A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Preprocessing and Keyword Extraction
2.3. Test Article Selection and Evaluation Set
2.4. Embedding Models
2.5. Similarity Computation
2.6. Evaluation Metrics
2.6.1. Shannon Entropy
2.6.2. Gini Coefficient
2.7. Software and Environment
2.8. Study Focus and Limitations
3. Results
3.1. Case Study
- Article-Level Similarity: For each model, we sought the top ten articles (out of those 5000) most similar to the test article and identified the journals where they were published.
- Journal-Level Similarity: We computed the average similarity of each journal’s articles to the test article, and then ranked the journals accordingly.
3.1.1. Article-Level Findings
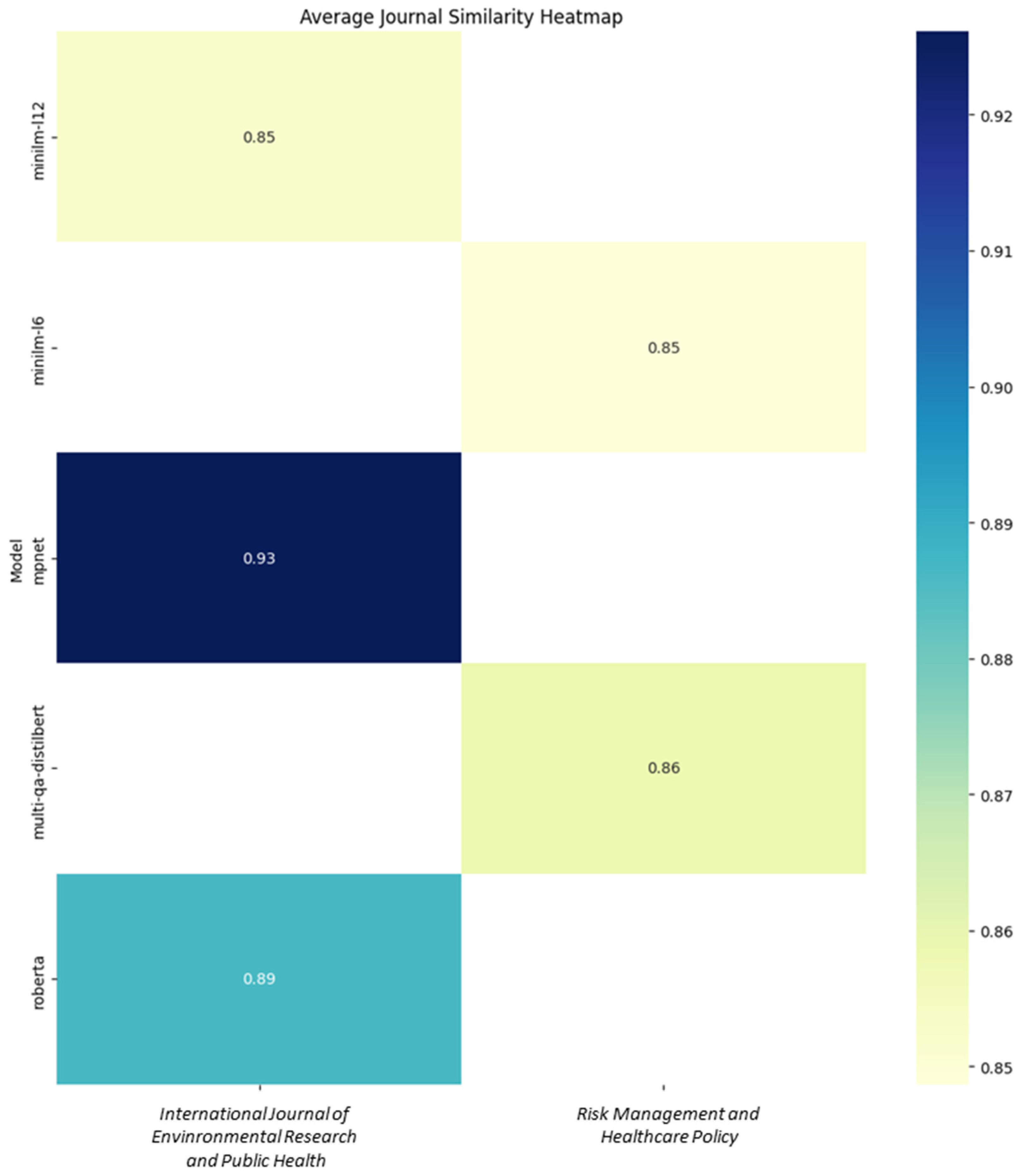
3.1.2. Journal-Level Suggestions
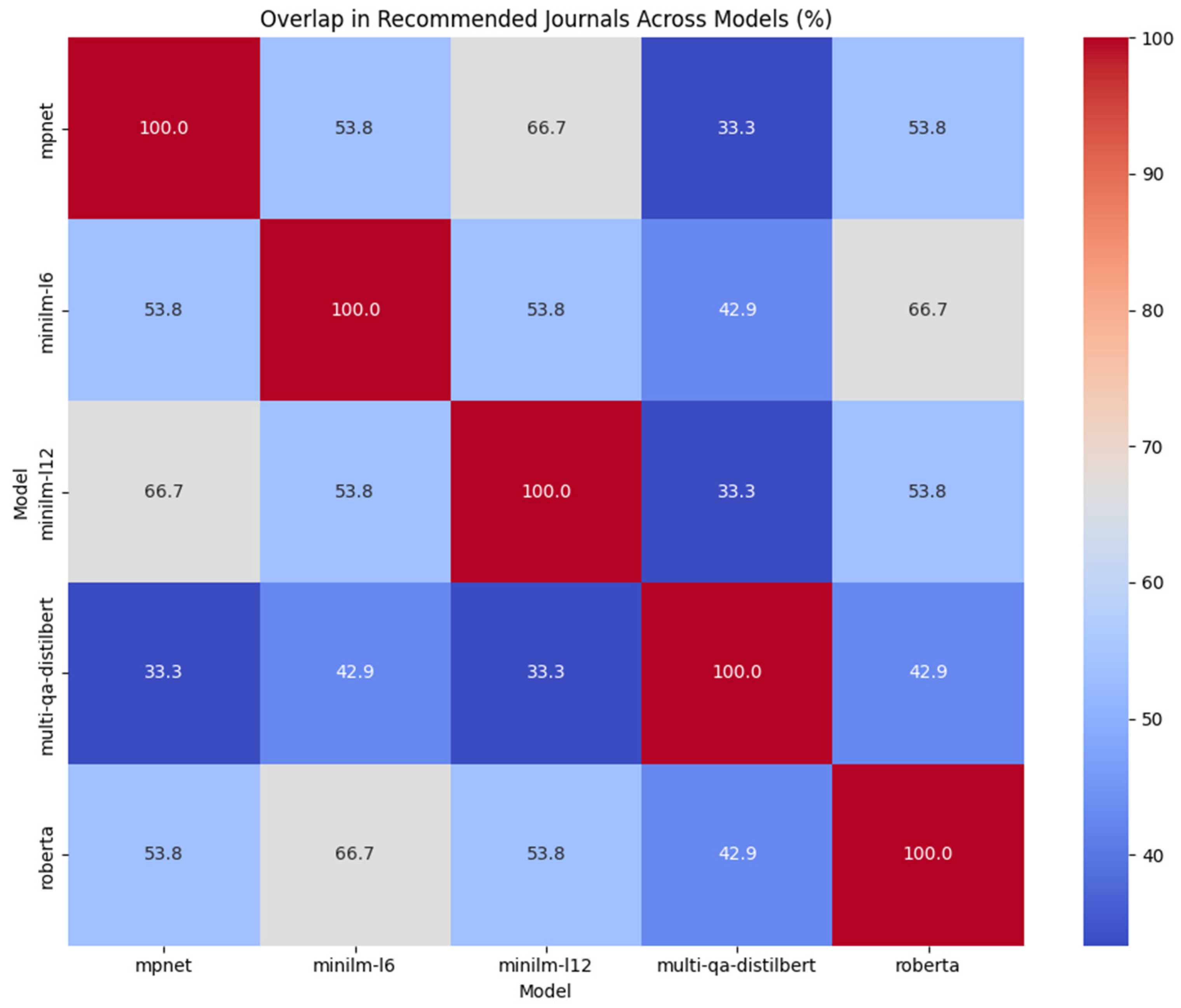
3.1.3. Overlap Among Model Recommendations
3.2. Quantitative Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Welch, S.J. Selecting the Right Journal for Your Submission. J. Thorac. Dis. 2012, 4, 336–338. [Google Scholar] [CrossRef]
- Nicholas, D.; Herman, E.; Clark, D.; Boukacem-Zeghmouri, C.; Rodríguez-Bravo, B.; Abrizah, A.; Watkinson, A.; Xu, J.; Sims, D.; Serbina, G.; et al. Choosing the ‘Right’ Journal for Publication: Perceptions and Practices of Pandemic-era Early Career Researchers. Learn. Publ. 2022, 35, 605–616. [Google Scholar] [CrossRef]
- Larsen, P.O.; von Ins, M. The Rate of Growth in Scientific Publication and the Decline in Coverage Provided by Science Citation Index. Scientometrics 2010, 84, 575–603. [Google Scholar] [CrossRef]
- Kreutz, C.K.; Schenkel, R. Scientific Paper Recommendation Systems: A Literature Review of Recent Publications. Int. J. Digit. Libr. 2022, 23, 335–369. [Google Scholar] [CrossRef]
- Park, D.H.; Kim, H.K.; Choi, I.Y.; Kim, J.K. A Literature Review and Classification of Recommender Systems Research. Expert. Syst. Appl. 2012, 39, 10059–10072. [Google Scholar] [CrossRef]
- Qaiser, S.; Ali, R. Text Mining: Use of TF-IDF to Examine the Relevance of Words to Documents. Int. J. Comput. Appl. 2018, 181, 25–29. [Google Scholar] [CrossRef]
- Wang, D.; Liang, Y.; Xu, D.; Feng, X.; Guan, R. A Content-Based Recommender System for Computer Science Publications. Knowl. Based Syst. 2018, 157, 1–9. [Google Scholar] [CrossRef]
- Medvet, E.; Bartoli, A.; Piccinin, G. Publication Venue Recommendation Based on Paper Abstract. In Proceedings of the 2014 IEEE 26th International Conference on Tools with Artificial Intelligence, Limassol, Cyprus, 10–12 November 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 1004–1010. [Google Scholar]
- Yang, Z.; Davison, B.D. Venue Recommendation: Submitting Your Paper with Style. In Proceedings of the 2012 11th International Conference on Machine Learning and Applications, Boca Raton, FL, USA, 12–15 December 2012; IEEE: Piscataway, NJ, USA; pp. 681–686. [Google Scholar]
