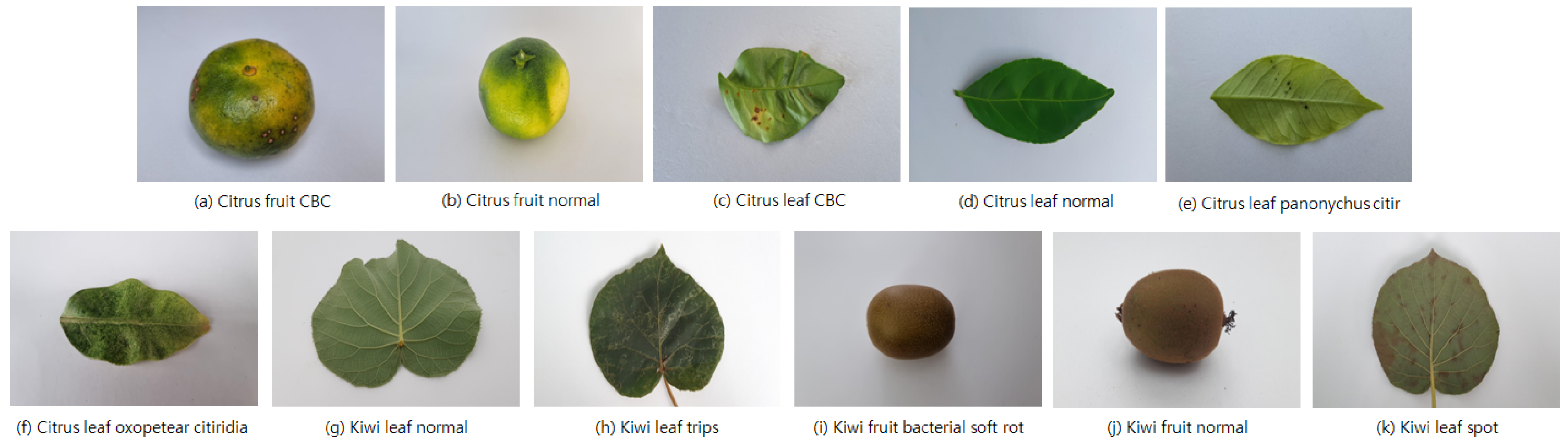
1. Introduction
Sustainable agricultural ecosystems that promote economic growth and environmental protection require a paradigm shift toward green-technology-based approaches. In this regard, the integration of ICT with AI plays a pivotal role in advancing sustainable green growth within agricultural systems [
1,
2]. Specifically, AI-driven methods for crop pest and disease detection can automate pest management in agricultural ecosystems and significantly reduce the usage of chemical pesticides, thus preventing environmental pollution and enhancing resource efficiency. According to the 21st Century Guidebook to Fungi [
3], approximately 16% of global crops are afflicted by pests and diseases, with most agricultural areas currently relying on pesticides for pest control. Moreover, a study published in Nature Geoscience [
4] reported that the 92 chemical substances found in pesticides used across 168 countries have contaminated 64% of the agricultural land. Notably, the countries with the largest shares of contaminated land are those considered the breadbaskets of Asia, which are responsible for a substantial portion of the world’s food supply.
It is crucial to reduce the use of traditional pesticides and apply machine learning technologies capable of automatically recognizing and analyzing pest patterns to resolve these issues. Machine learning, particularly deep learning, is an example of radical digital innovation in that it enables a shift from the fixed generation patterns of power plants, originally designed to supply base load power, to more flexible generation patterns [
5]. With the increase in computing resources, research on pest and disease recognition using deep learning is being actively conducted [
6]. Deep learning models automatically extract features from images during the training process, thus achieving high performance, necessitating large datasets. However, most current research has involved small datasets and been limited to a few types of crops infected by pests. Moreover, rather than effectively extracting various pest features, the focus has predominantly been on creating models capable of classifying three to five diseases in the same crop species. Particularly in Asia, crops affected by pests display a range of symptoms such as browning, spotting, and fine-thread formations; yet, research accurately identifying and recognizing these characteristics remains inadequate. While deep learning models optimized for maximum performance can achieve high accuracy for up to 10 types of diseases, they struggle to maintain this performance for a more extensive array of disease types [
7].
