2.9000 | 5 |
| 111 | twonorm | 7400 | 20 | 2 | 0.0027 | 3700 |
| 112 | vertebral-column-2clases | 310 | 6 | 2 | 0.0194 | 155 |
| 113 | vertebral-column-3clases | 310 | 6 | 3 | 0.0194 | 103 |
| 114 | wall-following | 5456 | 24 | 4 | 0.0044 | 1364 |
| 115 | waveform | 5000 | 21 | 3 | 0.0042 | 1667 |
| 116 | waveform-noise | 5000 | 40 | 3 | 0.0080 | 1667 |
| 117 | wine | 178 | 13 | 3 | 0.0730 | 59 |
| 118 | wine-quality-red | 1599 | 11 | 6 | 0.0069 | 266 |
| 119 | wine-quality-white | 4898 | 11 | 7 | 0.0022 | 700 |
| 120 | yeast | 1484 | 8 | 10 | 0.0054 | 148 |
| 121 | zoo | 101 | 16 | 7 | 0.1584 | 14 |
Appendix B. Detailed Results per Dataset
| ID | Prot-HDCL | Prot-HDCL | AM-HDCL | MM-HDCL | AM-HDCL | MM-HDCL | AM | MM |
|---|---|---|---|---|---|---|---|---|
| 1 | 54.10 (±0.21) | 54.03 (±0.24) | 60.46 (±0.82) | 61.80 (±0.91) | 59.70 (±0.50) | 62.61 (±0.56) | 0.2 | 0.1 |
| 2 | 96.42 (±0.97) | 96.25 (±0.90) | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 0 | 0 |
| 3 | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 100.00 (±0.00) | 0 | 0 |
| 4 | 75.12 (±0.38) | 75.55 (±1.55) | 85.08 (±0.20) | 84.54 (±0.46) | 85.08 (±0.20) | 84.54 (±0.46) | 0 | 0 |
| 5 | 55.50 (±1.51) | 47.50 (±3.17) | 97.30 (±0.82) | 95.80 (±1.40) | 97.70 (±0.82) | 96.60 (±0.84) | 0.02 | 0.02 |
| 6 | 59.89 (±1.53) | 61.59 (±1.54) | 71.70 (±0.92) | 61.59 (±1.54) | 71.70 (±0.92) | 61.75 (±1.33) | 0 | 0.02 |
| 7 | 77.60 (±2.80) | 48.40 (±7.88) | 72.40 (±6.10) | 76.00 (±3.77) | 72.40 (±5.80) | 77.60 (±6.31) | 0.02 | 0.04 |
| 8 | 66.96 (±0.59) | 61.84 (±0.98) | 92.98 (±0.86) | 95.95 (±0.54) | 92.98 (±0.86) | 95.95 (±0.54) | 0 | 0 |
| 9 | 85.63 (±3.02) | 79.38 (±10.64) | 80.00 (±2.64) | 80.63 (±8.56) | 75.00 (±6.59) | 81.88 (±6.22) | 0.2 | 0.4 |
| 10 | 64.01 (±0.64) | 63.64 (±2.08) | 89.17 (±0.30) | 89.89 (±0.26) | 90.15 (±0.15) | 90.02 (±0.19) | 0.04 | 0.02 |
| 11 | 65.88 (±0.44) | 65.57 (±0.86) | 76.24 (±0.77) | 78.64 (±0.37) | 79.97 (±0.50) | 79.41 (±0.62) | 0.2 | 0.2 |
| 12 | 71.94 (±0.60) | 66.02 (±1.38) | 68.70 (±1.20) | 73.03 (±1.90) | 75.85 (±1.08) | 74.61 (±0.35) | 0.2 | 0.4 |
| 13 | 96.83 (±0.09) | 96.74 (±0.11) | 96.97 (±0.22) | 97.50 (±0.18) | 97.46 (±0.09) | 97.71 (±0.00) | 0.12 | 0.15 |
| 14 | 94.61 (±0.36) | 94.51 (±0.35) | 95.48 (±0.33) | 94.42 (±0.74) | 96.57 (±0.27) | 96.71 (±0.31) | 0.02 | 0.04 |
| 15 | 61.68 (±1.28) | 54.29 (±3.51) | 74.95 (±2.88) | 77.45 (±2.55) | 76.63 (±0.89) | 82.09 (±0.88) | 0.3 | 0.3 |
| 16 | 65.00 (±0.50) | 64.81 (±0.67) | 69.13 (±1.89) | 70.96 (±1.68) | 67.40 (±3.38) | 66.92 (±3.21) | 0.12 | 0.1 |
| 17 | 68.96 (±0.50) | 76.86 (±1.47) | 92.59 (±0.58) | 91.45 (±0.99) | 92.59 (±0.58) | 91.45 (±0.99) | 0 | 0 |
| 18 | 51.69 (±0.29) | 49.06 (±1.02) | 77.59 (±2.03) | 73.01 (±1.57) | 77.59 (±2.03) | 73.78 (±0.89) | 0 | 0.12 |
| 19 | 77.37 (±0.45) | 76.32 (±1.82) | 93.05 (±0.25) | 93.12 (±0.31) | 91.34 (±0.16) | 93.12 (±0.31) | 0.06 | 0 |
| 20 | 23.27 (±0.10) | 22.36 (±0.43) | 33.23 (±0.72) | 31.18 (±0.72) | 36.71 (±0.41) | 33.62 (±0.74) | 0.2 | 0.1 |
| 21 | 83.30 (±0.61) | 82.73 (±1.17) | 97.42 (±0.17) | 95.54 (±0.11) | 97.06 (±0.21) | 95.54 (±0.11) | 0.02 | 0 |
| 22 | 61.47 (±0.00) | 61.47 (±0.00) | 59.45 (±0.28) | 59.98 (±0.93) | 62.22 (±0.27) | 61.08 (±0.19) | 0.12 | 0.7 |
| 23 | 67.07 (±1.86) | 66.63 (±1.91) | 81.15 (±1.36) | 77.31 (±4.52) | 82.98 (±1.34) | 80.72 (±0.86) | 0.08 | 0.15 |
| 24 | 80.28 (±1.50) | 73.61 (±2.50) | 76.88 (±2.41) | 98.07 (±0.39) | 84.98 (±2.65) | 89.20 (±2.61) | 0.06 | 0.02 |
| 25 | 65.88 (±1.29) | 67.14 (±6.70) | 81.93 (±0.40) | 80.14 (±0.31) | 81.93 (±0.40) | 80.14 (±0.31) | 0 | 0 |
| 26 | 46.24 (±0.38) | 46.13 (±0.48) | 50.69 (±0.42) | 52.53 (±1.29) | 52.81 (±0.21) | 51.02 (±0.50) | 0.1 | 0.25 |