- Beel, J.; Gipp, B.; Langer, S.; Breitinger, C. Research-Paper Recommender Systems: A Literature Survey. Int. J. Digit. Libr. 2016, 17, 305–338. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-Training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Liu, Q.; Kusner, M.J.; Blunsom, P. A Survey on Contextual Embeddings. arXiv 2020, arXiv:2003.07278. [Google Scholar]
- Noh, J.; Kavuluru, R. Improved Biomedical Word Embeddings in the Transformer Era. J. Biomed. Inform. 2021, 120, 103867. [Google Scholar] [CrossRef]
- Michail, S.; Ledet, J.W.; Alkan, T.Y.; İnce, M.N.; Günay, M. A Journal Recommender for Article Submission Using Transformers. Scientometrics 2023, 128, 1321–1336. [Google Scholar] [CrossRef]
- Galli, C.; Donos, N.; Calciolari, E. Performance of 4 Pre-Trained Sentence Transformer Models in the Semantic Query of a Systematic Review Dataset on Peri-Implantitis. Information 2024, 15, 68. [Google Scholar] [CrossRef]
- Stankevičius, L.; Lukoševičius, M. Extracting Sentence Embeddings from Pretrained Transformer Models. Appl. Sci. 2024, 14, 8887. [Google Scholar] [CrossRef]
- Siino, M. All-Mpnet at Semeval-2024 Task 1: Application of Mpnet for Evaluating Semantic Textual Relatedness. In Proceedings of the 18th International Workshop on Semantic Evaluation (SemEval-2024), Mexico City, Mexico, 20–21 June 2024; pp. 379–384. [Google Scholar]
- Yin, C.; Zhang, Z. A Study of Sentence Similarity Based on the All-Minilm-L6-v2 Model With “Same Semantics, Different Structure” After Fine Tuning. In Proceedings of the 2024 2nd International Conference on Image, Algorithms and Artificial Intelligence (ICIAAI 2024), Singapore, 9–11 August 2024; Atlantis Press: Dordrecht, The Netherlands, 2024; pp. 677–684. [Google Scholar]
- He, Y.; Yuan, M.; Chen, J.; Horrocks, I. Language Models as Hierarchy Encoders. Adv. Neural Inf. Process Syst. 2025, 37, 14690–14711. [Google Scholar]
- Bistarelli, S.; Cuccarini, M. BERT-Based Questions Answering on Close Domains: Preliminary Report. In Proceedings of the Proceedings of the 39th Italian Conference on Computational Logic, Rome, Italy, 26–28 June 2024. [Google Scholar]
- Wijanto, M.C.; Yong, H.-S. Combining Balancing Dataset and SentenceTransformers to Improve Short Answer Grading Performance. Appl. Sci. 2024, 14, 4532. [Google Scholar] [CrossRef]
- Issa, B.; Jasser, M.B.; Chua, H.N.; Hamzah, M. A Comparative Study on Embedding Models for Keyword Extraction Using KeyBERT Method. In Proceedings of the 2023 IEEE 13th International Conference on System Engineering and Technology (ICSET), Shah Alam, Malaysia, 2 October 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 40–45. [Google Scholar]
- Jin, Q.; Leaman, R.; Lu, Z. PubMed and beyond: Biomedical Literature Search in the Age of Artificial Intelligence. EBioMedicine 2024, 100, 104988. [Google Scholar] [CrossRef] [PubMed]
- Mckinney, W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; Facebook, Z.D.; Research, A.I.; Lin, Z.; Desmaison, A.; Antiga, L.; et al. Automatic Differentiation in PyTorch. In Proceedings of the 31st Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Godden, J.W.; Bajorath, J. Analysis of Chemical Information Content Using Shannon Entropy. Rev. Comput. Chem. 2007, 23, 263–289. [Google Scholar]
