Robust Clinical Querying with Local LLMs: Lexical Challenges in NL2SQL and Retrieval-Augmented QA on EHRs
Abstract
1. Introduction
- NL2SQL: Natural language to SQL generation;
- RAG-QA: Retrieval-augmented generation for question answering;
- SQL-EC: Generated SQL error classification.
2. Literature Review
2.1. Recent Developments of Clinical NL2SQL
2.2. Retrieval-Augmented Generation in Clinical Environments
2.3. Lexical Challenges in Clinical Environments
2.4. Integration Challenges and Policy Considerations
2.5. Synthesis and Key Takeaways
3. Methodology
3.1. Language Models Under Test
3.2. Task 1: Natural Language to SQL Generation (NL2SQL)
- Syntactic Validity: The generated SQL query must execute without syntax errors, meaning it is successfully parsed and run by the SQLite engine.
- Semantic Correctness: The query must return results consistent with the intended meaning of the natural language prompt.
- Adherence to Constraints: The query must respect the structure and integrity constraints defined by the underlying database schema.
- The result set must contain the same data values as the reference query;
- The result set must include the same number of rows;
- The order of rows is disregarded—all result sets are sorted before comparison. For instance, when evaluating a query such as “return the top 5 diagnoses,” the results are ordered alphabetically before comparison, even if the query lacks an explicit ORDER BY clause.
- Input tokens: The total number of tokens in the prompt, encompassing the natural language query, schema details, instructions, and other contextual elements.
- Output tokens: The number of tokens the model generates as part of the SQL response.
- Pricing structure: Each model’s specific cost per input and output token.
- Job description: A brief but explicit statement defining the model’s role as a SQL query generator within the medical domain. This component anchors the model’s objective and ensures domain-specific alignment.
- Instructions: A comprehensive list of behavioral constraints to shape the SQL outputs. These include formatting directives, SQL dialect restrictions (specific to SQLite), avoidance of placeholders, and preferences for readability and syntactic simplicity. Such instructions minimize ambiguity and promote consistency.
- Schema information: A detailed presentation of the database schema, including table names, field types, and descriptions. This context helps the model to generate syntactically correct and semantically coherent SQL queries.
- 1-shot example query: A single illustrative example that pairs a user query with its corresponding SQL translation. This demonstrates expected output structure and style, helping to condition the model’s subsequent generations.
- Data preview: A selection of sample rows from key database tables, offering empirical insight into data content and structure. This component further enhances the model’s ability to align query logic with real-world data patterns.
3.3. Task 2: Retrieval-Augmented Question Answering (RAG-QA)
3.3.1. RAG-QA Metrics
| Reference answer: | “Patient presented with severe chest pain and shortness of breath” |
| Generated answer: | “Patient reported severe chest pain and breathing difficulty” |
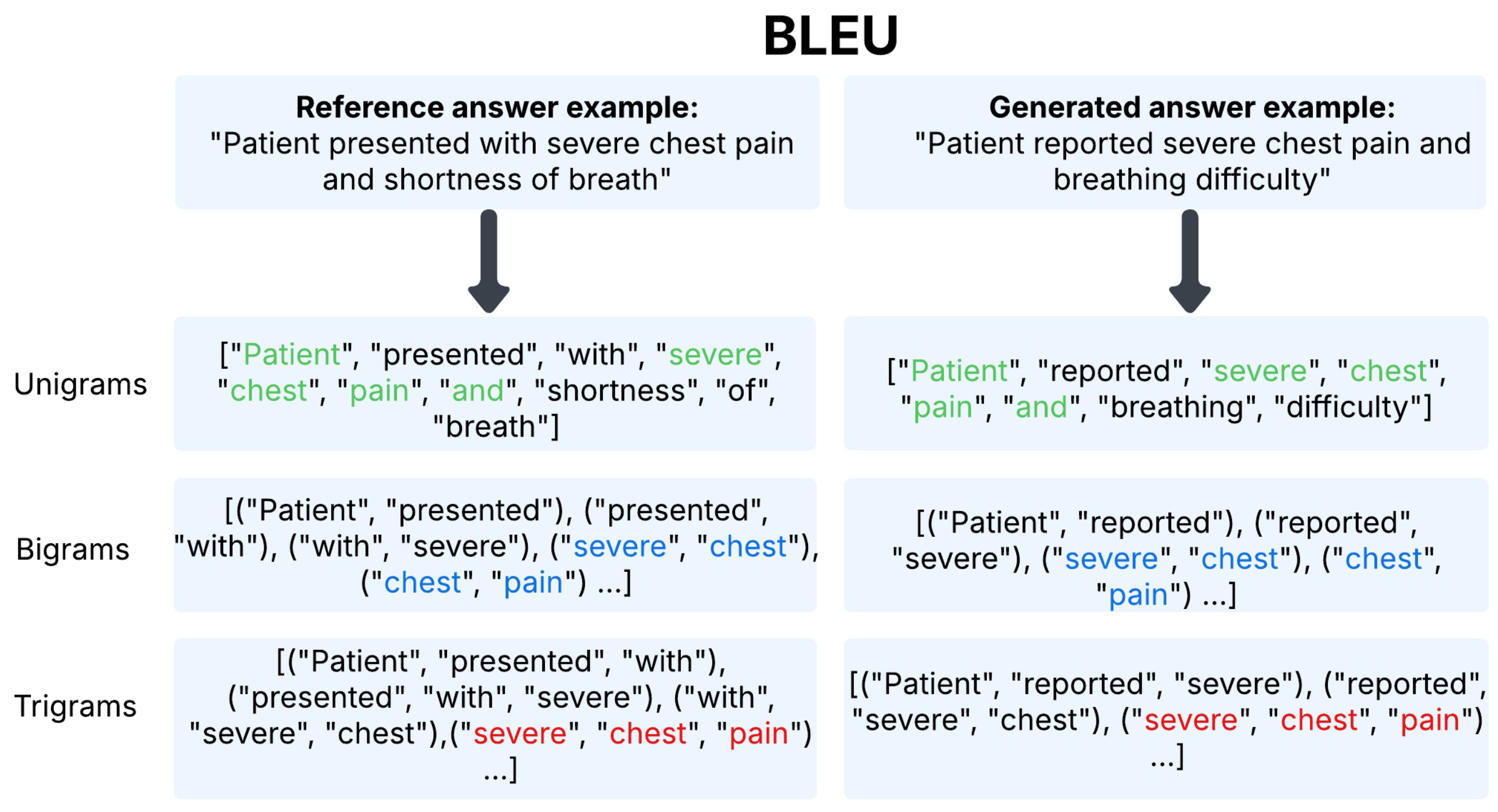
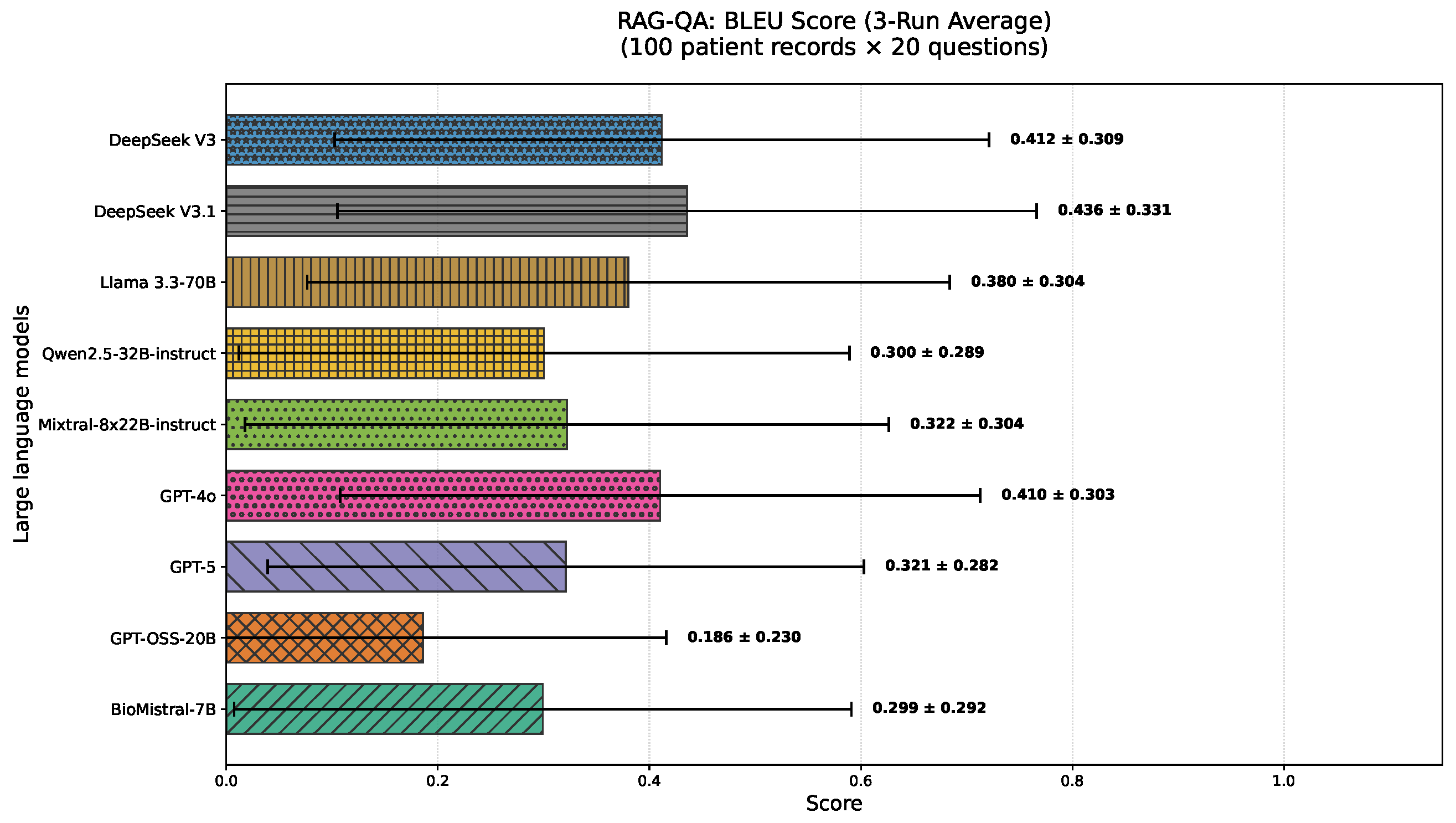
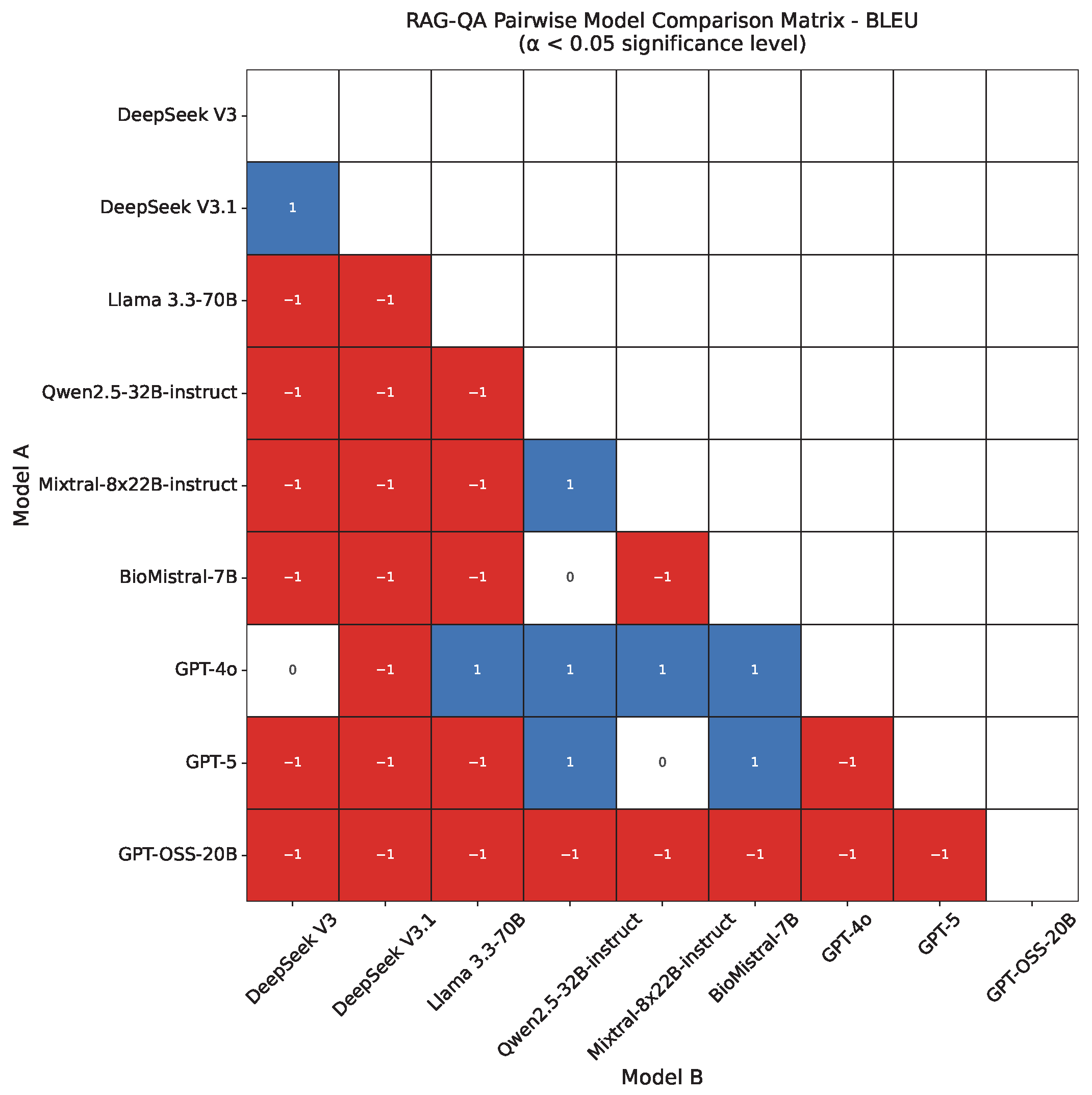
- BLEU [121] (Bilingual Evaluation Understudy) evaluates the generated text quality via n-gram overlap with a reference. We report BLEU-4, capturing matches up to 4-g.Figure 2 presents tokenized versions of the reference and generated answers for the example pair, with overlapping unigrams, bigrams, and trigrams highlighted in green, blue, and red, respectively, to illustrate lexical similarity.We employ the BLEU-4 scoring function from NLTK [126], which incorporates n-gram precision up to and includes a brevity penalty. Summary statistics for n-gram matches in our example are given in Table 5, with the complete BLEU computation provided in Appendix C.1.