In this paper, we propose a data augmentation method that enables deep learning models to effectively extract patterns of pests and diseases, thereby addressing these issues and enabling the development of sustainable green technologies. We compared and evaluated six different deep learning models, including convolution and transformer models, for the recognition of 24 diseases across five distinct crops, creating a comprehensive pest and disease classification model. The data used in these models were augmented with over 60,000 new images, combining publicly available data from PlantVillage [
8] with data on citrus and kiwi varieties collected in Asia. Our comprehensive pest and disease classification model was utilized in meticulous experiments with data augmentation techniques, subdivided into detailed categories of geometric transformations and color space transformations. The results demonstrate that considering the color distribution is crucial as the diversity in data patterns increases, as evidenced by the experimental outcomes and feature maps.
The contributions of this work are threefold:
We collected private data on citrus and kiwi varieties and enhanced the validity of our experimental results by including the PlantVillage dataset, a public dataset. The constructed dataset comprises a total of 60,165 images, representing a large-scale dataset; however, the classes are imbalanced. We addressed the data bias problem by employing stratified cross-validation for verification.
The data used in the experiments included 24 diseases across five types of crops. Some of these diseases are common to different crops or are the same disease affecting multiple crops. We developed a data augmentation method combining geometric and color space transformations designed to enable models to efficiently extract data patterns even for diseases across different domains.
To validate the performance of the data augmentation methods, we compared and evaluated six deep learning models, including convolution-based and transformer-based models. The experimental results confirmed the prominence of the disease patterns in the data through feature maps, emphasizing the importance of color distribution.
The remainder of this paper is organized as follows:
Section 2 provides a brief review of the related works.
Section 3 discusses the data acquisition methods, preprocessing steps, and data augmentation techniques used in this study.