- Vajapeyam, S. Understanding Shannon’s Entropy Metric for Information. arXiv 2014, arXiv:1405.2061. [Google Scholar]
- Dorfman, R. A Formula for the Gini Coefficient. Rev. Econ. Stat. 1979, 61, 146. [Google Scholar] [CrossRef]
- Bisong, E. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Apress: Berkeley, CA, USA, 2019; pp. 59–64. ISBN 978-1-4842-4470-8. [Google Scholar]
- Chapman, B.; Chang, J. Biopython: Python Tools for Computational Biology. ACM Sigbio Newsl. 2000, 20, 15–19. [Google Scholar] [CrossRef]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32. Available online: https://proceedings.neurips.cc/paper/2019/hash/bdbca288fee7f92f2bfa9f7012727740-Abstract.html (accessed on 14 January 2025).
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Kiersnowska, Z.; Lemiech-Mirowska, E.; Ginter-Kramarczyk, D.; Kruszelnicka, I.; Michałkiewicz, M.; Marczak, M. Problems of Clostridium Difficile Infection (CDI) in Polish Healthcare Units. Ann. Agric. Environ. Med. 2021, 28, 224–230. [Google Scholar] [CrossRef]
- Khanafer, N.; Voirin, N.; Barbut, F.; Kuijper, E.; Vanhems, P. Hospital Management of Clostridium Difficile Infection: A Review of the Literature. J. Hosp. Infect. 2015, 90, 91–101. [Google Scholar] [CrossRef]
- Legenza, L.; Barnett, S.; Rose, W.; Safdar, N.; Emmerling, T.; Peh, K.H.; Coetzee, R. Clostridium Difficile Infection Perceptions and Practices: A Multicenter Qualitative Study in South Africa. Antimicrob. Resist. Infect. Control 2018, 7, 125. [Google Scholar] [CrossRef]
- Nordling, S.; Anttila, V.J.; Norén, T.; Cockburn, E. The Burden of Clostridium Difficile (CDI) Infection in Hospitals, in Denmark, Finland, Norway And Sweden. Value Health 2014, 17, A670. [Google Scholar] [CrossRef]
- Luo, R.; Barlam, T.F. Ten-Year Review of Clostridium Difficile Infection in Acute Care Hospitals in the USA, 2005–2014. J. Hosp. Infect. 2018, 98, 40–43. [Google Scholar] [CrossRef]
- Xu, X.; Xie, J.; Sun, J.; Cheng, Y. Factors Affecting Authors’ Manuscript Submission Behaviour: A Systematic Review. Learn. Publ. 2023, 36, 285–298. [Google Scholar] [CrossRef]
- Gaston, T.E.; Ounsworth, F.; Senders, T.; Ritchie, S.; Jones, E. Factors Affecting Journal Submission Numbers: Impact Factor and Peer Review Reputation. Learn. Publ. 2020, 33, 154–162. [Google Scholar] [CrossRef]
- Worth, P.J. Word Embeddings and Semantic Spaces in Natural Language Processing. Int. J. Intell. Sci. 2023, 13, 1–21. [Google Scholar] [CrossRef]
- Yao, Z.; Sun, Y.; Ding, W.; Rao, N.; Xiong, H. Dynamic Word Embeddings for Evolving Semantic Discovery. In Proceedings of the WSDM 2018—Proceedings of the 11th ACM International Conference on Web Search and Data Mining, Los Angeles, CA, USA, 5–9 February 2018; ACM: New York, NY, USA, 2018; pp. 673–681. [Google Scholar] [CrossRef]