- ROUGE-L [122] (Recall-Oriented Understudy for Gisting Evaluation) measures sequence-level similarity via the longest common subsequence (LCS), which captures non-contiguous token matches that preserve order. Higher LCS values reflect closer alignment in information structure. We use the ROUGE-L F1 score from Google’s ROUGE implementation, harmonizing LCS-based precision and recall.Figure 3 illustrates an LCS-based comparison between a reference and a generated answer example. Tokens retained in order are visually highlighted in pink to demonstrate the LCS.A summary of token-level statistics for LCS matching is provided inline, with the complete ROUGE-L F1 derivation given in Appendix C.2.
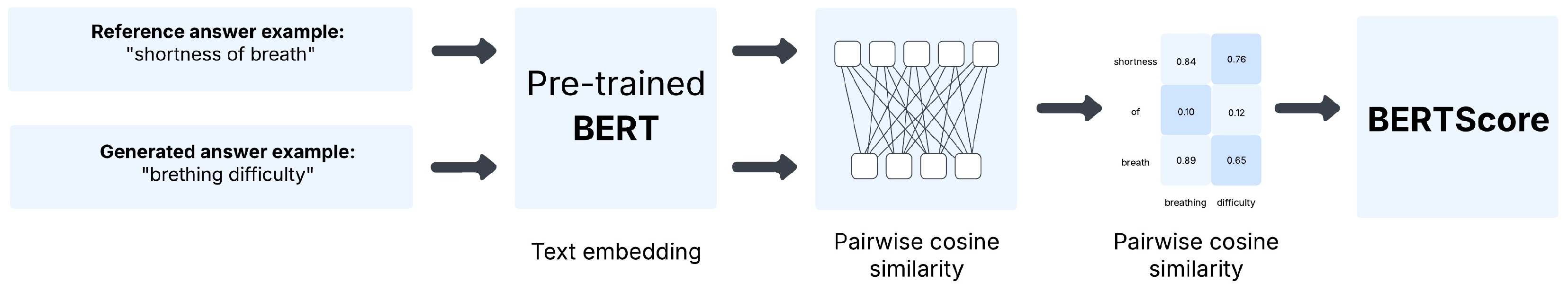
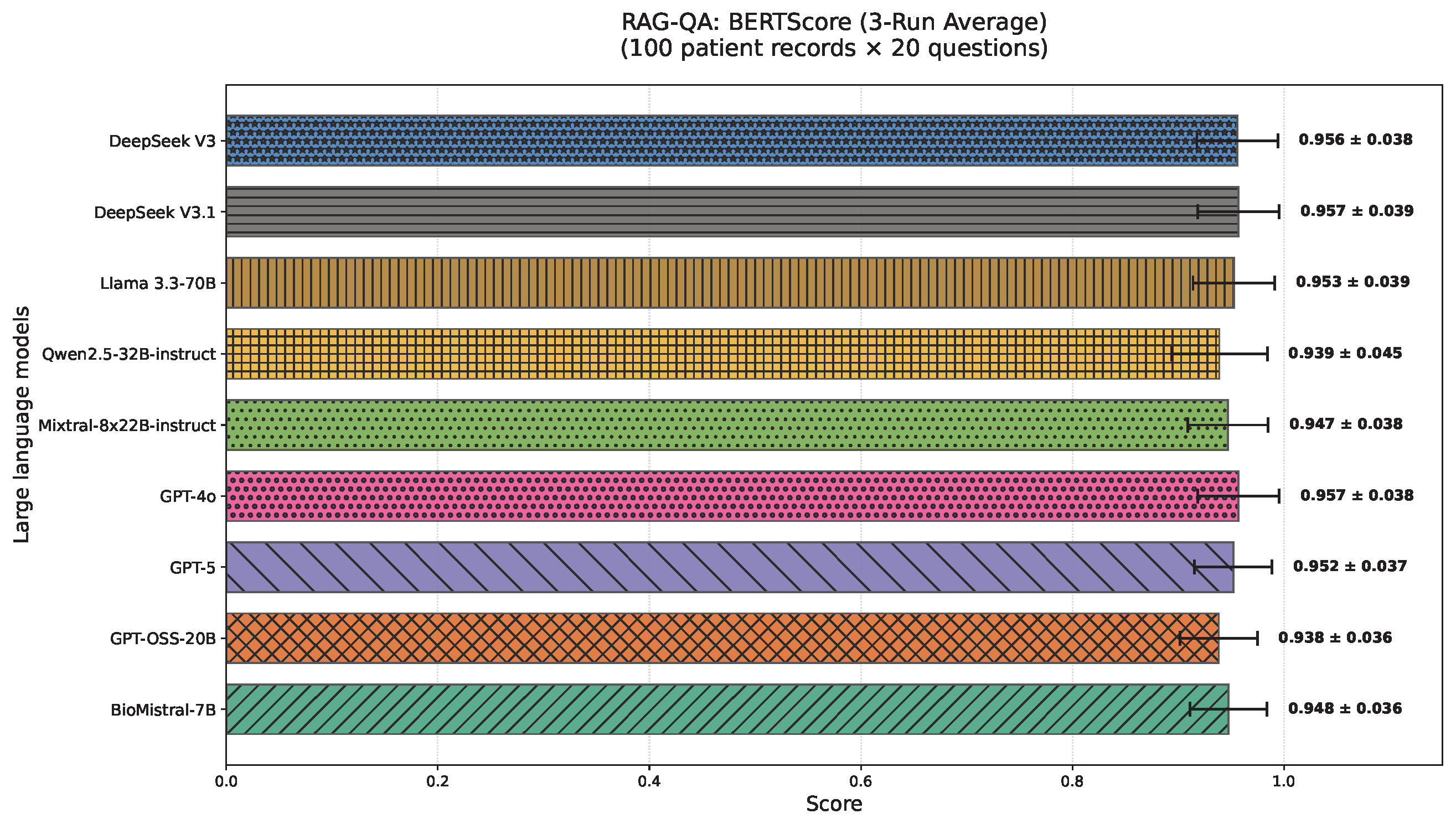
- BERTScore [123] measures semantic similarity between generated and reference texts using contextual embeddings from pre-trained transformer models. Unlike lexical metrics, it aligns each token with its most semantically similar counterpart (in both directions) via cosine similarity.We report BERTScore F1, which integrates the precision and recall of token alignments, capturing semantic equivalence. This is particularly valuable in clinical contexts where lexical variation is quite common.Figure 4 illustrates this with a comparison between “shortness of breath” and “breathing difficulty”, showing that BERTScore captures semantic equivalence beyond surface token overlap.The formal mathematical definition and computation of BERTScore are provided in Appendix C.3.
- Job description: It defines the model’s role as a health administrator responding to clinical questions based on patient documentation. This sets the scope and tone for the expected outputs.
- 1-shot example: A single illustrative query–response pair demonstrates the expected style, format, and specificity of the model’s answers.
- RAG Documentation: It is an excerpt of a patient record retrieved in the RAG retrieval and search step. This context provides the factual basis for the model’s response and constrains it to a grounded answer.
- Output format: Instructions require responses to be returned as plain text, without any formatting or markup, ensuring clarity and consistency.
3.4. Task 3: SQL Error Classification (SQL-EC)
- Deterministic classification: The first stage involved applying a rule-based error classifier (SQL-EC) to systematically categorize erroneous SQL generations. The classifier compared each generated SQL query against the ground truth from the MIMICSQL dataset and assigned one or more error categories deterministically. Each error log included the original natural language question, the corresponding reference query, and the incorrect query produced by the model. If no specific pattern matched, the classifier assigned the query to the OTHER category. This ensured consistency across all models and enabled multifaceted labeling when multiple types of errors were present.
- Manual annotation: In the second stage, we manually reviewed cases that the classifier had assigned to the OTHER category. These manual checks ensured that residual errors were correctly interpreted and aligned with the predefined taxonomy, avoiding under-classification due to limitations of the deterministic rules.