| 27 | 87.12 (±0.28) | 86.80 (±0.74) | 84.16 (±1.07) | 86.89 (±0.57) | 87.05 (±0.44) | 87.50 (±0.65) | 0.5 | 0.3 |
| 28 | 66.88 (±0.78) | 66.37 (±1.67) | 77.13 (±1.22) | 75.96 (±0.87) | 77.64 (±1.26) | 76.84 (±1.16) | 0.08 | 0.15 |
| 29 | 97.28 (±0.16) | 97.20 (±0.17) | 97.53 (±0.29) | 92.39 (±0.86) | 97.42 (±0.27) | 97.47 (±0.28) | 0.04 | 0.08 |
| 30 | 81.21 (±0.48) | 81.36 (±0.39) | 80.91 (±1.55) | 78.26 (±1.75) | 85.61 (±0.00) | 85.68 (±0.24) | 0.2 | 0.4 |
| 31 | 85.33 (±0.54) | 51.34 (±2.76) | 81.93 (±1.25) | 83.93 (±1.25) | 85.83 (±0.90) | 85.27 (±0.58) | 0.15 | 0.15 |
| 32 | 83.13 (±0.43) | 82.19 (±0.42) | 93.72 (±0.12) | 93.98 (±0.61) | 91.80 (±0.51) | 93.98 (±0.61) | 0.08 | 0 |
| 33 | 84.58 (±0.29) | 84.31 (±0.24) | 89.26 (±0.54) | 88.93 (±0.35) | 85.09 (±0.26) | 85.65 (±0.74) | 0.2 | 0.06 |
| 34 | 72.50 (±1.35) | 76.00 (±1.76) | 84.00 (±1.49) | 85.30 (±0.67) | 86.90 (±0.57) | 86.30 (±0.67) | 0.15 | 0.25 |
| 35 | 47.81 (±1.12) | 34.11 (±1.87) | 55.36 (±1.25) | 57.71 (±1.34) | 57.50 (±1.82) | 59.06 (±1.67) | 0.04 | 0.04 |
| 36 | 53.44 (±1.34) | 54.15 (±1.57) | 67.41 (±2.08) | 68.44 (±1.21) | 67.41 (±2.08) | 69.86 (±1.36) | 0 | 0.02 |
| 37 | 66.55 (±0.64) | 66.94 (±1.26) | 71.41 (±0.81) | 70.66 (±1.53) | 70.43 (±0.80) | 74.01 (±0.00) | 0.3 | 0.7 |
| 38 | 50.00 (±0.00) | 50.00 (±0.00) | 49.29 (±6.02) | 39.29 (±0.00) | 59.64 (±13.26) | 67.14 (±10.49) | 0.02 | 0.08 |
| 39 | 55.20 (±0.98) | 49.05 (±1.28) | 54.93 (±1.15) | 55.23 (±1.84) | 59.97 (±0.64) | 59.67 (±0.96) | 0.4 | 0.4 |
| 40 | 85.58 (±0.25) | 85.31 (±0.38) | 78.49 (±0.58) | 78.66 (±1.61) | 85.68 (±0.22) | 85.24 (±0.25) | 0.3 | 0.4 |
| 41 | 33.39 (±1.02) | 7.18 (±0.71) | 35.32 (±1.69) | 40.48 (±1.51) | 39.27 (±2.19) | 40.73 (±2.56) | 0.06 | 0.12 |
| 42 | 31.00 (±1.00) | 30.15 (±0.88) | 31.30 (±1.70) | 31.30 (±2.53) | 31.35 (±2.84) | 31.30 (±2.53) | 0.04 | 0 |
| 43 | 78.59 (±0.45) | 76.86 (±1.02) | 82.56 (±0.95) | 83.21 (±1.27) | 83.46 (±0.84) | 84.81 (±0.80) | 0.02 | 0.12 |
| 44 | 51.68 (±0.38) | 51.67 (±1.07) | 51.52 (±0.55) | 53.18 (±1.42) | 51.82 (±0.17) | 53.91 (±1.37) | 0.06 | 0.02 |
| 45 | 76.91 (±1.56) | 77.06 (±2.21) | 82.94 (±1.73) | 83.68 (±0.83) | 87.94 (±0.93) | 87.50 (±1.59) | 0.15 | 0.25 |
| 46 | 60.12 (±0.58) | 59.61 (±1.46) | 71.92 (±1.29) | 70.96 (±1.06) | 71.92 (±0.00) | 72.07 (±0.26) | 0.4 | 0.7 |
| 47 | 84.60 (±0.38) | 84.11 (±0.86) | 90.17 (±0.99) | 90.87 (±1.53) | 90.20 (±1.11) | 91.39 (±0.50) | 0.08 | 0.06 |
| 48 | 69.69 (±0.45) | 69.77 (±0.57) | 89.94 (±0.93) | 93.21 (±0.94) | 94.40 (±0.73) | 94.09 (±0.42) | 0.08 | 0.25 |
| 49 | 92.77 (±0.72) | 92.23 (±0.97) | 96.08 (±0.70) | 97.70 (±0.47) | 96.28 (±0.57) | 96.76 (±0.53) | 0.06 | 0.06 |
| 50 | 63.16 (±1.37) | 63.60 (±1.33) | 71.08 (±0.98) | 71.21 (±1.08) | 74.20 (±0.25) | 73.33 (±0.48) | 0.25 | 0.12 |
| 51 | 84.58 (±3.43) | 61.67 (±2.64) | 79.17 (±3.40) | 72.08 (±3.95) | 79.17 (±3.40) | 79.17 (±2.78) | 0 | 0.1 |
| 52 | 65.09 (±0.41) | 64.84 (±0.60) | 69.54 (±0.59) | 65.44 (±0.98) | 69.54 (±0.59) | 65.44 (±0.98) | 0 | 0 |
| 53 | 57.11 (±0.79) | 57.58 (±1.00) | 73.97 (±1.37) | 76.69 (±0.85) | 76.14 (±1.68) | 76.69 (±0.85) | 0.04 | 0 |
| 54 | 77.33 (±0.44) | 51.73 (±3.24) | 90.51 (±0.47) | 89.21 (±0.64) | 89.61 (±0.58) | 89.21 (±0.64) | 0.02 | 0 |
| 55 | 51.56 (±2.21) | 41.88 (±4.22) | 47.50 (±4.84) | 50.31 (±4.76) | 44.06 (±2.31) | 48.13 (±4.93) | 0.06 | 0.4 |
| 56 | 85.47 (±0.80) | 67.70 (±1.85) | 86.28 (±0.90) | 87.77 (±0.67) | 86.82 (±1.40) | 89.12 (±1.08) | 0.08 | 0.2 |
| 57 | 75.01 (±0.35) | 74.76 (±0.40) | 84.89 (±0.16) | 82.81 (±0.64) | 84.89 (±0.16) | 82.59 (±0.59) | 0 | 0.02 |
| 58 | 79.64 (±0.17) | 79.75 (±0.38) | 80.07 (±0.66) | 79.00 (±0.62) | 81.07 (±0.27) | 79.00 (±0.62) | 0.3 | 0 |