- Si, Y.; Wang, J.; Xu, H.; Roberts, K. Enhancing Clinical Concept Extraction with Contextual Embeddings. J. Am. Med. Inform. Assoc. 2019, 26, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, L.; Keith, B. A Systematic Literature Review on Word Embeddings. In Trends and Applications in Software Engineering, Proceedings of the 7th International Conference on Software Process Improvement (CIMPS 2018); Guadalajara, Mexico, 17–19 October 2018, Springer: Berlin/Heidelberg, Germany, 2019; Volume 7, pp. 132–141. [Google Scholar]
- Lu, W.-H.; Iuli, R.; Strano-Paul, L.; Chandran, L. Renaissance School of Medicine at Stony Brook University. Acad. Med. 2020, 95, S362–S366. [Google Scholar] [CrossRef]
- Gottlieb, M.; Alerhand, S.; Long, B. Response to: “POCUS to Confirm Intubation in a Trauma Setting”. West. J. Emerg. Med. 2020, 22, 400. [Google Scholar] [CrossRef]
- He, L.; Zhang, Q.; Li, Z.; Shen, L.; Zhang, J.; Wang, P.; Wu, S.; Zhou, T.; Xu, Q.; Chen, X.; et al. Incorporation of Urinary Neutrophil Gelatinase-Associated Lipocalin and Computed Tomography Quantification to Predict Acute Kidney Injury and In-Hospital Death in COVID-19 Patients. Kidney Dis. 2021, 7, 120–130. [Google Scholar] [CrossRef]
- Scheffers, L.E.; Berg, L.E.M.V.; Ismailova, G.; Dulfer, K.; Takkenberg, J.J.M.; Helbing, W.A. Physical Exercise Training in Patients with a Fontan Circulation: A Systematic Review. Eur. J. Prev. Cardiol. 2021, 28, 1269–1278. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Lai, D.; Lu, D.; Lan, Z.; Kang, J.; Xu, Y.; Cai, S. Vitamin D Status Is Negatively Related to Insulin Resistance and Bone Turnover in Chinese Non-Osteoporosis Patients With Type 2 Diabetes: A Retrospective Cross-Section Research. Front. Public. Health 2022, 9, 727132. [Google Scholar] [CrossRef]
- Gayathri, A.; Ramachandran, R.; Narasimhan, M. Segmental Neurofibromatosis with Lisch Nodules. Med. J. Armed Forces India 2023, 79, 356–359. [Google Scholar] [CrossRef]
- Rice, P.E.; Nimphius, S. When Task Constraints Delimit Movement Strategy: Implications for Isolated Joint Training in Dancers. Front. Sports Act. Living 2020, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Marques, E.S.; de Oliveira, A.G.e.S.; Faerstein, E. Psychometric Properties of a Modified Version of Brazilian Household Food Insecurity Measurement Scale—Pró-Saúde Study. Cien Saude Colet. 2021, 26, 3175–3185. [Google Scholar] [CrossRef]
- Kumar, P.; Desai, C.; Das, A. Unilateral Linear Capillaritis. Indian. Dermatol. Online J. 2021, 12, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Archer, T.; Aziz, I.; Kurien, M.; Knott, V.; Ball, A. Prioritisation of Lower Gastrointestinal Endoscopy during the COVID-19 Pandemic: Outcomes of a Novel Triage Pathway. Frontline Gastroenterol. 2022, 13, 225–230. [Google Scholar] [CrossRef]
- Ortiz-Martínez, Y.; Fajardo-Rivero, J.E.; Vergara-Retamoza, R.; Vergel-Torrado, J.A.; Esquiaqui-Rangel, V. Chronic Obstructive Pulmonary Disease in Latin America and the Caribbean: Mapping the Research by Bibliometric Analysis. Indian J. Tuberc. 2022, 69, 262–263. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Maclean, J.G. Anterior Hip Ultrasound: A Useful Technique in Developmental Dysplasia of the Hips. Ultrasound 2021, 29, 179–186. [Google Scholar] [CrossRef]