| Algorithm 1: Pseudocode of the proposed rule-based SQL error classification framework for SQL-EC task |
 |
4. Results and Discussion
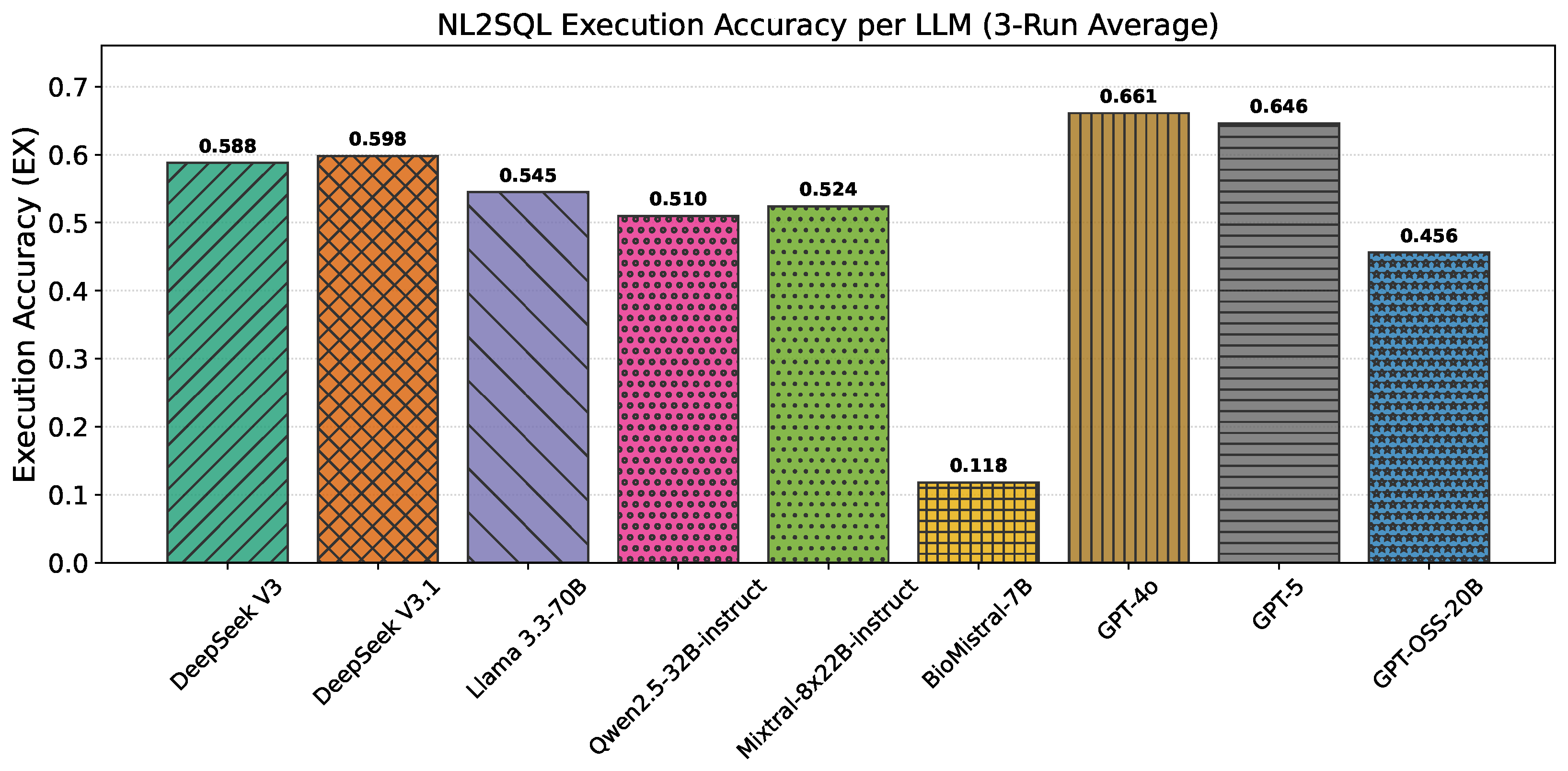
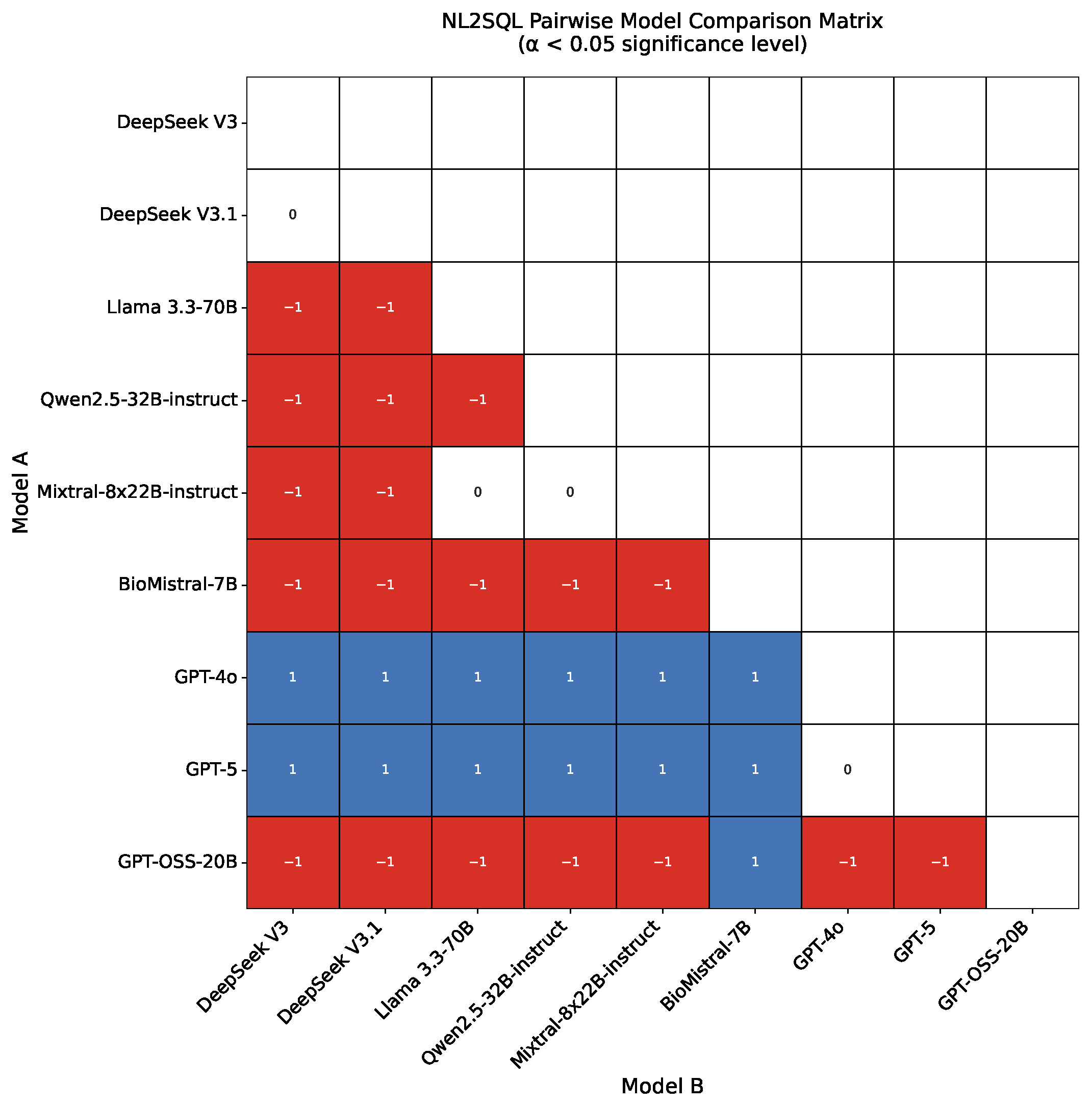
4.1. NL2SQL Results
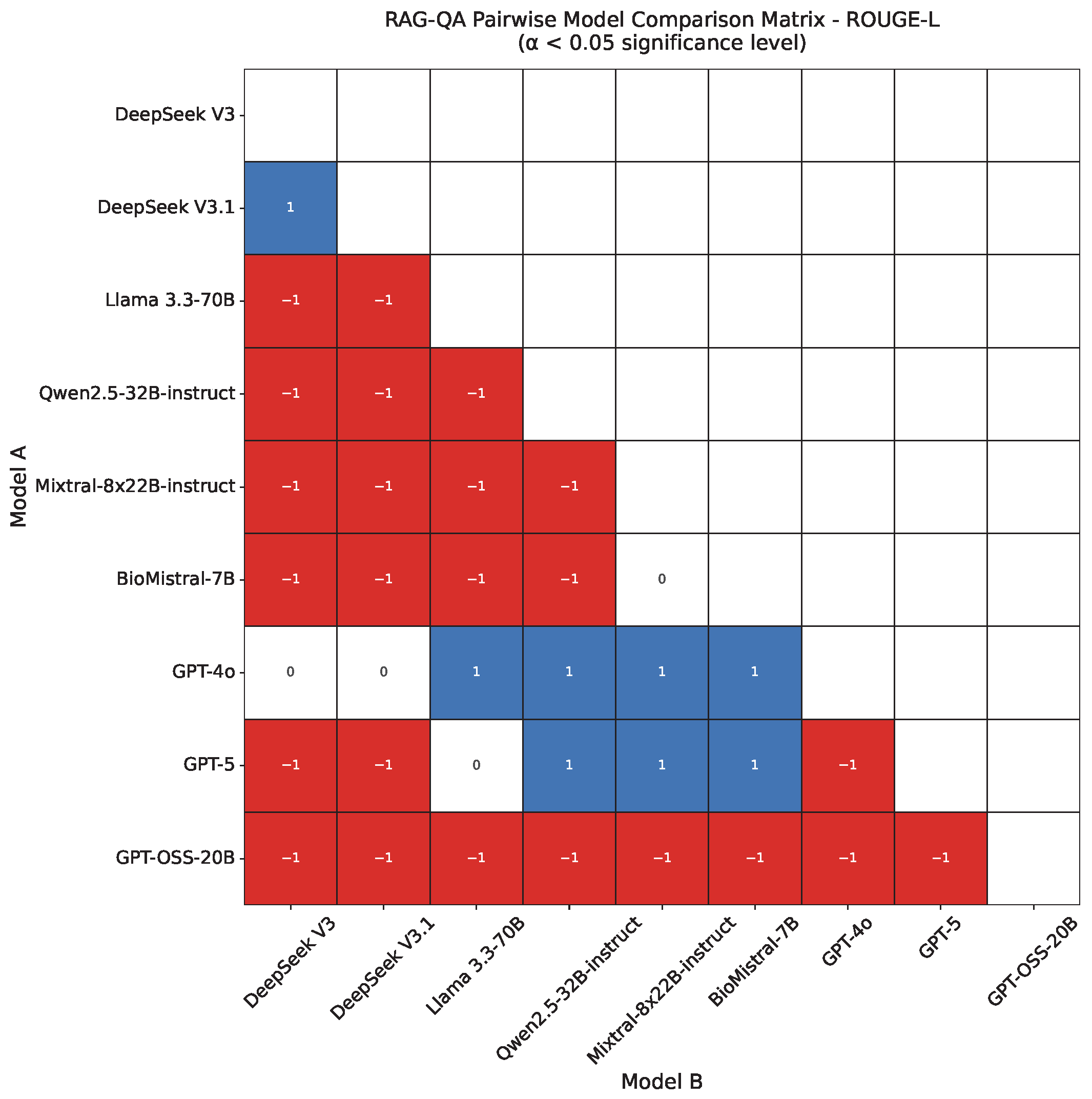
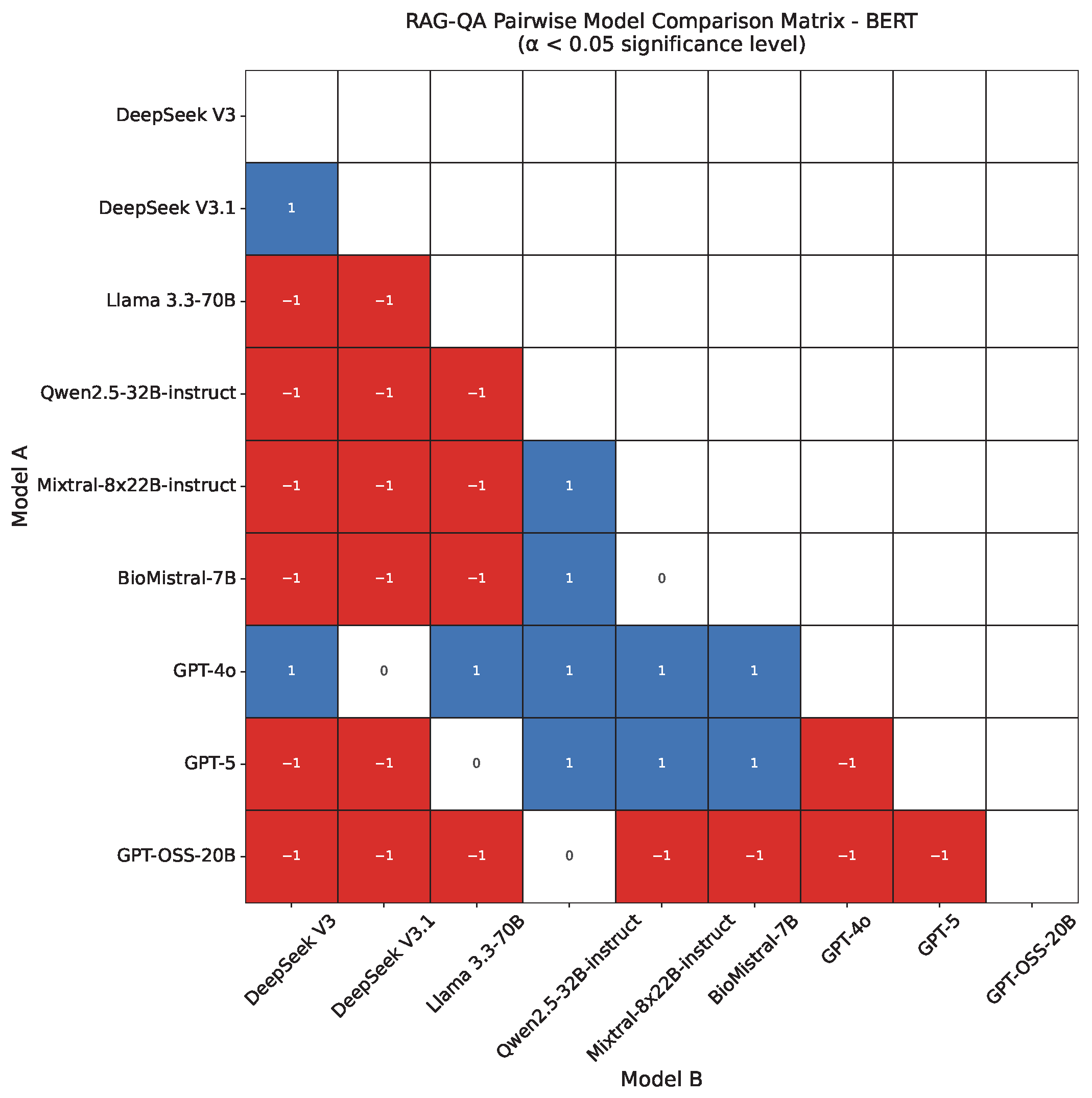
4.2. RAG-QA Results
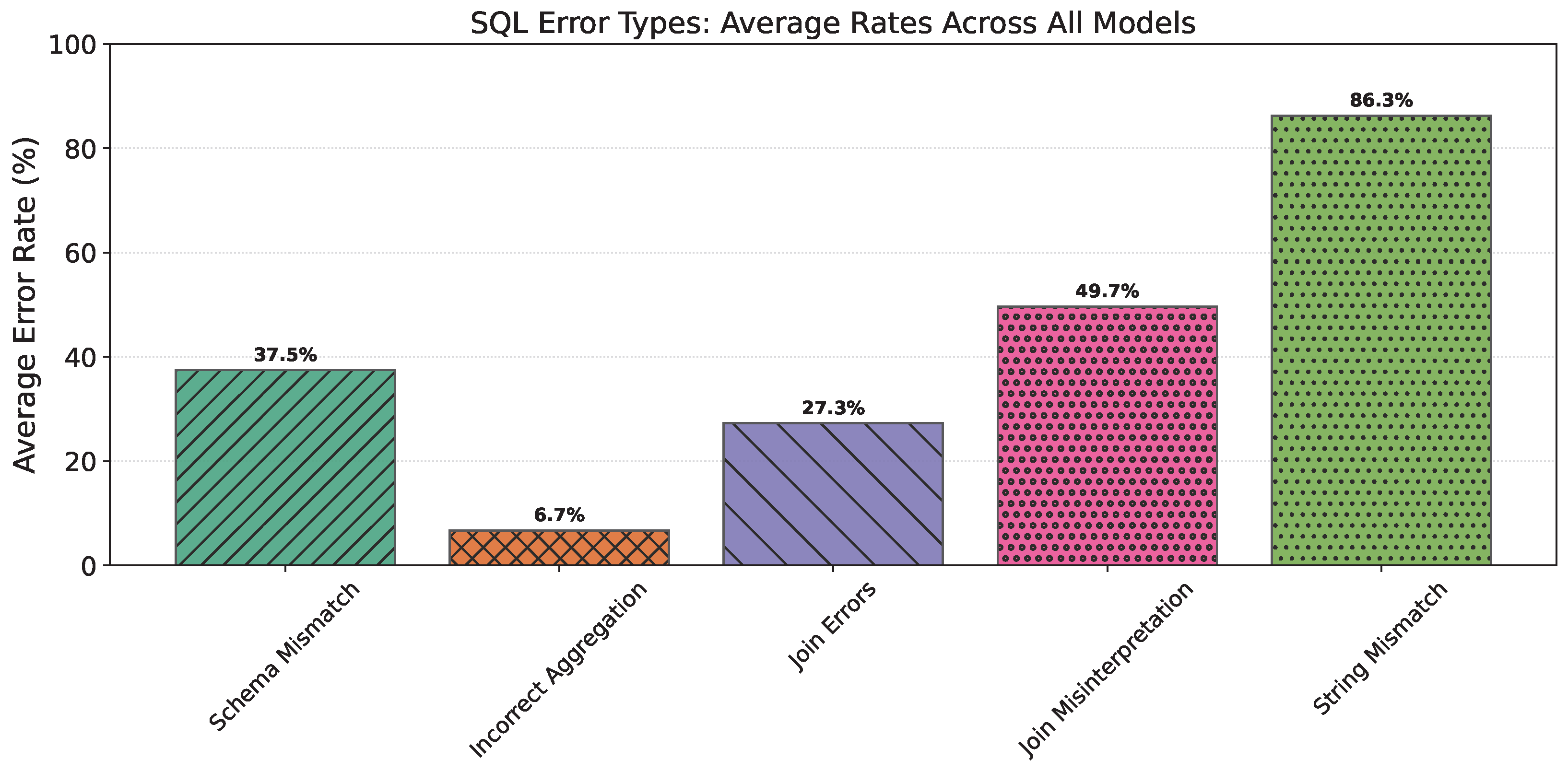
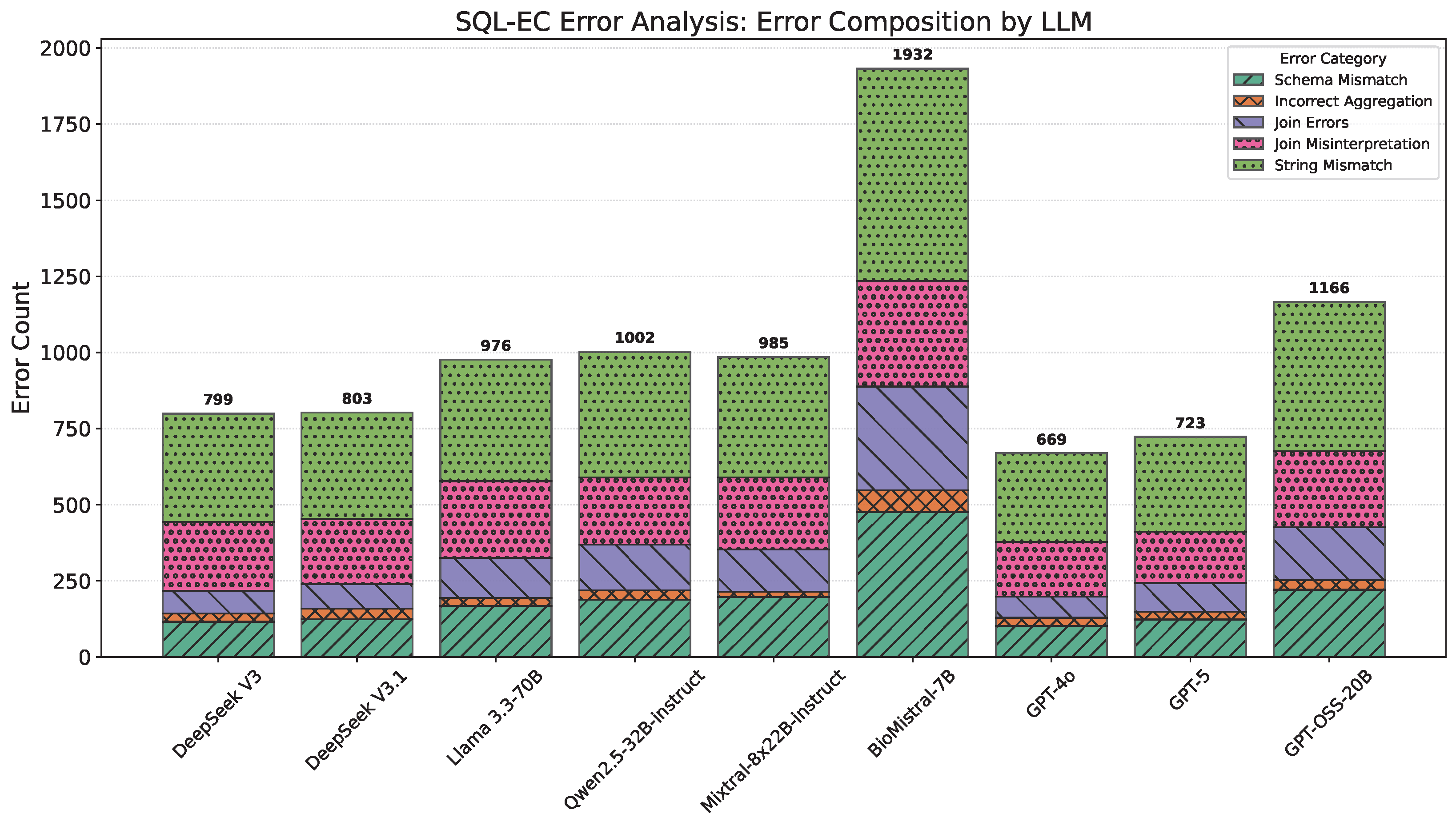
4.3. SQL-EC Results
5. Limitations and Future Directions
5.1. Limitations
5.2. Future Directions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. NL2SQL—Prompt Structure (Excerpt)
Appendix A.1. NL2SQL Job Description and Example
- Job Description:You are an advanced language model specialized in generating SQL queries for electronic health record (EHR) databases. Given a natural language question and the relevant schema, return a syntactically correct SQL query using SQLite syntax. All table and column names must be double-quoted. No table aliases are allowed, and comments should be excluded from the output.
- Example:
- Example Query:
- User: List the number of female patients above 60 diagnosed with hypertension.
- Expected Output:
- SELECT COUNT(DISTINCT ‘‘demographic’’."SUBJECT_ID")
- FROM "demographic"
- JOIN "diagnoses" ON "demographic"."HADM_ID" = "diagnoses"."HADM_ID"
- WHERE LOWER("demographic"."GENDER") = ’female’
- AND "demographic"."AGE" > 60
- AND LOWER("diagnoses"."SHORT_TITLE") LIKE ’%hypertension%’;
Appendix A.2. NL2SQL—SQL Schema (Excerpt)
-- demographic: patient-level information
CREATE TABLE demographic (
SUBJECT_ID INTEGER,
HADM_ID INTEGER,
AGE INTEGER,
GENDER TEXT,
ADMISSION_TYPE TEXT,
INSURANCE TEXT
);
-- diagnoses: diagnostic codes per admission
CREATE TABLE diagnoses (
SUBJECT_ID INTEGER,
HADM_ID INTEGER,
ICD9_CODE TEXT,
SHORT_TITLE TEXT,
LONG_TITLE TEXT
);
Appendix A.3. NL2SQL—Data Preview (Excerpt)