Section 4 introduces the proposed network architecture for crop disease classification, while
Section 5 presents the corresponding experimental results. In the final two sections, we present the visualizations of the feature maps based on the experimental results and discuss potential future research directions.
4. Network Architecture
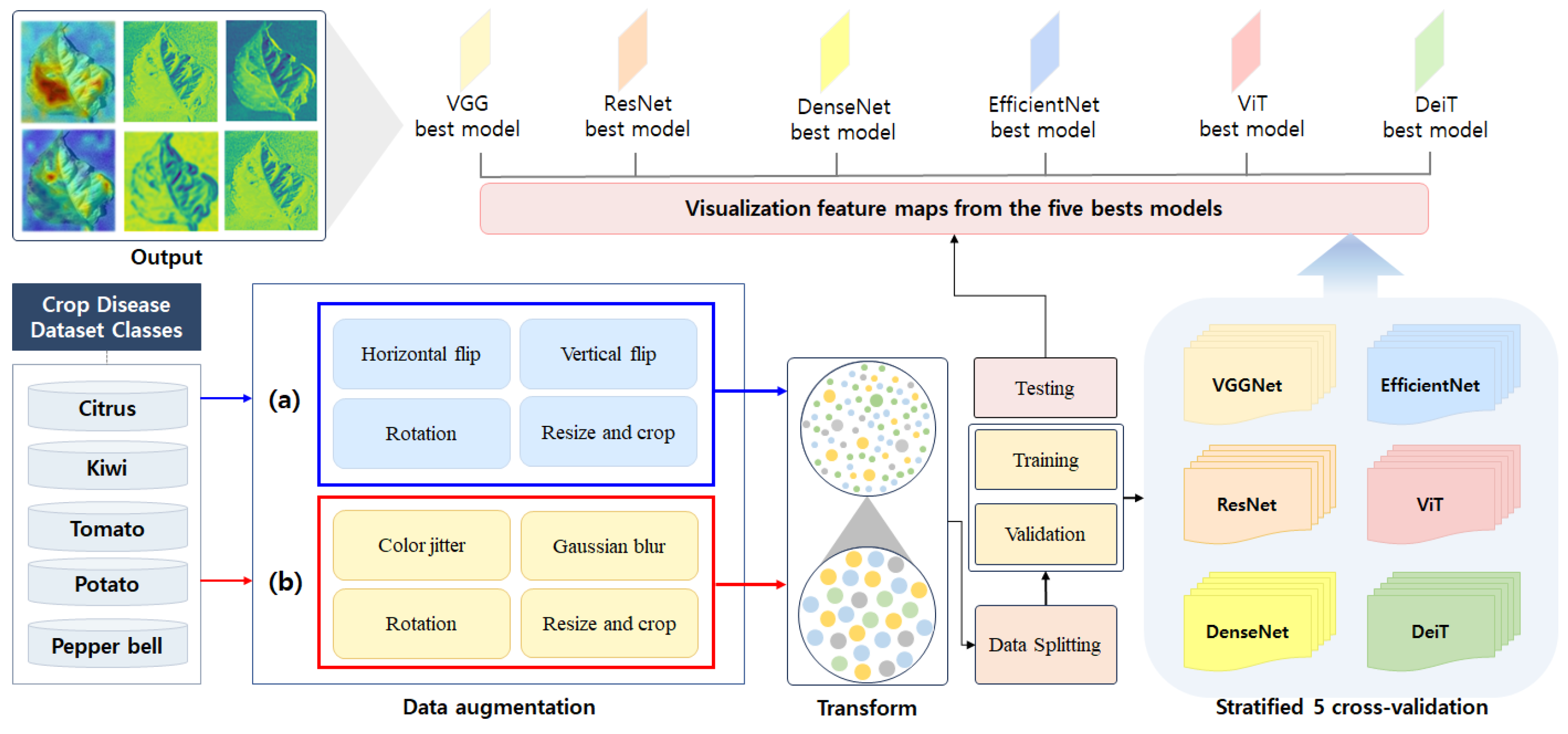
This section describes the network architecture for crop disease recognition proposed in this paper. This network architecture saves the weights of the model that yield the highest performance and uses them for model testing and feature map visualization. The network architecture is depicted in
Figure 6. It is divided into three main sections: data transformation, model training, and model testing with feature map visualization. During the data transformation process, it is essential to standardize the sizes of the images originating from different sources. The images in the public dataset are formatted at
pixels, while those in the private dataset are
pixels in size. Therefore, after resizing the input images to
, we conducted experiments by dividing the data augmentation methods into strategy (a) and strategy (b) based on the disease patterns in the images. The methods employed for data augmentation included noise removal, color transformations, and geometric transformations. The criteria for selecting strategies (a) and (b) are explained in detail in
Section 3.
In the model training stage, the network was trained using the VGGNet, ResNet, DenseNet, EfficientNet, ViT, and DeiT models pretrained on the ImageNet dataset. As the pretrained models had 1000 output nodes, this study modified the model architecture by removing the existing output layer of each pretrained model and replacing it with 24 new output layers to match the constructed dataset. Furthermore, the model training followed a fine-tuning approach in which the pretrained model’s architectures were utilized and trained with the new dataset. Training and validation were conducted using stratified k-fold cross-validation, considering the data distribution. Stratified k-fold cross-validation helped alleviate the dataset imbalance issue. The weights of the model that achieved the highest F1 score during the training process were validated using the test dataset and utilized to extract feature maps. The experimental results of the network structure are explained in
Section 5. The workflow of the network architecture is summarized in Algorithm 1.