- Habigt, M.A.; Gesenhues, J.; Ketelhut, M.; Hein, M.; Duschner, P.; Rossaint, R.; Mechelinck, M. In Vivo Evaluation of Two Adaptive Starling-like Control Algorithms for Left Ventricular Assist Devices. Biomed. Eng. Biomed. Tech. 2021, 66, 257–266. [Google Scholar] [CrossRef]
- Sachser, N.; Zimmermann, T.D.; Hennessy, M.B.; Kaiser, S. Sensitive Phases in the Development of Rodent Social Behavior. Curr. Opin. Behav. Sci. 2020, 36, 63–70. [Google Scholar] [CrossRef]
- Dominguez-Colman, L.; Mehta, S.U.; Mansourkhani, S.; Sehgal, N.; Alvarado, L.A.; Mariscal, J.; Tonarelli, S. Teaching Psychiatric Emergencies Using Simulation: An Experience During the Boot Camp. Med. Sci. Educ. 2020, 30, 1481–1486. [Google Scholar] [CrossRef]
- Roozenbeek, J.; Maertens, R.; McClanahan, W.; van der Linden, S. Disentangling Item and Testing Effects in Inoculation Research on Online Misinformation: Solomon Revisited. Educ. Psychol. Meas. 2021, 81, 340–362. [Google Scholar] [CrossRef]
- Tumyan, G.; Mantha, Y.; Gill, R.; Feldman, M. Acute Sterile Meningitis as a Primary Manifestation of Pituitary Apoplexy. AACE Clin. Case Rep. 2021, 7, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Alade, O.; Ajoloko, E.; Dedeke, A.; Uti, O.; Sofola, O. Self-Reported Halitosis and Oral Health Related Quality of Life in Adolescent Students from a Suburban Community in Nigeria. Afr. Health Sci. 2020, 20, 2044–2049. [Google Scholar] [CrossRef]
- Evans, T.J.; Davidson, H.C.; Low, J.M.; Basarab, M.; Arnold, A. Antibiotic Usage and Stewardship in Patients with COVID-19: Too Much Antibiotic in Uncharted Waters? J. Infect. Prev. 2021, 22, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.K.; He, Y.; Howlader, P.; Wang, W.; Yin, H. The N-Glycan Processing Enzymes β-D-N-Acetylhexosaminidase Are Involved in Ripening-Associated Softening in Strawberry Fruit. J. Food Sci. Technol. 2021, 58, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Falsaperla, R.; Saporito, M.A.N.; Pisani, F.; Mailo, J.; Pavone, P.; Ruggieri, M.; Suppiej, A.; Corsello, G. Ocular Motor Paroxysmal Events in Neonates and Infants: A Review of the Literature. Pediatr. Neurol. 2021, 117, 4–9. [Google Scholar] [CrossRef]
- Almutairi, A.F.; Albesher, N.; Aljohani, M.; Alsinanni, M.; Turkistani, O.; Salam, M. Association of Oral Parafunctional Habits with Anxiety and the Big-Five Personality Traits in the Saudi Adult Population. Saudi Dent. J. 2021, 33, 90–98. [Google Scholar] [CrossRef]
- Lu, Z.; Yang, Y.-J.; Ni, W.-X.; Li, M.; Zhao, Y.; Huang, Y.-L.; Luo, D.; Wang, X.; Omary, M.A.; Li, D. Aggregation-Induced Phosphorescence Sensitization in Two Heptanuclear and Decanuclear Gold–Silver Sandwich Clusters. Chem. Sci. 2021, 12, 702–708. [Google Scholar] [CrossRef]
- Morgun, A.V.; Salmin, V.V.; Boytsova, E.B.; Lopatina, O.L.; Salmina, A.B. Molecular Mechanisms of Proteins—Targets for SARS-CoV-2. Sovrem Tekhnologii Med. 2020, 12, 98–108. [Google Scholar] [CrossRef]