TABLE: DEMOGRAPHIC
SUBJECT_ID,HADM_ID,AGE,GENDER,ADMISSION_TYPE,INSURANCE
10001,123456,65,FEMALE,EMERGENCY,Medicare
10002,123457,58,MALE,URGENT,Private
TABLE: DIAGNOSES
SUBJECT_ID,HADM_ID,ICD9_CODE,SHORT_TITLE,LONG_TITLE
10001,123456,4019,Hypertension,Essential hypertension, unspecified
10002,123457,2500,Diabetes Mellitus,Type 2 diabetes without complications
Appendix A.4. NL2SQL—Example of Prompted Query
- Case number: 05cbfa2dbd0e6f14a3b99380a97d21e0Question: What number of patients had the lab test named fibrin degradation products and self paid the costs?Ground Truth SQL:
SELECT Count(DISTINCT demographic."subject_id")
FROM demographic
INNER JOIN lab ON demographic.hadm_id = lab.hadm_id
WHERE demographic."insurance" = "self pay"
AND lab."label" = "Fibrin Degradation Products"
-
Model Responses:DeepSeek-V3:
SELECT Count(DISTINCT demographic."subject_id")
FROM demographic
INNER JOIN lab ON demographic.hadm_id = lab.hadm_id
WHERE Lower(lab."label") = "fibrin degradation products"
AND demographic."insurance" = "self pay"
-
Llama3.3-70b:
SELECT Count(DISTINCT demographic."subject_id")
FROM demographic
INNER JOIN lab ON demographic."subject_id" = lab."subject_id"
WHERE Lower(lab."label") = ’fibrin degradation products’
AND demographic."insurance" = ’Self Pay’
-
GPT-4o:
SELECT Count(DISTINCT demographic."subject_id")
FROM demographic
INNER JOIN lab ON demographic.hadm_id = lab.hadm_id
WHERE Lower(lab."label") = ’fibrin degradation products’
AND Lower(demographic."insurance") = ’self pay’
Appendix B. RAG-QA—Prompt Structure
Appendix B.1. RAG-QA—Job Description and Example
- Job Description:You are a health administrator that will respond to user question about a patient in a concise and informative way. For that purpose you will be provided an excerpt from the documentation on the patient.Example Question-Answer Pair:
- Q: What is the patient’s chief complaint?