| 59 | 80.73 (±0.65) | 80.78 (±1.25) | 86.61 (±0.40) | 87.85 (±0.25) | 87.66 (±0.19) | 87.85 (±0.25) | 0 | 0 |
| 60 | 83.75 (±2.00) | 85.38 (±1.42) | 85.96 (±1.77) | 88.85 (±2.18) | 89.90 (±1.38) | 88.75 (±1.57) | 0.15 | 0.3 |
| 61 | 84.18 (±0.34) | 81.88 (±3.76) | 91.86 (±0.33) | 92.07 (±1.14) | 92.78 (±0.36) | 92.07 (±1.14) | 0.04 | 0 |
| 62 | 63.84 (±0.37) | 60.76 (±0.97) | 79.91 (±1.09) | 77.87 (±1.10) | 81.25 (±1.01) | 77.87 (±1.10) | 0.02 | 0 |
| 63 | 80.90 (±0.76) | 80.97 (±0.83) | 94.31 (±1.14) | 90.02 (±1.60) | 88.17 (±0.92) | 86.46 (±1.19) | 0.12 | 0.2 |
| 64 | 88.68 (±0.71) | 77.01 (±1.77) | 93.01 (±0.71) | 92.94 (±0.78) | 92.94 (±0.93) | 92.15 (±0.57) | 0.08 | 0.25 |
| 65 | 87.06 (±0.48) | 87.53 (±0.91) | 99.78 (±0.05) | 99.83 (±0.08) | 99.99 (±0.01) | 99.94 (±0.04) | 0.02 | 0.02 |
| 66 | 65.15 (±0.47) | 64.98 (±1.25) | 84.33 (±0.51) | 74.12 (±3.00) | 83.34 (±1.52) | 74.12 (±3.00) | 0.02 | 0 |
| 67 | 75.56 (±0.12) | 75.66 (±0.21) | 97.11 (±0.31) | 96.09 (±0.30) | 97.11 (±0.31) | 96.09 (±0.30) | 0 | 0 |
| 68 | 83.17 (±0.19) | 66.98 (±1.83) | 93.63 (±0.45) | 88.72 (±0.56) | 94.68 (±0.17) | 91.49 (±0.26) | 0.04 | 0.12 |
| 69 | 56.91 (±0.20) | 57.16 (±0.31) | 73.63 (±1.62) | 74.31 (±0.75) | 73.63 (±1.62) | 72.03 (±0.73) | 0 | 0.08 |
| 70 | 83.70 (±0.24) | 83.76 (±0.45) | 90.84 (±0.46) | 91.50 (±0.47) | 91.93 (±0.35) | 92.06 (±0.28) | 0.04 | 0.02 |
| 71 | 59.45 (±0.85) | 59.40 (±1.43) | 60.96 (±1.57) | 68.00 (±2.27) | 74.29 (±0.83) | 70.68 (±2.26) | 0.08 | 0.2 |
| 72 | 72.45 (±0.52) | 72.70 (±0.79) | 90.86 (±0.51) | 90.83 (±0.39) | 88.84 (±0.49) | 89.67 (±0.69) | 0.08 | 0.06 |
| 73 | 89.00 (±0.22) | 89.03 (±0.36) | 94.00 (±0.19) | 94.75 (±0.34) | 95.89 (±0.21) | 95.39 (±0.28) | 0.04 | 0.04 |
| 74 | 69.87 (±0.52) | 71.14 (±1.61) | 96.65 (±0.11) | 96.88 (±0.15) | 97.15 (±0.02) | 97.16 (±0.00) | 0.1 | 0.04 |
| 75 | 73.75 (±0.93) | 73.89 (±1.52) | 95.27 (±0.25) | 96.22 (±0.15) | 95.48 (±0.17) | 96.22 (±0.15) | 0.02 | 0 |
| 76 | 76.58 (±0.78) | 76.53 (±0.96) | 89.34 (±0.70) | 90.20 (±1.20) | 89.34 (±0.70) | 90.51 (±0.55) | 0 | 0.02 |
| 77 | 78.79 (±0.19) | 78.68 (±0.45) | 95.34 (±0.12) | 95.05 (±0.16) | 95.99 (±0.18) | 95.35 (±0.41) | 0.04 | 0.04 |
| 78 | 67.93 (±0.39) | 67.92 (±0.46) | 73.66 (±1.30) | 73.95 (±1.01) | 74.70 (±0.30) | 74.24 (±0.37) | 0.12 | 0.15 |
| 79 | 84.90 (±0.46) | 83.17 (±0.82) | 93.17 (±1.24) | 91.44 (±1.39) | 92.50 (±1.68) | 93.27 (±1.01) | 0.08 | 0.06 |
| 80 | 62.50 (±1.01) | 54.23 (±2.91) | 71.83 (±2.13) | 74.90 (±1.95) | 73.08 (±1.01) | 74.13 (±2.62) | 0.2 | 0.25 |
| 81 | 59.89 (±1.08) | 59.46 (±1.45) | 59.67 (±3.01) | 54.67 (±1.54) | 68.59 (±0.95) | 65.11 (±1.66) | 0.25 | 0.25 |
| 82 | 89.10 (±1.29) | 91.20 (±0.63) | 88.70 (±1.49) | 87.70 (±1.34) | 88.80 (±0.79) | 88.70 (±0.48) | 0.12 | 0.4 |
| 83 | 54.62 (±1.09) | 50.10 (±0.84) | 61.92 (±2.65) | 62.88 (±1.58) | 71.25 (±0.84) | 71.83 (±1.57) | 0.4 | 0.5 |
| 84 | 49.44 (±2.22) | 53.67 (±2.67) | 55.72 (±3.34) | 56.72 (±3.73) | 71.11 (±0.00) | 71.11 (±0.00) | 0.5 | 0.4 |
| 85 | 79.34 (±0.42) | 78.89 (±0.61) | 79.08 (±0.63) | 69.21 (±1.16) | 79.08 (±0.63) | 73.83 (±1.30) | 0 | 0.04 |
| 86 | 51.68 (±0.33) | 51.88 (±0.67) | 42.11 (±1.00) | 53.56 (±1.63) | 47.51 (±0.78) | 53.56 (±1.63) | 0.02 | 0 |
| 87 | 70.98 (±0.49) | 71.13 (±0.64) | 74.77 (±0.43) | 69.53 (±0.87) | 74.77 (±0.43) | 73.19 (±1.28) | 0 | 0.02 |
| 88 | 51.02 (±2.48) | 0.68 (±0.59) | 47.27 (±2.35) | 47.95 (±2.61) | 70.34 (±0.99) | 70.23 (±0.72) | 0.4 | 0.7 |
| 89 | 41.65 (±0.54) | 35.30 (±0.69) | 45.03 (±1.23) | 49.09 (±1.55) | 52.23 (±0.83) | 52.65 (±1.23) | 0.25 | 0.25 |
| 90 | 83.22 (±0.47) | 62.87 (±4.17) | 98.07 (±0.08) | 98.30 (±0.09) | 98.09 (±0.07) | 98.11 (±0.07) | 0.08 | 0.4 |