- Do Nascimento, P.A.; Kogawa, A.C.; Salgado, H.R.N. Current Status of Vancomycin Analytical Methods. J. AOAC Int. 2020, 103, 755–769. [Google Scholar] [CrossRef]
- Booth, V. Deuterium Solid State NMR Studies of Intact Bacteria Treated With Antimicrobial Peptides. Front. Med. Technol. 2021, 2, 621572. [Google Scholar] [CrossRef]
- Nguyen, H.; Haeney, O.; Galletly, C. The Characteristics of Older Homicide Offenders: A Systematic Review. Psychiatry Psychol. Law 2022, 29, 413–430. [Google Scholar] [CrossRef]
- Duseja, A.; Chahal, G.; Jain, A.; Mehta, M.; Ranjan, A.; Grover, V. Association between Nonalcoholic Fatty Liver Disease and Inflammatory Periodontal Disease: A Case-Control Study. J. Indian. Soc. Periodontol. 2021, 25, 47. [Google Scholar] [CrossRef]
- Kim, K.Y.; Lee, E.; Cho, J. Factors Affecting Healthcare Utilization among Patients with Single and Multiple Chronic Diseases. Iran. J. Public. Health 2020, 49, 2367–2375. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Shangguan, X.; Li, M.; Wang, Y.; Xi, D.; Sun, W.; Fan, J.; Shao, K.; Peng, X. Ex Vivo Identification of Circulating Tumor Cells in Peripheral Blood by Fluorometric “Turn on” Aptamer Nanoparticles. Chem. Sci. 2021, 12, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Rathour, M.; Sharma, A.; Kaur, A.; Upadhyay, S.K. Genome-Wide Characterization and Expression and Co-Expression Analysis Suggested Diverse Functions of WOX Genes in Bread Wheat. Heliyon 2020, 6, e05762. [Google Scholar] [CrossRef] [PubMed]
- Hepatobiliary Disease Study Group; Chinese Society of Gastroenterology; Chinese Medical Association. Consensus for Management of Portal Vein Thrombosis in Liver Cirrhosis (2020, Shanghai). J. Dig. Dis. 2021, 22, 176–186. [Google Scholar] [CrossRef]
- Bansal, L.K.; Gupta, S.; Gupta, A.K.; Chaudhary, P. Thyroid Tuberculosis. Indian J. Tuberc. 2021, 68, 272–278. [Google Scholar] [CrossRef]
- Eble, J. Implications of John Kavanaugh’s Philosophy of the Human Person as Embodied Reflexive Consciousness for Conscientious Decision-Making in Brain Death. Linacre Q. 2021, 88, 71–81. [Google Scholar] [CrossRef]
- Rivera, A.; González-Pozega, C.; Ibarra, G.; Fernandez-Ibarburu, B.; García-Ruano, Á.; Vallejo-Valero, A. Punctual Breast Implant Rupture Following Lipofilling: Only a Myth? Breast Care 2021, 16, 544–547. [Google Scholar] [CrossRef]
- Álvarez Hernández, P.; de la Mata Llord, J. Expanded Mesenchymal Stromal Cells in Knee Osteoarthritis: A Systematic Literature Review. Reumatol. Clínica (Engl. Ed.) 2022, 18, 49–55. [Google Scholar] [CrossRef]
- Keche, P.N.; Davange, N.A.; Gawarle, S.H.; Ganguly, S. A Study of Anatomical Variations in the Nasal Cavity in Cases of Chronic Rhinosinusitis. Indian J. Otolaryngol. Head. Neck Surg. 2022, 74, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Igbokwe, M.; Adebayo, O.; Ogunsuji, O.; Popoola, G.; Babalola, R.; Oiwoh, S.O.; Makinde, A.M.; Adeniyi, A.M.; Kanmodi, K.; Umar, W.F.; et al. An Exploration of Profile, Perceptions, Barriers, and Predictors of Research Engagement among Resident Doctors. Perspect. Clin. Res. 2022, 13, 106–113. [Google Scholar] [CrossRef]