- A: The patient’s chief complaint is chronic pelvic pain that
- has worsened over the past 12 months,
- with increased menstrual flow duration, fatigue, bloating, and nausea.
Appendix B.2. RAG-QA—Documentation
Appendix B.3. RAG-QA—Output
Appendix C. RAG Metric Computation Details
Appendix C.1. BLEU Computation Details
Appendix C.2. ROUGE-L Computation Details
Appendix C.3. BERTScore Computation Details
References
- Garets, D.; Davis, M. Electronic Medical Records vs. Electronic Health Records: Yes, There is a Difference; Policy White Paper; HIMSS Analytics: Chicago, IL, USA, 2006; Volume 1. [Google Scholar]
- Kim, M.K.; Rouphael, C.; McMichael, J.; Welch, N.; Dasarathy, S. Challenges in and Opportunities for Electronic Health Record-Based Data Analysis and Interpretation. Gut Liver 2024, 18, 201–208. [Google Scholar] [CrossRef]
- Shabestari, O.; Roudsari, A. Challenges in data quality assurance for electronic health records. In Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Lewis, A.E.; Weiskopf, N.; Abrams, Z.B.; Foraker, R.; Lai, A.M.; Payne, P.R.; Gupta, A. Electronic health record data quality assessment and tools: A systematic review. J. Am. Med. Inform. Assoc. 2023, 30, 1730–1740. [Google Scholar] [CrossRef]
- Upadhyay, S.; Hu, H.F. A qualitative analysis of the impact of electronic health records (EHR) on healthcare quality and safety: Clinicians’ lived experiences. Health Serv. Insights 2022, 15, 11786329211070722. [Google Scholar] [CrossRef]
- Campanella, P.; Lovato, E.; Marone, C.; Fallacara, L.; Mancuso, A.; Ricciardi, W.; Specchia, M.L. The impact of electronic health records on healthcare quality: A systematic review and meta-analysis. Eur. J. Public Health 2016, 26, 60–64. [Google Scholar] [CrossRef]
- Tapuria, A.; Porat, T.; Kalra, D.; Dsouza, G.; Xiaohui, S.; Curcin, V. Impact of patient access to their electronic health record: Systematic review. Inform. Health Soc. Care 2021, 46, 194–206. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef]
- Huang, K.; Altosaar, J.; Ranganath, R. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv 2019, arXiv:1904.05342. [Google Scholar]
- Bolton, E.; Hall, D.; Yasunaga, M.; Lee, T.; Manning, C.; Liang, P. Stanford CRFM Introduces PubMedGPT 2.7B. 2022. Available online: https://hai.stanford.edu/news/stanford-crfm-introduces-pubmedgpt-27b (accessed on 31 March 2025).
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large language models encode clinical knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Singhal, K.; Tu, T.; Gottweis, J.; Sayres, R.; Wulczyn, E.; Amin, M.; Hou, L.; Clark, K.; Pfohl, S.R.; Cole-Lewis, H.E.A. Toward expert-level medical question answering with large language models. Nat. Med. 2025, 31, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Leonte, K.G.; Chen, M.L.; Torous, J.B.; Linos, E.; Pinto, A.; Rodriguez, C.I. Large language models outperform mental and medical health care professionals in identifying obsessive-compulsive disorder. npj Digit. Med. 2024, 7, 193. [Google Scholar] [CrossRef]
- Hou, Y.; Bert, C.; Gomaa, A.; Lahmer, G.; Höfler, D.; Weissmann, T.; Voigt, R.; Schubert, P.; Schmitter, C.; Depardon, A.; et al. Fine-tuning a local LLaMA-3 large language model for automated privacy-preserving physician letter generation in radiation oncology. Front. Artif. Intell. 2025, 7, 1493716. [Google Scholar] [CrossRef]
- Kuckelman, I.J.; Wetley, K.; Yi, P.H.; Ross, A.B. Translating musculoskeletal radiology reports into patient-friendly summaries using ChatGPT-4. Skelet. Radiol. 2024, 53, 1621–1624. [Google Scholar] [CrossRef]
- Mustafa, A.; Naseem, U.; Azghadi, M.R. Large language models vs human for classifying clinical documents. Int. J. Med. Inform. 2025, 195, 105800. [Google Scholar] [CrossRef]
- Ong, J.C.L.; Jin, L.; Elangovan, K.; Lim, G.Y.S.; Lim, D.Y.Z.; Sng, G.G.R.; Ke, Y.; Tung, J.Y.M.; Zhong, R.J.; Koh, C.M.Y.; et al. Development and testing of a novel large language model-based clinical decision support systems for medication safety in 12 clinical specialties. arXiv 2024, arXiv:2402.01741. [Google Scholar] [CrossRef]
- Liu, S.; McCoy, A.B.; Wright, A. Improving large language model applications in biomedicine with retrieval-augmented generation: A systematic review, meta-analysis, and clinical development guidelines. J. Am. Med. Inform. Assoc. 2025, 32, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Dennstädt, F.; Hastings, J.; Putora, P.M.; Schmerder, M.; Cihoric, N. Implementing large language models in healthcare while balancing control, collaboration, costs and security. npj Digit. Med. 2025, 8, 143. [Google Scholar] [CrossRef]
- Wada, A.; Tanaka, Y.; Nishizawa, M.; Yamamoto, A.; Akashi, T.; Hagiwara, A.; Hayakawa, Y.; Kikuta, J.; Shimoji, K.; Sano, K.; et al. Retrieval-augmented generation elevates local LLM quality in radiology contrast media consultation. npj Digit. Med. 2025, 8, 395. [Google Scholar] [CrossRef]
- Mortensen, G.A.; Zhu, R. Early Alzheimer’s Detection Through Voice Analysis: Harnessing Locally Deployable LLMs via ADetectoLocum, a privacy-preserving diagnostic system. AMIA Summits Transl. Sci. Proc. 2025, 2025, 365. [Google Scholar] [PubMed]
- Lorencin, I.; Tankovic, N.; Etinger, D. Optimizing Healthcare Efficiency with Local Large Language Models. In Intelligent Human Systems Integration (IHSI 2025): Integrating People and Intelligent Systems; AHFE International Open Access: Rome, Italy, 2025; Volume 160. [Google Scholar]
- Wiest, I.C.; Leßmann, M.E.; Wolf, F.; Ferber, D.; Van Treeck, M.; Zhu, J.; Ebert, M.P.; Westphalen, C.B.; Wermke, M.; Kather, J.N. Anonymizing medical documents with local, privacy preserving large language models: The LLM-Anonymizer. medRxiv 2024. [Google Scholar] [CrossRef]
- Hong, Z.; Yuan, Z.; Zhang, Q.; Chen, H.; Dong, J.; Huang, F.; Huang, X. Next-Generation Database Interfaces: A Survey of LLM-based Text-to-SQL. arXiv 2025, arXiv:2406.08426. [Google Scholar] [CrossRef]
- Malešević, A.; Čartolovni, A. Healthcare digitalization: Insights from Croatia’s experience and lessons from the COVID-19 pandemic. In Digital Healthcare, Digital Transformation and Citizen Empowerment in Asia-Pacific and Europe for a Healthier Society; Academic Press (Elsevier): Cambridge, MA, USA, 2025; pp. 459–473. [Google Scholar]
- Mastilica, M.; Kušec, S. Croatian healthcare system in transition, from the perspective of users. BMJ 2005, 331, 223–226. [Google Scholar] [CrossRef]
- Björnberg, A.; Phang, A.Y. Euro Health Consumer Index 2018 Report. 2018. Available online: https://santesecu.public.lu/dam-assets/fr/publications/e/euro-health-consumer-index-2018/euro-health-consumer-index-2018.pdf (accessed on 30 March 2025).
- OECD. Croatia Country Health Profile 2023; OECD Publishing: Paris, France, 2023. [Google Scholar]
- Džakula, A.; Vočanec, D.; Banadinović, M.; Vajagić, M.; Lončarek, K.; Lovrenčić, I.L.; Radin, D.; Rechel, B. Croatia: Health System Summary, 2024; European Observatory on Health Systems and Policies, WHO Regional Office for Europe: Copenhagen, Denmark, 2024. [Google Scholar]
- Zhong, V.; Xiong, C.; Socher, R. Seq2sql: Generating structured queries from natural language using reinforcement learning. arXiv 2017, arXiv:1709.00103. [Google Scholar] [CrossRef]
- Xu, X.; Liu, C.; Song, D. SQLNet: Generating Structured Queries From Natural Language Without Reinforcement Learning. arXiv 2017, arXiv:1711.04436. [Google Scholar] [CrossRef]
- Roberts, K.; Patra, B.G. A semantic parsing method for mapping clinical questions to logical forms. In Proceedings of the AMIA 2018 Annual Symposium Proceedings, San Francisco, CA, USA, 3–7 November 2018; American Medical Informatics Association (AMIA): Washington, DC, USA, 2018; Volume 2017, p. 1478. [Google Scholar]
- Zhu, X.; Li, Q.; Cui, L.; Liu, Y. Large language model enhanced text-to-sql generation: A survey. arXiv 2024, arXiv:2410.06011. [Google Scholar]
- Wang, P.; Shi, T.; Reddy, C.K. Text-to-SQL Generation for Question Answering on Electronic Medical Records. In Proceedings of the WWW ’20: The Web Conference 2020, Taipei Taiwan, 20–24 April 2020; Association for Computing Machinery (ACM): New York, NY, USA, 2020; pp. 350–361. [Google Scholar]
- Pan, Y.; Wang, C.; Hu, B.; Xiang, Y.; Wang, X.; Chen, Q.; Chen, J.; Du, J. A BERT-based generation model to transform medical texts to SQL queries for electronic medical records: Model development and validation. JMIR Med. Inform. 2021, 9, e32698. [Google Scholar] [CrossRef]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. arXiv 2020, arXiv:2005.14165. [Google Scholar] [CrossRef]
- Chen, M.; Tworek, J.; Jun, H.; Yuan, Q.; de Oliveira Pinto, H.P.; Kaplan, J.; Edwards, H.; Burda, Y.; Joseph, N.; Brockman, G.; et al. Evaluating Large Language Models Trained on Code. arXiv 2021, arXiv:2107.03374. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, R.; Yang, K.; Yasunaga, M.; Wang, D.; Li, Z.; Ma, J.; Li, I.; Yao, Q.; Roman, S.; et al. Spider: A Large-Scale Human-Labeled Dataset for Complex and Cross-Domain Semantic Parsing and Text-to-SQL Task. arXiv 2019, arXiv:1809.08887. [Google Scholar]
- Li, J.; Hui, B.; Qu, G.; Yang, J.; Li, B.; Li, B.; Wang, B.; Qin, B.; Geng, R.; Huo, N.; et al. Can llm already serve as a database interface? a big bench for large-scale database grounded text-to-sqls. Adv. Neural Inf. Process. Syst. 2023, 36, 42330–42357. [Google Scholar]
- Liu, X.; Shen, S.; Li, B.; Ma, P.; Jiang, R.; Zhang, Y.; Fan, J.; Li, G.; Tang, N.; Luo, Y. A Survey of NL2SQL with Large Language Models: Where are we, and where are we going? arXiv 2024, arXiv:2408.05109. [Google Scholar] [CrossRef]
- Guo, J.; Zhan, Z.; Gao, Y.; Xiao, Y.; Lou, J.G.; Liu, T.; Zhang, D. Towards Complex Text-to-SQL in Cross-Domain Database with Intermediate Representation. arXiv 2019, arXiv:1905.08205. [Google Scholar]
- Hwang, W.; Yim, J.; Park, S.; Seo, M. A comprehensive exploration on wikisql with table-aware word contextualization. arXiv 2019, arXiv:1902.01069. [Google Scholar]
- OpenAI. GPT-3.5. 2023. Available online: https://platform.openai.com/docs/models/gpt-3.5-turbo (accessed on 6 September 2025).