| 91 | 89.62 (±0.52) | 89.90 (±0.60) | 92.36 (±0.58) | 92.12 (±0.61) | 92.88 (±0.63) | 93.32 (±0.48) | 0.06 | 0.04 |
| 92 | 81.60 (±0.39) | 81.06 (±0.69) | 90.20 (±0.23) | 90.68 (±0.43) | 92.31 (±0.25) | 92.99 (±0.25) | 0.08 | 0.04 |
| 93 | 81.81 (±0.60) | 78.86 (±0.49) | 86.97 (±0.80) | 85.00 (±1.30) | 86.57 (±0.65) | 86.94 (±1.55) | 0.2 | 0.12 |
| 94 | 82.29 (±0.33) | 82.25 (±0.74) | 92.36 (±0.44) | 92.13 (±0.36) | 92.36 (±0.44) | 92.13 (±0.36) | 0 | 0 |
| 95 | 69.68 (±0.73) | 63.33 (±3.00) | 64.25 (±2.02) | 68.71 (±1.29) | 59.73 (±0.53) | 63.23 (±1.57) | 0.7 | 0.4 |
| 96 | 44.17 (±1.75) | 52.19 (±3.66) | 51.55 (±3.66) | 58.66 (±6.89) | 51.55 (±3.66) | 59.25 (±3.45) | 0 | 0.15 |
| 97 | 54.68 (±0.90) | 55.00 (±2.77) | 66.41 (±1.08) | 65.87 (±1.23) | 66.85 (±0.41) | 68.39 (±1.15) | 0.3 | 0.3 |
| 98 | 70.14 (±0.68) | 69.60 (±1.57) | 75.12 (±0.97) | 77.34 (±0.58) | 76.76 (±0.97) | 77.34 (±0.58) | 0.04 | 0 |
| 99 | 84.81 (±0.50) | 84.59 (±0.43) | 82.61 (±0.95) | 86.90 (±1.00) | 87.84 (±0.40) | 87.31 (±0.63) | 0.15 | 0.3 |
| 100 | 85.43 (±0.41) | 85.57 (±0.61) | 96.70 (±0.23) | 96.55 (±0.28) | 95.44 (±0.29) | 96.55 (±0.28) | 0.02 | 0 |
| 101 | 77.75 (±0.35) | 77.81 (±0.64) | 83.98 (±0.88) | 87.49 (±1.16) | 83.98 (±0.88) | 87.49 (±1.16) | 0 | 0 |
| 102 | 76.49 (±0.94) | 75.98 (±2.99) | 99.66 (±0.10) | 99.59 (±0.13) | 99.66 (±0.10) | 99.59 (±0.13) | 0 | 0 |
| 103 | 50.63 (±0.95) | 50.79 (±0.76) | 71.37 (±1.34) | 68.87 (±1.14) | 71.37 (±1.34) | 67.63 (±1.01) | 0 | 0.2 |
| 104 | 53.61 (±0.41) | 53.02 (±0.85) | 69.97 (±1.15) | 68.28 (±0.62) | 71.02 (±0.75) | 68.28 (±0.62) | 0.06 | 0 |
| 105 | 91.60 (±0.83) | 91.55 (±0.47) | 96.55 (±0.25) | 97.32 (±0.58) | 98.08 (±0.50) | 98.57 (±0.33) | 0.04 | 0.02 |
| 106 | 49.87 (±0.42) | 51.18 (±0.87) | 54.34 (±2.50) | 53.42 (±2.52) | 54.74 (±1.34) | 56.71 (±1.61) | 0.06 | 0.25 |
| 107 | 66.75 (±2.50) | 64.06 (±7.99) | 93.92 (±0.65) | 96.45 (±0.18) | 93.91 (±0.28) | 96.45 (±0.18) | 0.04 | 0 |
| 108 | 69.45 (±0.73) | 69.59 (±0.60) | 97.56 (±0.28) | 97.54 (±0.29) | 97.91 (±0.21) | 97.81 (±0.33) | 0.02 | 0.02 |
| 109 | 75.50 (±0.00) | 74.48 (±0.70) | 77.84 (±0.04) | 78.83 (±0.17) | 77.95 (±0.27) | 78.83 (±0.17) | 0.3 | 0 |
| 110 | 87.50 (±0.00) | 87.50 (±0.00) | 87.50 (±0.00) | 62.50 (±0.00) | 87.50 (±0.00) | 62.50 (±0.00) | 0 | 0 |
| 111 | 96.72 (±0.14) | 95.73 (±1.16) | 96.76 (±0.12) | 97.50 (±0.11) | 97.47 (±0.06) | 97.50 (±0.11) | 0.04 | 0 |
| 112 | 71.14 (±0.56) | 72.24 (±0.80) | 82.56 (±0.98) | 83.67 (±0.97) | 82.56 (±0.98) | 83.67 (±0.97) | 0 | 0 |
| 113 | 76.56 (±0.76) | 76.95 (±0.53) | 81.95 (±1.06) | 81.46 (±1.03) | 81.95 (±1.06) | 82.56 (±0.87) | 0 | 0.02 |
| 114 | 61.89 (±0.31) | 61.87 (±1.05) | 93.96 (±0.52) | 78.58 (±1.77) | 93.96 (±0.52) | 82.31 (±0.49) | 0 | 0.15 |
| 115 | 79.99 (±0.29) | 79.61 (±0.71) | 84.03 (±0.44) | 84.96 (±0.57) | 86.02 (±0.30) | 85.75 (±0.29) | 0.06 | 0.1 |
| 116 | 79.43 (±0.31) | 81.64 (±1.01) | 85.43 (±0.28) | 85.43 (±0.67) | 85.89 (±0.30) | 85.43 (±0.67) | 0.06 | 0 |
| 117 | 97.67 (±0.18) | 96.82 (±0.72) | 96.88 (±0.40) | 98.24 (±0.63) | 98.30 (±0.60) | 98.75 (±0.59) | 0.04 | 0.02 |
| 118 | 42.56 (±0.82) | 29.13 (±1.41) | 59.11 (±0.63) | 60.30 (±0.52) | 60.51 (±0.51) | 60.31 (±0.37) | 0.12 | 0.1 |
| 119 | 34.27 (±0.41) | 30.92 (±0.88) | 53.56 (±0.66) | 53.63 (±0.42) | 55.50 (±0.21) | 55.13 (±0.41) | 0.06 | 0.06 |
| 120 | 50.06 (±0.53) | 49.82 (±1.06) | 54.89 (±1.21) | 57.93 (±0.83) | 58.14 (±0.81) | 57.99 (±1.01) | 0.25 | 0.12 |
| 121 | 96.40 (±0.52) | 93.10 (±1.29) | 99.00 (±0.00) | 98.60 (±0.70) | 98.80 (±0.42) | 98.90 (±0.32) | 0.3 | 0.3 |
| mean | 70.92 (±16.05) | 67.43 (±19.35) | 79.00 (±16.55) | 78.89 (±16.32) | 80.49 (±15.40) | 80.27 (±15.25) |
References