- Marraccini, M.E.; Pittleman, C. Returning to School Following Hospitalization for Suicide-Related Behaviors: Recognizing Student Voices for Improving Practice. School Psych. Rev. 2022, 51, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Howe, N. Podcast Extra: Coronavirus—Science in the Pandemic. Nature 2020. online ahead of print. [Google Scholar] [CrossRef]
- Jain, V.; Sybil, D.; Premchandani, S.; Krishna, M.; Singh, S. Intraoral Bone Regeneration Using Stem Cells-What a Clinician Needs to Know: Based on a 15-Year MEDLINE Search. Front. Dent. 2021, 18, 40. [Google Scholar] [CrossRef]
- Joo, J.Y.; Liu, M.F. Culturally Tailored Interventions for Ethnic Minorities: A Scoping Review. Nurs. Open 2021, 8, 2078–2090. [Google Scholar] [CrossRef]
- Jarmul, S.; Dangour, A.D.; Green, R.; Liew, Z.; Haines, A.; Scheelbeek, P.F. Climate Change Mitigation through Dietary Change: A Systematic Review of Empirical and Modelling Studies on the Environmental Footprints and Health Effects of ‘Sustainable Diets’. Environ. Res. Lett. 2020, 15, 123014. [Google Scholar] [CrossRef]
- Tezuka, M.; Mizusawa, H.; Tsukada, M.; Mimura, Y.; Shimizu, T.; Kobayashi, A.; Takahashi, Y.; Maejima, T. Severe Necrosis of the Glans Penis Associated with Calciphylaxis Treated by Partial Penectomy. IJU Case Rep. 2020, 3, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Reeder, J.A.; Reynolds, T.R.; Gilbert, B.W. Successful Use of Aspirin, Apixaban, and Viscoelastography in a Patient with Severe COVID Disease and Allergy to Porcine Products. Hosp. Pharm. 2022, 57, 135–137. [Google Scholar] [CrossRef]
- Alzer, H.; Kalbouneh, H.; Alsoleihat, F.; Abu Shahin, N.; Ryalat, S.; Alsalem, M.; Alahmad, H.; Tahtamouni, L. Age of the Donor Affects the Nature of in Vitro Cultured Human Dental Pulp Stem Cells. Saudi Dent. J. 2021, 33, 524–532. [Google Scholar] [CrossRef]
- Patel, A.; Muthukrishnan, I.; Kurian, A.; Amalchandra, J.; Sampathirao, N.; Simon, S. Multicentric Primary Diffuse Large B-Cell Lymphoma in Genitourinary Tract Detected on 18F-Fluorodeoxyglucose Positron Emission Tomography with Computed Tomography: An Uncommon Presentation of a Common Malignancy. World J. Nucl. Med. 2021, 20, 117–120. [Google Scholar] [CrossRef]
- Bittmann, L. Quantum Grothendieck Rings as Quantum Cluster Algebras. J. Lond. Math. Soc. 2021, 103, 161–197. [Google Scholar] [CrossRef] [PubMed]
- da Silva Antas, J.; de Holanda, A.K.G.; de Sousa Andrade, A.; de Araujo, A.M.S.; de Carvalho Costa, I.G.; de Lima, S.K.M.; da Fonseca Abrantes Sarmen, P.L. Curva de Aprendizado de Anastomose Arteriovenosa Com Uso de Simulador de Baixo Custo. J. Vasc. Bras. 2020, 19, e20190144. [Google Scholar] [CrossRef] [PubMed]
| Model | Embedding Dimension | Approx. Parameter Count | Pretraining/Fine-Tuning Corpus | Pooling Strategy |
|---|---|---|---|---|
| mpnet | 68 | ~110 M | Trained on a >1 B sentence dataset and large-scale general text corpora using the MPNet architecture. | Mean pooling |