- OpenAI. GPT-4 Technical Report. arXiv 2023, arXiv:2303.08774. [Google Scholar] [CrossRef]
- Anthropic. Claude 2. 2023. Available online: https://www.anthropic.com/index/claude-2 (accessed on 6 September 2025).
- DeepMind, G. Gemini 1.5: Unlocking Multimodal Long Context Understanding. 2024. Available online: https://blog.google/technology/ai/google-gemini-next-generation-model-february-2024/ (accessed on 6 September 2025).
- OpenAI. Introducing GPT-5 for Developers. 2025. Available online: https://openai.com/index/introducing-gpt-5-for-developers/ (accessed on 6 September 2025).
- Attrach, R.A.; Moreira, P.; Fani, R.; Umeton, R.; Celi, L.A. Conversational LLMs Simplify Secure Clinical Data Access, Understanding, and Analysis. arXiv 2025, arXiv:2507.01053. [Google Scholar]
- Anthropic. Introducing the Model Context Protocol. 2024. Available online: https://www.anthropic.com/news/model-context-protocol (accessed on 10 September 2025).
- Chadha, I.K.; Gupta, A.; Sarkar, S.; Tomer, M.; Rathee, T. Performance Evaluation of Open-Source LLMs for Text-to-SQL Conversion in Healthcare Data. In Proceedings of the 2025 International Conference on Pervasive Computational Technologies (ICPCT), Greater Noida, India, 8–9 February 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 606–609. [Google Scholar]
- OpenAI. gpt-oss-120b & gpt-oss-20b Model Card. arXiv 2025, arXiv:2508.10925. [Google Scholar]
- Liu, A.; Feng, B.; Xue, B.; Wang, B.; Wu, B.; Lu, C.; Zhao, C.; Deng, C.; Zhang, C.; Ruan, C.; et al. DeepSeek-V3 Technical Report. arXiv 2025, arXiv:2412.19437. [Google Scholar]
- Tarbell, R.; Choo, K.K.R.; Dietrich, G.; Rios, A. Towards understanding the generalization of medical text-to-sql models and datasets. In Proceedings of the AMIA Annual Symposium Proceedings, San Francisco, CA, USA, 9–13 November 2024; American Medical Informatics Association (AMIA): Washington, DC, USA, 2024; Volume 2023, p. 669. [Google Scholar]
- Rahman, S.; Jiang, L.Y.; Gabriel, S.; Aphinyanaphongs, Y.; Oermann, E.K.; Chunara, R. Generalization in healthcare ai: Evaluation of a clinical large language model. arXiv 2024, arXiv:2402.10965. [Google Scholar] [CrossRef]
- Chen, C.; Yu, J.; Chen, S.; Liu, C.; Wan, Z.; Bitterman, D.; Wang, F.; Shu, K. ClinicalBench: Can LLMs Beat Traditional ML Models in Clinical Prediction? arXiv 2024, arXiv:2411.06469. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, Z.; Jin, Q.; Chen, F.; Fang, Y.; Liu, Y.; Rousseau, J.F.; Xu, Z.; Lu, Z.; Weng, C.; et al. Leveraging long context in retrieval augmented language models for medical question answering. npj Digit. Med. 2025, 8, 239. [Google Scholar] [CrossRef]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; Yih, W.t.; Rocktäschel, T.; et al. Retrieval-augmented generation for knowledge-intensive nlp tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Amugongo, L.M.; Mascheroni, P.; Brooks, S.; Doering, S.; Seidel, J. Retrieval augmented generation for large language models in healthcare: A systematic review. PLoS Digit. Health 2025, 4, e0000877. [Google Scholar] [CrossRef]
- Alkhalaf, M.; Yu, P.; Yin, M.; Deng, C. Applying generative AI with retrieval augmented generation to summarize and extract key clinical information from electronic health records. J. Biomed. Inform. 2024, 156, 104662. [Google Scholar] [CrossRef]
- Myers, S.; Dligach, D.; Miller, T.A.; Barr, S.; Gao, Y.; Churpek, M.; Mayampurath, A.; Afshar, M. Evaluating Retrieval-Augmented Generation vs. Long-Context Input for Clinical Reasoning over EHRs. arXiv 2025, arXiv:2508.14817. [Google Scholar]
- Long, C.; Subburam, D.; Lowe, K.; Dos Santos, A.; Zhang, J.; Hwang, S.; Saduka, N.; Horev, Y.; Su, T.; Côté, D.W.; et al. ChatENT: Augmented large language model for expert knowledge retrieval in otolaryngology–head and neck surgery. Otolaryngol.–Head Neck Surg. 2024, 171, 1042–1051. [Google Scholar] [CrossRef]
- Ge, J.; Sun, S.; Owens, J.; Galvez, V.; Gologorskaya, O.; Lai, J.C.; Pletcher, M.J.; Lai, K. Development of a liver disease–specific large language model chat interface using retrieval-augmented generation. Hepatology 2024, 80, 1158–1168. [Google Scholar] [CrossRef]
- Rouhollahi, A.; Homaei, A.; Sahu, A.; Harari, R.E.; Nezami, F.R. RAGnosis: Retrieval-Augmented Generation for Enhanced Medical Decision Making. medRxiv 2025. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Yang, S.Y.; Miao, C. Medrag: Enhancing retrieval-augmented generation with knowledge graph-elicited reasoning for healthcare copilot. In Proceedings of the ACM on Web Conference 2025, Sydney, Australia, 28 April–2 May 2025; Association for Computing Machinery (ACM): New York, NY, USA, 2025; pp. 4442–4457. [Google Scholar]
- Soni, S.; Gayen, S.; Demner-Fushman, D. Overview of the archehr-qa 2025 shared task on grounded question answering from electronic health records. In Proceedings of the 24th Workshop on Biomedical Language Processing, Vienna, Austria, 1 August 2025; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2025; pp. 396–405. [Google Scholar]
- Yu, H.; Gan, A.; Zhang, K.; Tong, S.; Liu, Q.; Liu, Z. Evaluation of retrieval-augmented generation: A survey. In Proceedings of the CCF Conference on Big Data, Shenyang, China, 28–30 August 2025; Springer: Singapore, 2025; pp. 102–120. [Google Scholar]
- Gargari, O.K.; Habibi, G. Enhancing medical AI with retrieval-augmented generation: A mini narrative review. Digit. Health 2025, 11, 20552076251337177. [Google Scholar] [CrossRef]
- Xiong, G.; Jin, Q.; Lu, Z.; Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Proceedings of the Findings of the Association for Computational Linguistics, Bangkok, Thailand, 11–16 August 2024; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2024; pp. 6233–6251. [Google Scholar]
- Wu, J.; Zhu, J.; Qi, Y. Medical Graph RAG: Towards Safe Medical Large Language Model via Graph Retrieval-Augmented Generation. arXiv 2024, arXiv:2408.04187. [Google Scholar] [CrossRef]
- Jadhav, S.; Shanbhag, A.G.; Joshi, S.; Date, A.; Sonawane, S. Maven at MEDIQA-CORR 2024: Leveraging RAG and Medical LLM for Error Detection and Correction in Medical Notes. In Proceedings of the Clinical Natural Language Processing Workshop, Mexico City, Mexico, 24 April 2024; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2024. [Google Scholar]
- Jayatilake, D.C.; Oyibo, S.O.; Jayatilake, D. Interpretation and misinterpretation of medical abbreviations found in patient medical records: A cross-sectional survey. Cureus 2023, 15, e44735. [Google Scholar] [CrossRef] [PubMed]
- SNOMED CT: Myocardial infarction (Concept ID 22298006). 2025. Available online: https://bioportal.bioontology.org/ontologies/SNOMEDCT?p=classes&conceptid=22298006 (accessed on 16 January 2025).
- Cheung, S.; Hoi, S.; Fernandes, O.; Huh, J.; Kynicos, S.; Murphy, L.; Lowe, D. Audit on the use of dangerous abbreviations, symbols, and dose designations in paper compared to electronic medication orders: A multicenter study. Ann. Pharmacother. 2018, 52, 332–337. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health (NIH) Exhibit in NIH Grants Policy Statement (NIH GPS). 2024. Available online: https://grants.nih.gov/grants/policy/nihgps/html5/section_1/1.1_abbreviations.htm (accessed on 6 September 2025).
- Do Not Use: Dangerous Abbreviations, Symbols, and Dose Designations—2025 Update. 2025. Available online: https://ismpcanada.ca/resource/do-not-use-list/ (accessed on 6 September 2025).
- Marecková, E.; Cervenỳ, L. Latin as the language of medical terminology: Some remarks on its role and prospects. Swiss Med. Wkly. 2002, 132, 581. [Google Scholar] [CrossRef]
- Lysanets, Y.V.; Bieliaieva, O.M. The use of Latin terminology in medical case reports: Quantitative, structural, and thematic analysis. J. Med. Case Rep. 2018, 12, 45. [Google Scholar] [CrossRef]
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef]
- Liu, H.; Lussier, Y.A.; Friedman, C. A study of abbreviations in the UMLS. In Proceedings of the AMIA Symposium, Portland, OR, USA, 6–10 November 2001; American Medical Informatics Association (AMIA): Washington, DC, USA, 2001; p. 393. [Google Scholar]
- Xu, R.; Jiang, P.; Luo, L.; Xiao, C.; Cross, A.; Pan, S.; Sun, J.; Yang, C. A Survey on Unifying Large Language Models and Knowledge Graphs for Biomedicine and Healthcare. In Proceedings of the 31st ACM SIGKDD Conference on Knowledge Discovery and Data Mining V.2, Toronto, ON, Canada, 3–7 August 2025; Association for Computing Machinery (ACM): New York, NY, USA, 2025; pp. 6195–6205. [Google Scholar]
- Nazi, Z.A.; Hristidis, V.; McLean, A.L.; Meem, J.A.; Chowdhury, M.T.A. Ontology-Guided Query Expansion for Biomedical Document Retrieval using Large Language Models. arXiv 2025, arXiv:2508.11784. [Google Scholar] [CrossRef]
- Fan, Y.; Xue, K.; Li, Z.; Zhang, X.; Ruan, T. An LLM-based Framework for Biomedical Terminology Normalization in Social Media via Multi-Agent Collaboration. In Proceedings of the 31st International Conference on Computational Linguistics, Abu Dhabi, United Arab Emirates, 19–24 January 2025; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2025; pp. 10712–10726. [Google Scholar]
- Borchert, F.; Llorca, I.; Schapranow, M.P. Improving biomedical entity linking for complex entity mentions with LLM-based text simplification. Database 2024, 2024, baae067. [Google Scholar] [CrossRef]
- Remy, F.; Demuynck, K.; Demeester, T. BioLORD-2023: Semantic textual representations fusing llm and clinical knowledge graph insights. arXiv 2023, arXiv:2311.16075. [Google Scholar] [CrossRef]
- Tertulino, R.; Antunes, N.; Morais, H. Privacy in electronic health records: A systematic mapping study. J. Public Health 2024, 32, 435–454. [Google Scholar] [CrossRef]
- Mirzaei, T.; Amini, L.; Esmaeilzadeh, P. Clinician voices on ethics of LLM integration in healthcare: A thematic analysis of ethical concerns and implications. BMC Med. Inform. Decis. Mak. 2024, 24, 250. [Google Scholar] [CrossRef] [PubMed]
- Rathod, V.; Nabavirazavi, S.; Zad, S.; Iyengar, S.S. Privacy and security challenges in large language models. In Proceedings of the 2025 IEEE 15th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 6–8 January 2025; IEEE: Piscataway, NJ, USA, 2025; pp. 00746–00752. [Google Scholar]
- Yao, Y.; Duan, J.; Xu, K.; Cai, Y.; Sun, Z.; Zhang, Y. A survey on large language model (llm) security and privacy: The good, the bad, and the ugly. High-Confid. Comput. 2024, 4, 100211. [Google Scholar] [CrossRef]
- Gemma Team. Gemma: Open Models Based on Gemini Research and Technology. arXiv 2024, arXiv:2403.08295. [Google Scholar] [CrossRef]
- Gemma Team. Mistral 7B. arXiv 2023, arXiv:2310.06825. [Google Scholar] [CrossRef]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and Efficient Foundation Language Models. arXiv 2023, arXiv:2302.13971. [Google Scholar] [CrossRef]
- Kukreja, S.; Kumar, T.; Purohit, A.; Dasgupta, A.; Guha, D. A literature survey on open source large language models. In Proceedings of the 2024 7th International Conference on Computers in Management and Business, Singapore, 12–14 January 2024; Association for Computing Machinery (ACM): New York, NY, USA, 2024; pp. 133–143. [Google Scholar]
- Plaat, A.; Wong, A.; Verberne, S.; Broekens, J.; van Stein, N.; Bäck, T. Reasoning with large language models, a survey. CoRR 2024. [Google Scholar] [CrossRef]
- Floratou, A.; Psallidas, F.; Zhao, F.; Deep, S.; Hagleither, G.; Tan, W.; Cahoon, J.; Alotaibi, R.; Henkel, J.; Singla, A.; et al. Nl2sql is a solved problem… not! In Proceedings of the Conference of Innovative Data Systems Research (CIDR), Chaminade, HI, USA, 14–17 January 2024; CIDR Foundation: San Mateo, CA, USA, 2024. [Google Scholar]
- Tai, C.Y.; Chen, Z.; Zhang, T.; Deng, X.; Sun, H. Exploring chain-of-thought style prompting for text-to-sql. arXiv 2023, arXiv:2305.14215. [Google Scholar]
- Ali Alkamel, S. DeepSeek and the Power of Mixture of Experts (MoE). Available online: https://dev.to/sayed_ali_alkamel/deepseek-and-the-power-of-mixture-of-experts-moe-ham (accessed on 1 April 2025).