- Xu, W.; Gupta, S.; Morris, J.; Shen, X.; Imani, M.; Aksanli, B.; Rosing, T. Tri-HD: Energy-Efficient On-Chip Learning with In-Memory Hyperdimensional Computing. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2024, 44, 525–539. [Google Scholar] [CrossRef]
- Kang, J.; Xu, W.; Bittremieux, W.; Moshiri, N.; Rosing, T.Š. DRAM-Based Acceleration of Open Modification Search in Hyperdimensional Space. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2024, 43, 2592–2605. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference and Prediction, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Reed, S.K. Pattern recognition and categorization. Cogn. Psychol. 1972, 3, 382–407. [Google Scholar] [CrossRef]
- Kanerva, P. Hyperdimensional computing: An introduction to computing in distributed representation with high-dimensional random vectors. Cogn. Comput. 2009, 1, 139–159. [Google Scholar] [CrossRef]
- Rahimi, A.; Kanerva, P.; Benini, L.; Rabaey, J.M. Efficient biosignal processing using hyperdimensional computing: Network templates for combined learning and classification of ExG signals. Proc. IEEE 2019, 107, 123–143. [Google Scholar] [CrossRef]
- Vdovychenko, R.; Tulchinsky, V. Increasing the Semantic Storage Density of Sparse Distributed Memory. Cybern. Syst. Anal. 2022, 58, 331–342. [Google Scholar] [CrossRef]
- Vdovychenko, R.; Tulchinsky, V. Sparse Distributed Memory for Sparse Distributed Data; Springer: Cham, Switzerland, 2023; Volume 542, pp. 74–81. [Google Scholar] [CrossRef]
- Widdows, D.; Cohen, T. Reasoning with vectors: A continuous model for fast robust inference. Log. J. IGPL 2015, 23, 141–173. [Google Scholar] [CrossRef]
- Imani, M.; Bosch, S.; Javaheripi, M.; Rouhani, B.; Wu, X.; Koushanfar, F.; Rosing, T. SemiHD: Semi-supervised learning using hyperdimensional computing. In Proceedings of the 2019 IEEE/ACM International Conference on Computer-Aided Design (ICCAD), Westminster, CO, USA, 4–7 November 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Imani, M.; Salamat, S.; Khaleghi, B.; Samragh, M.; Koushanfar, F.; Rosing, T. SparseHD: Algorithm-hardware co-optimization for efficient high-dimensional computing. In Proceedings of the 27th IEEE International Symposium on Field-Programmable Custom Computing Machines, FCCM 2019, San Diego, CA, USA, 28 April–1 May 2019; IEEE: New York, NY, USA, 2019; pp. 190–198. [Google Scholar] [CrossRef]
- Morris, J.; Imani, M.; Bosch, S.; Thomas, A.; Shu, H.; Rosing, T. CompHD: Efficient hyperdimensional computing using model compression. In Proceedings of the IEEE/ACM International Symposium on Low Power Electronics and Design, Lausanne, Switzerland, 29–31 July 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Zou, Z.; Kim, Y.; Imani, F.; Alimohamadi, H.; Cammarota, R.; Imani, M. Scalable edge-based hyperdimensional learning system with brain-like neural adaptation. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, St. Louis, MO, USA, 14–19 November 2021; IEEE Computer Society: New York, NY, USA, 2021; pp. 1–15. [Google Scholar] [CrossRef]
- Duan, S.; Liu, Y.; Ren, S.; Xu, X. LeHDC: Learning-based hyperdimensional computing classifier. In Proceedings of the Design Automation Conference, San Francisco, CA, USA, 10–14 July 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Pale, U.; Teijeiro, T.; Atienza, D. Multi-centroid hyperdimensional computing approach for epileptic seizure detection. Front. Neurol. 2022, 13, 816294. [Google Scholar] [CrossRef]
- Wang, J.; Huang, S.; Imani, M. DistHD: A learner-aware dynamic encoding method for hyperdimensional classification. In Proceedings of the Proceedings—Design Automation Conference, San Francisco, CA, USA, 9–13 July 2023; IEEE: New York, NY, USA, 2023; Volume 2023, pp. 1–6. [Google Scholar] [CrossRef]