| minilm-l6 | 384 | ~22 M | Distilled from a teacher model on a >1 B paraphrase dataset, emphasizing efficiency with fewer layers (six layers). | Mean pooling |
| minilm-l12 | 384 | ~33 M | Distilled similarly to minilm-l6 but retains more layers (12 layers), trained on the same broad paraphrase/entailment dataset. | Mean pooling |
| multi-qa-distilbert | 768 | ~66 M | Fine-tuned on 215 M QA datasets to produce universal question-answer embeddings. | Mean pooling |
| roberta | 768 | ~82 M | Based on DistilRoBERTa-base, further tuned on a large paraphrase corpus (>1 B sentence pairs) for enhanced sentence embedding. | Mean pooling |
| Model | Top Article | Top Avg Journal | Avg Journal Similarity | Journal in Top 10 | Encoding Time |
|---|---|---|---|---|---|
| minilm-l12 | Journal of Preventive Medicine and Hygiene | International journal of environmental research and public health | 0.852531 | False | 9” |
| minilm-l6 | Journal of Hospital Infection | Risk management and healthcare policy | 0.848661 | False | 9” |
| mpnet | International Journal of Environmental Research and Public Health | International journal of environmental research and public health | 0.926116 | False | 98” |
| multi-qa-distilbert | European Review for Medical and Pharmacological Sciences | Risk management and healthcare policy | 0.858523 | False | 54” |
| roberta | Journal of Hospital Infection | International journal of environmental research and public health | 0.886216 | False | 52” |
| Model | Top Article | MaxSimilarity | MeanSim | Shannon Entropy | Gini | Journal in Top 10 |
|---|---|---|---|---|---|---|
| minilm-l12 | 0.799 | 0.765548 | 0.67 ± 0.04 | 3.24 | 0.036077 | 12.5 |
| minilm-l6 | 0.775 | 0.746310 | 0.66 ± 0.04 | 3.24 | 0.034712 | 14.5 |
| mpnet | 0.831 | 0.794203 | 0.71 ± 0.04 | 3.24 | 0.029997 | 14.5 |
| multi-qa-distilbert | 0.755 | 0.717609 | 0.63 ± 0.04 | 3.24 | 0.034632 | 12.5 |
| roberta | 0.803 | 0.773727 | 0.68 ± 0.04 | 3.24 | 0.033623 | 12.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colangelo, M.T.; Meleti, M.; Guizzardi, S.; Calciolari, E.; Galli, C. A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata. Big Data Cogn. Comput. 2025, 9, 67. https://doi.org/10.3390/bdcc9030067
Colangelo MT, Meleti M, Guizzardi S, Calciolari E, Galli C. A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata. Big Data and Cognitive Computing. 2025; 9(3):67. https://doi.org/10.3390/bdcc9030067
Chicago/Turabian StyleColangelo, Maria Teresa, Marco Meleti, Stefano Guizzardi, Elena Calciolari, and Carlo Galli. 2025. "A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata" Big Data and Cognitive Computing 9, no. 3: 67. https://doi.org/10.3390/bdcc9030067
APA StyleColangelo, M. T., Meleti, M., Guizzardi, S., Calciolari, E., & Galli, C. (2025). A Comparative Analysis of Sentence Transformer Models for Automated Journal Recommendation Using PubMed Metadata. Big Data and Cognitive Computing, 9(3), 67. https://doi.org/10.3390/bdcc9030067