- DeepSeek-V3.1 Release. 2025. Available online: https://api-docs.deepseek.com/news/news250821 (accessed on 10 September 2025).
- Open Source Initiative. MIT License. 1988. Available online: https://opensource.org/licenses/MIT (accessed on 10 September 2025).
- Meta Platforms, Inc. Meta LLaMA 3 Community License. 2024. Available online: https://www.llama.com/llama3/license/ (accessed on 10 September 2025).
- Team Qwen. Qwen2.5 Technical Report. arXiv 2025, arXiv:2412.15115. [Google Scholar]
- Apache Software Foundation. Apache License, Version 2.0. 2004. Available online: http://www.apache.org/licenses/LICENSE-2.0 (accessed on 10 September 2025).
- Mistral AI. Cheaper, Better, Faster, Stronger: Mixtral 8x22B. Release Blog Post. 2024. Available online: https://mistral.ai/news/mixtral-8x22b (accessed on 10 September 2025).
- Labrak, Y.; Bazoge, A.; Morin, E.; Gourraud, P.A.; Rouvier, M.; Dufour, R. Biomistral: A collection of open-source pretrained large language models for medical domains. arXiv 2024, arXiv:2402.10373. [Google Scholar]
- OpenAI. GPT-4o. 2024. Available online: https://openai.com/index/hello-gpt-4o/ (accessed on 22 April 2025).
- OpenAI. Introducing GPT-5. 2025. Available online: https://openai.com/index/introducing-gpt-5/ (accessed on 7 August 2025).
- Tanković, N.; Šajina, R.; Lorencin, I. Transforming Medical Data Access: The Role and Challenges of Recent Language Models in SQL Query Automation. Algorithms 2025, 18, 124. [Google Scholar] [CrossRef]
- Xiao, G.; Lin, J.; Seznec, M.; Wu, H.; Demouth, J.; Han, S. SmoothQuant: Accurate and Efficient Post-Training Quantization for Large Language Models. arXiv 2024, arXiv:2211.10438. [Google Scholar]
- Frantar, E.; Ashkboos, S.; Hoefler, T.; Alistarh, D. GPTQ: Accurate Post-Training Quantization for Generative Pre-trained Transformers. arXiv 2023, arXiv:2210.17323. [Google Scholar]
- Fireworks AI. How Fireworks Evaluates Quantization Precisely and Interpretably. Blog Post. 2024. Available online: https://fireworks.ai/blog/fireworks-quantization (accessed on 11 September 2025).
- Fireworks AI. LLM Inference Performance Benchmarking (Part 1). Blog Post. 2023. Available online: https://fireworks.ai/blog/llm-inference-performance-benchmarking-part-1 (accessed on 11 September 2025).
- Sandrini, P. Beyond the Cloud: Assessing the Benefits and Drawbacks of Local LLM Deployment for Translators. arXiv 2025, arXiv:2507.23399. [Google Scholar] [CrossRef]
- Chen, K.; Zhou, X.; Lin, Y.; Feng, S.; Shen, L.; Wu, P. A survey on privacy risks and protection in large language models. J. King Saud Univ. Comput. Inf. Sci. 2025, 37, 163. [Google Scholar] [CrossRef]
- Hamburg Commissioner for Data Protection and Freedom of Information. Discussion Paper: Large Language Models and Personal Data. Discussion paper, Hamburg Commissioner for Data Protection and Freedom of Information (HmbBfDI). 2024. Available online: https://datenschutz-hamburg.de/fileadmin/user_upload/HmbBfDI/Datenschutz/Informationen/240715_Discussion_Paper_Hamburg_DPA_KI_Models.pdf (accessed on 11 September 2025).
- Holtzman, A.; Buys, J.; Du, L.; Forbes, M.; Choi, Y. The Curious Case of Neural Text Degeneration. arXiv 2020, arXiv:1904.09751. [Google Scholar] [CrossRef]
- Hipp, R.D. SQLite. 2020. Available online: https://sqlite.org/ (accessed on 11 September 2025).
- Qin, B.; Hui, B.; Wang, L.; Yang, M.; Li, J.; Li, B.; Geng, R.; Cao, R.; Sun, J.; Si, L.; et al. A Survey on Text-to-SQL Parsing: Concepts, Methods, and Future Directions. arXiv 2022, arXiv:2208.13629. [Google Scholar]
- Noor, H. What Do You Mean? Using Large Language Models for Semantic Evaluation of NL2SQL Queries. 2025. Available online: https://uwspace.uwaterloo.ca/bitstreams/73520bc6-13dd-4586-a9c4-5b8ced8ddfc1/download (accessed on 9 September 2025).
- OpenAI. API Pricing. 2025. Available online: https://openai.com/api/pricing/ (accessed on 12 September 2025).
- Fireworks AI. Pricing. 2025. Available online: https://fireworks.ai/pricing (accessed on 12 September 2025).
- Qdrant Team. Qdrant—Vector Database. 2025. Available online: https://qdrant.tech/ (accessed on 11 September 2025).
- Papineni, K.; Roukos, S.; Ward, T.; Zhu, W.J. Bleu: A method for automatic evaluation of machine translation. In Proceedings of the 40th Annual Meeting of the Association for Computational Linguistics, Philadelphia, PA, USA, 6–12 July 2002; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2002; pp. 311–318. [Google Scholar]
- Lin, C.Y. Rouge: A package for automatic evaluation of summaries. In Proceedings of the Text Summarization Branches Out, Barcelona, Spain, 25–26 July 2004; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2004; pp. 74–81. [Google Scholar]
- Zhang, T.; Kishore, V.; Wu, F.; Weinberger, K.Q.; Artzi, Y. BERTScore: Evaluating Text Generation with BERT. In Proceedings of the International Conference on Learning Representations (ICLR), Addis Ababa, Ethiopia, 30 April 2020. [Google Scholar]
- OpenAI. Text Embedding 3 Small. 2023. Available online: https://platform.openai.com/docs/models/text-embedding-3-small (accessed on 12 September 2025).
- Malkov, Y.A.; Yashunin, D.A. Efficient and robust approximate nearest neighbor search using Hierarchical Navigable Small World graphs. arXiv 2018, arXiv:1603.09320. [Google Scholar] [CrossRef] [PubMed]
- Bird, S.; Klein, E.; Loper, E. Natural Language Processing with Python; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2009. [Google Scholar]
- Liu, X.; Shen, S.; Li, B.; Tang, N.; Luo, Y. NL2SQL-BUGs: A Benchmark for Detecting Semantic Errors in NL2SQL Translation. arXiv 2025, arXiv:2503.11984. [Google Scholar]
- Shen, J.; Wan, C.; Qiao, R.; Zou, J.; Xu, H.; Shao, Y.; Zhang, Y.; Miao, W.; Pu, G. A Study of In-Context-Learning-Based Text-to-SQL Errors. arXiv 2025, arXiv:2501.09310. [Google Scholar]
- Ning, Z.; Tian, Y.; Zhang, Z.; Zhang, T.; Li, T.J.J. Insights into natural language database query errors: From attention misalignment to user handling strategies. arXiv 2024, arXiv:2402.07304. [Google Scholar] [CrossRef]
- Ren, T.; Fan, Y.; He, Z.; Huang, R.; Dai, J.; Huang, C.; Jing, Y.; Zhang, K.; Yang, Y.; Wang, X.S. Purple: Making a large language model a better sql writer. In Proceedings of the 2024 IEEE 40th International Conference on Data Engineering (ICDE), Utrecht, The Netherlands, 13–16 May 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 15–28. [Google Scholar]
- Zhu, C.; Lin, Y.; Cai, Y.; Li, Y. SCoT2S: Self-Correcting Text-to-SQL Parsing by Leveraging LLMs. Comput. Speech Lang. 2026, 95, 101865. [Google Scholar] [CrossRef]
- Mitsopoulou, A.V. Towards More Robust Text-to-SQL Translation. 2024. Available online: https://pergamos.lib.uoa.gr/uoa/dl/object/3401060/file.pdf (accessed on 10 April 2025).