- Joshi, A.; Halseth, J.T.; Kanerva, P. Language Geometry Using Random Indexing. In Quantum Interaction; de Barros, J.A., Coecke, B., Pothos, E., Eds.; Springer: Cham, Switzerland, 2017; pp. 265–274. [Google Scholar]
- Schlegel, K.; Mirus, F.; Neubert, P.; Protzel, P. Multivariate time series analysis for driving style classification using neural networks and hyperdimensional computing. In Proceedings of the IEEE Intelligent Vehicles Symposium, Nagoya, Japan, 11–17 July 2021; IEEE: New York, NY, USA, 2021; Volume 2021, pp. 602–609. [Google Scholar] [CrossRef]
- Schlegel, K.; Neubert, P.; Protzel, P. HDC-MiniROCKET: Explicit time encoding in time series classification with hyperdimensional computing. In Proceedings of the International Joint Conference on Neural Network, Padua, Italy, 18–23 July 2022; pp. 1–8. [Google Scholar] [CrossRef]
- Schlegel, K.; Kleyko, D.; Brinkmann, B.H.; Nurse, E.S.; Gayler, R.W.; Neubert, P. Lessons from a challenge on forecasting epileptic seizures from non-cerebral signals. Nat. Mach. Intell. 2024, 6, 243–244. [Google Scholar] [CrossRef]
- Watkinson, N.; Givargis, T.; Joe, V.; Nicolau, A.; Veidenbaum, A. Detecting COVID-19 related pneumonia on CT scans using hyperdimensional computing. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Virtual, 1–5 November 2021; IEEE: New York, NY, USA, 2021; pp. 3970–3973. [Google Scholar] [CrossRef]
- Cumbo, F.; Cappelli, E.; Weitschek, E. A brain-inspired hyperdimensional computing approach for classifying massive DNA methylation data of cancer. Algorithms 2020, 13, 233. [Google Scholar] [CrossRef]
- Rahimi, A.; Kanerva, P.; Rabaey, J.M. A robust and energy-efficient classifier using brain-inspired hyperdimensional computing. In Proceedings of the International Symposium on Low Power Electronics and Design, San Francisco Airport, CA, USA, 8–10 August 2016; IEEE: New York, NY, USA, 2016; pp. 64–69. [Google Scholar] [CrossRef]
- Neubert, P.; Schubert, S.; Protzel, P. An introduction to hyperdimensional computing for robotics. KI-Kunstl. Intell. 2019, 33, 319–330. [Google Scholar] [CrossRef]
- Ge, L.; Parhi, K.K. Classification using hyperdimensional computing: A review. IEEE Circuits Syst. Mag. 2020, 20, 30–47. [Google Scholar] [CrossRef]
- Aygun, S.; Moghadam, M.S.; Najafi, M.H.; Imani, M. Learning from hypervectors: A survey on hypervector encoding. arXiv 2023, arXiv:2308.00685. [Google Scholar] [CrossRef]
- Vergés, P.; Heddes, M.; Nunes, I.; Givargis, T.; Nicolau, A. Classification Using Hyperdimensional Computing: A Review with Comparative Analysis; Research Square: Durham, NC, USA, 2023; pp. 1–49. [Google Scholar] [CrossRef]
- Plate, T.A. Holographic reduced representations. IEEE Trans. Neural Netw. 1995, 6, 623–641. [Google Scholar] [CrossRef]
- Plate, T.A. Holographic Reduced Representation: Distributed Representation for Cognitive Structures; CSLI Publications: Stanford, CA, USA, 2003. [Google Scholar]
- Gayler, R.W. Multiplicative binding, representation operators & analogy. In Proceedings of the Advances in Analogy Research: Integration of Theory and Data from the Cognitive, Computational, and Neural Sciences, Sofia, Bulgaria, July 1998; pp. 1–4. [Google Scholar]
- Kanerva, P. The spatter code for encoding concepts at many levels. In Proceedings of the International Conference on Artificial Neural Networks (ICANN), Sorrento, Italy, 26–29 May 1994; pp. 226–229. [Google Scholar] [CrossRef]
- Kanerva, P. Binary spatter-coding of ordered K-tuples. In Proceedings of the Artificial Neural Networks—ICANN, Bochum, Germany, 16–19 July 1996; pp. 869–873. [Google Scholar] [CrossRef]
- Laiho, M.; Poikonen, J.H.; Kanerva, P.; Lehtonen, E. High-dimensional computing with sparse vectors. In Proceedings of the IEEE Biomedical Circuits and Systems Conference: Engineering for Healthy Minds and Able Bodies, BioCAS 2015, Atlanta, GA, USA, 22–24 October 2015; IEEE: New York, NY, USA, 2015; pp. 1–4. [Google Scholar] [CrossRef]
- Imani, M.; Huang, C.; Kong, D.; Rosing, T. Hierarchical hyperdimensional computing for energy efficient classification. In Proceedings of the ACM/ESDA/IEEE Design Automation Conference (DAC), San Francisco, CA, USA, 24–29 June 2018; IEEE: New York, NY, USA, 2018; Volume Part F137710, pp. 1–6. [Google Scholar] [CrossRef]
- Rachkovskij, D.A.; Fedoseeva, T.V. On audio signals recognition by multilevel neural network. In Proceedings of the International Symposium on Neural Networks and Neural Computing—NEURONET’90, Prague, Czech Republic, 10–14 September 1990; pp. 281–283. [Google Scholar]
- Kussul, E.; Rachkovskij, D.A. On image texture recognition by associative-projective neurocomputer. In Proceedings of the ANNIE’91 Conference “Intelligent Engineering Systems Through Artificial Neural Networks”, St. Louis, MO, USA, 10–13 November 1991; pp. 453–458. [Google Scholar]
- Rachkovskij, D.A.; Slipchenko, S.V.; Misuno, I.S.; Kussul, E.M.; Baidyk, T.N. Sparse binary distributed encoding of numeric vectors. J. Autom. Inf. Sci. 2005, 37, 47–61. [Google Scholar]
- Rachkovskij, D.A.; Slipchenko, S.V.; Kussul, E.M.; Baidyk, T.N. Sparse binary distributed encoding of scalars. J. Autom. Inf. Sci. 2005, 37, 12–23. [Google Scholar]
- Imani, M.; Kong, D.; Rahimi, A.; Rosing, T. VoiceHD: Hyperdimensional computing for efficient speech recognition. In Proceedings of the IEEE International Conference on Rebooting Computing (ICRC), Washington, DC, USA, 8–9 November 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Kim, Y.; Imani, M.; Rosing, T.S. Efficient human activity recognition using hyperdimensional computing. In Proceedings of the 8th International Conference on the Internet of Things, Santa Barbara, CA, USA, 15–18 October 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Imani, M.; Messerly, J.; Wu, F.; Pi, W.; Rosing, T. A binary learning framework for hyperdimensional computing. In Proceedings of the 2019 Design, Automation and Test in Europe Conference and Exhibition (DATE), Florence, Italy, 25–29 March 2019; pp. 126–131. [Google Scholar] [CrossRef]
- Imani, M.; Bosch, S.; Datta, S.; Ramakrishna, S.; Salamat, S.; Rabaey, J.M.; Rosing, T. QuantHD: A quantization framework for hyperdimensional computing. IEEE Trans. Comput.-Aided Des. Integr. Circuits Syst. 2019, 39, 2268–2278. [Google Scholar] [CrossRef]
- Hsiao, Y.R.; Chuang, Y.C.; Chang, C.Y.; Wu, A.Y. Hyperdimensional computing with learnable projection for user adaptation framework. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations (AIAI), Crete, Greece, 25–27 June 2021; pp. 436–447. [Google Scholar] [CrossRef]
- Imani, M.; Morris, J.; Bosch, S.; Shu, H.; Micheli, G.D.; Rosing, T. AdaptHD: Adaptive efficient training for brain-inspired hyperdimensional computing. In Proceedings of the IEEE Biomedical Circuits and Systems Conference (BioCAS), Nara, Japan, 17–19 October 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Hernández-Cano, A.; Matsumoto, N.; Ping, E.; Imani, M. OnlineHD: Robust, efficient, and single-pass online learning using hyperdimensional system. In Proceedings of the Design, Automation and Test in Europe Conference and Exhibition (DATE), Grenoble, France, 1–5 February 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Chang, C.Y.; Wu, A.Y.A. Dynamic hyperdimensional computing for improving accuracy-energy efficiency trade-offs. In Proceedings of the IEEE Workshop on Signal Processing Systems, SiPS: Design and Implementation, Coimbra, Portugal, 20–22 October 2020; IEEE: New York, NY, USA, 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Vergés, P.; Givargis, T.; Nicolau, A. RefineHD: Accurate and efficient single-pass adaptive learning using hyperdimensional computing. In Proceedings of the IEEE International Conference on Rebooting Computing (ICRC), San Diego, CA, USA, 5–6 December 2023; pp. 1–8. [Google Scholar] [CrossRef]