- Kroll, H.; Kreutz, C.K.; Sackhoff, P.; Balke, W.T. Enriching Simple Keyword Queries for Domain-Aware Narrative Retrieval. In Proceedings of the 2023 ACM/IEEE Joint Conference on Digital Libraries (JCDL), Santa Fe, NM, USA, 26–30 June 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 143–154. [Google Scholar] [CrossRef]
- Banerjee, S.; Lavie, A. METEOR: An automatic metric for MT evaluation with improved correlation with human judgments. In Proceedings of the ACL Workshop on Intrinsic and Extrinsic Evaluation Measures for Machine Translation and/or Summarization, Ann Arbor, MI, USA, 1 June 2005; Association for Computational Linguistics (ACL): Stroudsburg, PA, USA, 2005; pp. 65–72. [Google Scholar]




| Model | Architecture | Params/Active (B) | Max. Ctx. (Tokens) | Availability |
|---|---|---|---|---|
| DeepSeek V3 | MoE (MLA) | 671/∼37 | 128k | OSS (MIT) |
| DeepSeek V3.1 | MoE (MLA) | 671/∼37 | 128k | OSS (MIT) |
| Llama 3.3-70B | Dense decoder-only | 70 | 128k | OSS (Llama 3 Comm.) |
| Qwen2.5-32B | Dense decoder-only (RoPE,GQA) | 32.5 | 131k | OSS (Apache-2.0) |
| Mixtral-8x22B | MoE (8 × 22B) | 141/39 | 64k | OSS (Apache-2.0) |
| BioMistral-7B | Dense decoder-only | 7 | 32k | OSS (Apache-2.0) |
| GPT-4o | Proprietary multimodal | n/a | 128k * | P (API) |
| GPT-5 | Unified proprietary † | n/a | 400k ‡ | P (API) |
| GPT-OSS-20B | MoE | 20.9/∼3.6 | 131k | OSS (Apache-2.0) |
| Natural Language QUESTION | Reference SQL Query |
|---|---|
| Tell me the number of patients who were diagnosed with unspecified cerebral artery occlusion with cerebral infarction. | SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID") FROM DEMOGRAPHIC INNER JOIN DIAGNOSES ON DEMOGRAPHIC.HADM_ID = DIAGNOSES.HADM_ID WHERE DIAGNOSES."SHORT_TITLE" = "Crbl art ocl NOS w infrc" |
| Find out the category of lab test for patient Stephanie Suchan. | SELECT LAB."CATEGORY" FROM DEMOGRAPHIC INNER JOIN LAB ON DEMOGRAPHIC.HADM_ID = LAB.HADM_ID WHERE DEMOGRAPHIC."NAME" = "Stephanie Suchan" |
| What is the number of patients with abdominal pain as their primary disease who were discharged to SNF? | SELECT COUNT(DISTINCT DEMOGRAPHIC."SUBJECT_ID") FROM DEMOGRAPHIC WHERE DEMOGRAPHIC."DISCHARGE_LOCATION" = "SNF" AND DEMOGRAPHIC."DIAGNOSIS" = "ABDOMINAL PAIN" |
| Category | Section | Example Snippet (Record ID: 4) |
|---|---|---|
| Generic Information | Patient Information | “Jane Doe, 40-year-old female nurse, divorced, registered April 1, 2024.” |
| Insurance Information | “HealthSecure, full coverage, deductible $1000, prior authorization obtained for pelvic MRI.” | |
| Emergency Contacts | “Primary contact: (555) 472-6833; Secondary contact: sister, (555) 987-6543.” | |
| Billing Information | “Electronic billing via HealthSecure portal, outstanding balance: none.” | |
| Medical Record | Chief Complaint | “Chronic pelvic pain worsening over 12 months; prolonged menstruation up to 10 days.” |
| History of Present Illness | “Progressively heavier bleeding, pain worsens with activity, occasional dizziness.” | |
| Past Medical History | “Anemia, hypertension (Losartan 50mg), appendectomy, 2 cesareans.” | |
| Gynecological History | “Irregular periods, fibroids diagnosed age 38, G2P2, last Pap smear normal.” | |
| Family History | “Mother with fibroids; sister with endometriosis; father with hypertension and diabetes.” | |
| Social History | “Non-smoker, occasional alcohol, moderate stress, 2 kg weight loss in 6 months.” | |
| Review of Systems | “Fatigue, dizziness, bloating, urinary frequency, mild lower back pain.” | |
| Physical Examination | “Mild pallor, BP 132/85, HR 87, enlarged uterus with irregular contours.” | |
| Diagnostic Studies | “Ultrasound: multiple fibroids (largest 4.8 cm); CBC: hemoglobin 10.2 g/dL.” | |
| Assessment and Treatment Plan | Assessment | “Likely symptomatic uterine fibroids; differential: adenomyosis, endometrial hyperplasia.” |
| Medical Management | “Ferrous sulfate 325 mg daily; Ibuprofen 400 mg as needed.” | |
| Surgical Consultation | “Referral for possible myomectomy or uterine artery embolization.” | |
| Lifestyle Modifications | “Increase iron-rich diet, encourage moderate exercise (e.g., yoga).” | |
| Follow-Up Plan | “Biopsy review in 1 week; monitor hemoglobin every 3 months.” | |
| Final Summary | “Chronic pelvic pain and menorrhagia due to fibroids, initial medical management plus surgical consult planned.” | |
| Evaluation Question |
|---|
| (1) What is the patient’s chief complaint? |
| (2) Who referred the patient for admission? |
| (3) What medications is the patient currently taking? |
| (4) What is the patient’s gynecological history? |
| (5) Has the patient experienced any recent surgical procedures? |
| (6) What is the patient’s preferred method of billing? |
| (7) What diagnostic studies have been performed? |
| (8) What is the attending physician’s name? |
| (9) What is the patient’s age? |
| (10) What is included in the patient’s treatment plan? |
| (11) What is the patient’s marital status? |
| (12) What is the patient’s occupation? |
| (13) Who are the patient’s emergency contacts and their contact information? |
| (14) What is the patient’s family medical history? |
| (15) What are the patient’s social habits including smoking, alcohol use, and exercise? |
| (16) What are the patient’s vital signs? |
| (17) What is the patient’s BMI and general physical appearance? |
| (18) What does the physical examination reveal? |
| (19) What is the patient’s assessment or diagnosis? |
| (20) What is the patient’s follow-up plan? |
| Unigrams (1-g) | Bigrams (2-g) | Trigrams (3-g) | 4-g | |
|---|---|---|---|---|
| Matches | 5 | 2 | 1 | 0 |
| Total generated | 8 | 7 | 6 | 5 |
| Precision | 5/8 = 0.625 | 2/7 ≈ 0.286 | 1/6 ≈ 0.167 | 0/5 = 0 |
| Error Category | Heuristic Rules for Classification |
|---|---|
| (1) Schema mismatch | Errors due to inconsistencies between a query and the database schema are often caused by ambiguous or imprecise language. Heuristic rules are as follows:
|
| (2) Incorrect aggregation usage | Errors in the application of aggregation operations are often due to misunderstandings of how to group or summarize data. Heuristic rules as follows:
|
| (3) Join errors | Errors are due to omitted joins, missing tables, or misconfigured join types, resulting in incomplete or misleading query results across tables. Heuristic rules are as follows:
|
| (4) Condition misinterpretation | Errors in logical expressions within WHERE or HAVING clauses lead to incorrect or overly broad results. Heuristic rules are as follows:
|
| (5) String mismatch | Errors in handling string values are often due to vocabulary differences, improper column use, or flawed comparison. Heuristic rules are as follows:
|
| Model | Total Obs. | True Obs. | Avg. Tokens In | Avg. Tokens Out |
|---|---|---|---|---|
| DeepSeek V3 | 3000 | 1764 | 2281.84 | 68.17 |
| DeepSeek V3.1 | 3000 | 1794 | 2307.84 | 71.53 |
| Llama 3.3-70B | 3000 | 1635 | 2174.07 | 60.68 |
| Qwen2.5-32B-instruct | 3000 | 1530 | 2425.20 | 63.29 |
| Mixtral-8x22B-instruct | 3000 | 1572 | 2979.29 | 92.97 |
| GPT-4o | 3000 | 1983 | 2134.92 | 58.11 |
| GPT-5 | 3000 | 1938 | 2158.91 | 59.58 |
| GPT-OSS-20B | 3000 | 1368 | 2221.92 | 126.12 |
| BioMistral-7B | 3000 | 354 | 2987.24 | 91.84 |
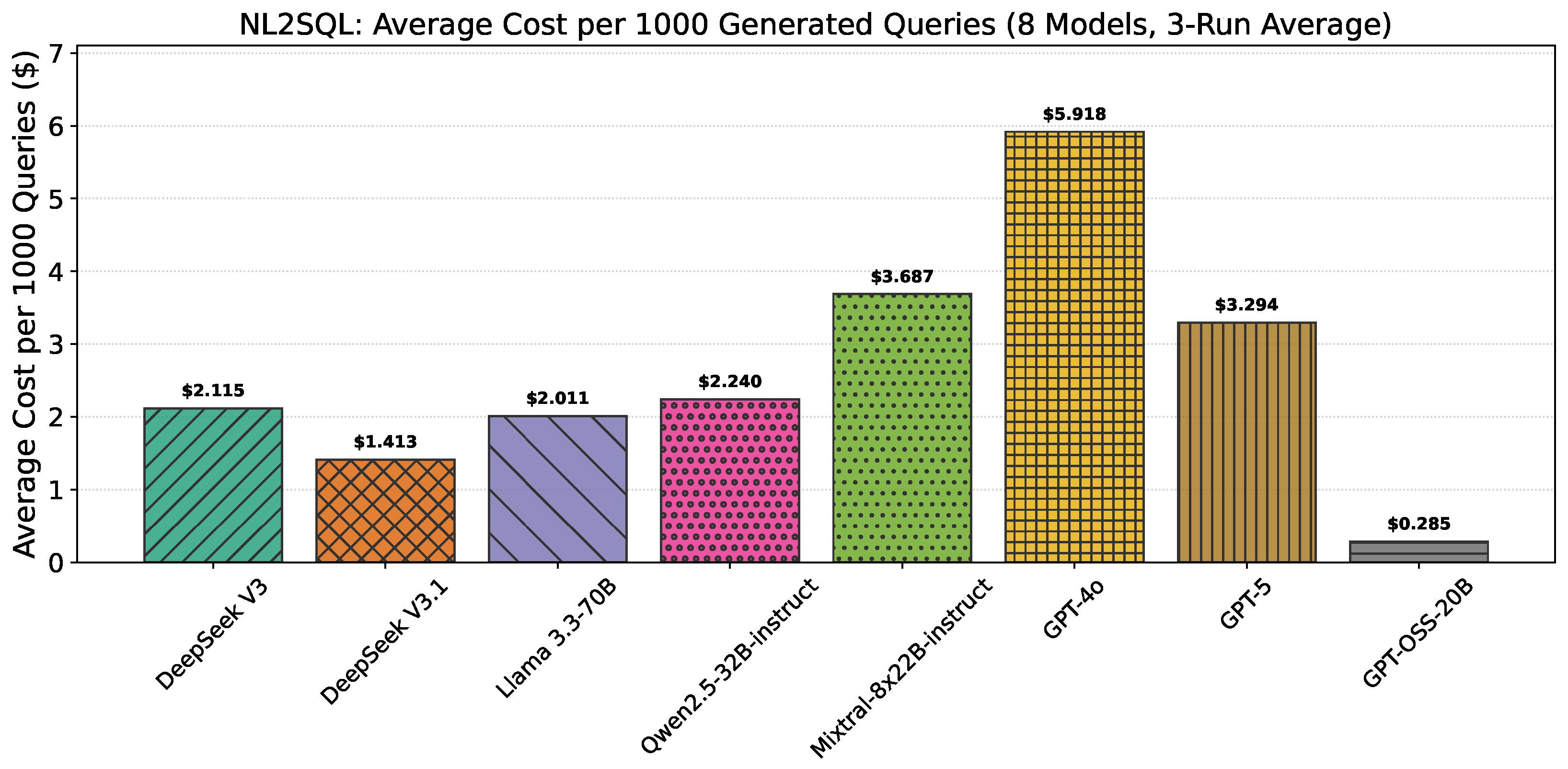
| Model | Price/1M Input Tokens ($) | Price/1M Output Tokens ($) | Avg. Price per Query ($) |
|---|---|---|---|
| DeepSeek V3 | 0.90 | 0.90 | 0.002115 |
| DeepSeek V3.1 | 0.56 | 1.68 | 0.001413 |
| Llama 3.3-70B | 0.90 | 0.90 | 0.002011 |
| Mixtral-8x22B-Instruct | 1.20 | 1.20 | 0.003687 |
| Qwen2.5-VL-32B-Instruct | 0.90 | 0.90 | 0.002240 |
| GPT-4o | 2.50 | 10.00 | 0.005918 |
| GPT-5 | 1.25 | 10.00 | 0.003294 |
| GPT-OSS-20B | 0.10 | 0.50 | 0.000285 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blašković, L.; Tanković, N.; Lorencin, I.; Baressi Šegota, S. Robust Clinical Querying with Local LLMs: Lexical Challenges in NL2SQL and Retrieval-Augmented QA on EHRs. Big Data Cogn. Comput. 2025, 9, 256. https://doi.org/10.3390/bdcc9100256
Blašković L, Tanković N, Lorencin I, Baressi Šegota S. Robust Clinical Querying with Local LLMs: Lexical Challenges in NL2SQL and Retrieval-Augmented QA on EHRs. Big Data and Cognitive Computing. 2025; 9(10):256. https://doi.org/10.3390/bdcc9100256
Chicago/Turabian StyleBlašković, Luka, Nikola Tanković, Ivan Lorencin, and Sandi Baressi Šegota. 2025. "Robust Clinical Querying with Local LLMs: Lexical Challenges in NL2SQL and Retrieval-Augmented QA on EHRs" Big Data and Cognitive Computing 9, no. 10: 256. https://doi.org/10.3390/bdcc9100256
APA StyleBlašković, L., Tanković, N., Lorencin, I., & Baressi Šegota, S. (2025). Robust Clinical Querying with Local LLMs: Lexical Challenges in NL2SQL and Retrieval-Augmented QA on EHRs. Big Data and Cognitive Computing, 9(10), 256. https://doi.org/10.3390/bdcc9100256