- Smets, L.; Leekwijck, W.V.; Tsang, I.J.; Latre, S. Training a hyperdimensional computing classifier using a threshold on its confidence. Neural Comput. 2023, 35, 2006–2023. [Google Scholar] [CrossRef]
- Kussul, E.; Baidyk, T.; Kasatkina, L.; Lukovich, V. Rosenblatt perceptrons for handwritten digit recognition. In Proceedings of the International Joint Conference on Neural Networks, Washington, DC, USA, 15–19 July 2001; pp. 1516–1520. [Google Scholar] [CrossRef]
- Kussul, E.; Baidyk, T. Improved method of handwritten digit recognition tested on MNIST database. Image Vis. Comput. 2004, 22, 971–981. [Google Scholar] [CrossRef]
- Rachkovskij, D.A. Linear classifiers based on binary distributed representations. Inf. Theor. Appl. 2007, 14, 270–274. [Google Scholar]
- Bartlett, P.; Freund, Y.; Lee, W.S.; Schapire, R.E. Boosting the margin: A new explanation for the effectiveness of voting methods. Ann. Stat. 1998, 26, 1651–1686. [Google Scholar] [CrossRef]
- Mohri, M.; Rostamizadeh, A.; Talwalkar, A. Foundations of Machine Learning; The MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D.; Fernández-Delgado, A. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Wainberg, M.; Alipanahi, B.; Frey, B.J. Are Random Forests Truly the Best Classifiers? J. Mach. Learn. Res. 2016, 17, 1–5. [Google Scholar]
- Kleyko, D.; Kheffache, M.; Frady, E.P.; Wiklund, U.; Osipov, E. Density encoding enables resource-efficient randomly connected neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 32, 3777–3783. [Google Scholar] [CrossRef]
- Frady, E.P.; Kleyko, D.; Sommer, F.T. Variable binding for sparse distributed representations: Theory and applications. IEEE Trans. Neural Netw. Learn. Syst. 2023, 34, 2191–2204. [Google Scholar] [CrossRef]
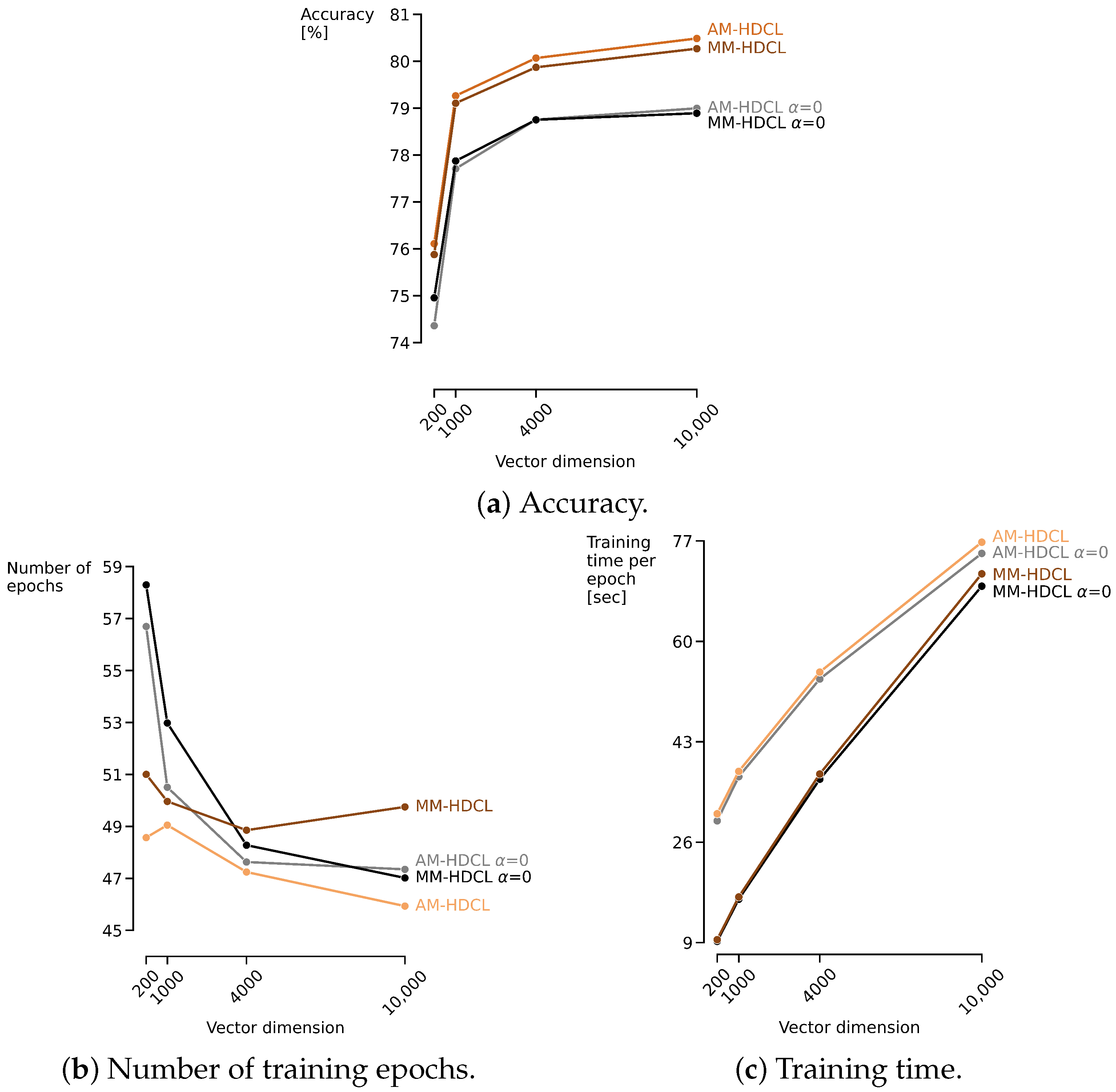


| D | 200 | 1000 | 4000 | 10,000 |
|---|---|---|---|---|
| Classifier | ||||
| Prot-HDCL | 66.36 (±16.28) | 69.70 (±15.96) | 70.66 (±15.98) | 70.92 (±16.05) |
| Prot-HDCL | 61.36 (±19.10) | 65.73 (±19.20) | 66.99 (±19.46) | 67.43 (±19.35) |
| AM-HDCL = 0 | 74.36 (±17.11) | 77.71 (±16.64) | 78.75 (±16.45) | 79.00 (±16.55) |
| MM-HDCL = 0 | 74.95 (±16.90) | 77.87 (±16.45) | 78.75 (±16.27) | 78.89 (±16.32) |
| AM-HDCL | 76.11 (±16.18) | 79.26 (±15.62) | 80.07 (±15.59) | 80.49 (±15.40) |
| MM-HDCL | 75.88 (±16.54) | 79.10 (±15.59) | 79.87 (±15.61) | 80.27 (±15.25) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smets, L.; Rachkovskij, D.; Osipov, E.; Van Leekwijck, W.; Volkov, O.; Latré, S. Margin-Based Training of HDC Classifiers. Big Data Cogn. Comput. 2025, 9, 68. https://doi.org/10.3390/bdcc9030068
Smets L, Rachkovskij D, Osipov E, Van Leekwijck W, Volkov O, Latré S. Margin-Based Training of HDC Classifiers. Big Data and Cognitive Computing. 2025; 9(3):68. https://doi.org/10.3390/bdcc9030068
Chicago/Turabian StyleSmets, Laura, Dmitri Rachkovskij, Evgeny Osipov, Werner Van Leekwijck, Olexander Volkov, and Steven Latré. 2025. "Margin-Based Training of HDC Classifiers" Big Data and Cognitive Computing 9, no. 3: 68. https://doi.org/10.3390/bdcc9030068
APA StyleSmets, L., Rachkovskij, D., Osipov, E., Van Leekwijck, W., Volkov, O., & Latré, S. (2025). Margin-Based Training of HDC Classifiers. Big Data and Cognitive Computing, 9(3), 68. https://doi.org/10.3390/bdcc9030068