| Algorithm 1 Network architecture. |
Input: Crop Classification Data x = input_data() x = data_augmentation(x) k_fold = initialize_k_fold() epochs = initialize_epochs() for fold in do train_data = stratified_xth_fold_train_data(x, fold) validation_data = stratified_xth_fold_validation_data(x, fold) models = load_model() for model in models do max_f1_score = 0 for epoch in do train_model(model, train_data) f1_score = validate_model(model, validation_data) if max_f1_score < f1_score then max_f1_score = f1_score end if end for end for save_states(model) show_feature_map(model) end for
|
VGGNet [
19], proposed by the Oxford University research team, highlights the critical role of network depth in improving CNN performance. By significantly deepening the architecture, VGGNet reduced the error rate from 16.4% to 7.3% in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) [
34]. A key innovation of VGGNet is the consistent use of a
filter size across all convolutional layers, optimizing computational efficiency and reducing the parameter space, enabling deeper and more expressive architectures. VGGNet16 consists of 13 convolutional layers and 3 fully connected layers, utilizing ReLU activation to accelerate training by addressing saturation issues. Dropout mitigates overfitting, and the final softmax layer outputs a probability distribution. These features showcase VGGNet’s ability to capture hierarchical representations while maintaining computational efficiency.
ResNet [
20] addresses the gradient vanishing and exploding problems in deep neural networks by introducing the residual block, which leverages skip connections to directly incorporate the input
x into the output. This reformulates the optimization objective to minimize the residual function
, preserving stable gradient flow during backpropagation and mitigating the vanishing gradient problem. Unlike plain architectures such as CNN, AlexNet [
35], and VGGNet, which degrade as depth increases, ResNet enables the construction of much deeper networks, reaching up to 152 layers, without performance loss due to its residual learning paradigm. Additionally, ResNet employs a bottleneck design with
convolutional layers to enhance computational efficiency while maintaining strong representational capacity. These innovations allow ResNet to surpass models like VGGNet and GoogleNet [
36] in both efficiency and predictive performance, verifying its impact on deep learning architecture design.
DenseNet [
21] is a neural network architecture that connects all layers directly via concatenation, enabling each layer to access the feature maps of all preceding layers. This design achieves superior performance with fewer parameters than ResNet. Unlike ResNet, which uses skip connections through element-wise addition, DenseNet employs concatenation, progressively increasing feature channels as layers are added. To manage this growth, DenseNet reduces the number of channels per layer and standardizes feature map dimensions for efficient concatenation. The architecture uses dense blocks to facilitate feature reuse and pooling operations. Bottleneck layers further enhance efficiency by limiting inputs to the
convolutional layers to
, where each layer generates
k feature maps. DenseNet supports 121-, 169-, 201-, and 264-layer configurations, providing deeper networks with greater parameter efficiency than ResNet.
EfficientNet [
22] is a state-of-the-art architecture for image classification that optimizes the balance between network depth, width, and input resolution to improve performance. Traditional models like VGGNet, GoogleNet, ResNet, and DenseNet primarily focus on increasing depth, with manual adjustments to width and resolution based on computational constraints. This heuristic approach often overlooks the interdependence of these dimensions. EfficientNet addresses this limitation by introducing a compound scaling method that systematically balances depth, width, and resolution, preventing the performance saturation observed in independent scaling. By using constants determined through grid search and user-defined computation budgets, EfficientNet scales performance proportionally to resources. This design allows EfficientNet to extract salient image features efficiently, maintaining parameter efficiency and enabling faster inference compared to earlier architectures.
ViT [
23] is a paradigm-shifting model that extends the transformer architecture, originally developed for natural language processing, to computer vision tasks. Departing from traditional CNN-based architectures, ViT employs transformers to overcome the limitations of conventional attention mechanisms, achieving state-of-the-art performance with modest computational overhead. The training pipeline involves segmenting an image into fixed-size patches, which are linearly embedded with positional encodings and fed into the transformer encoder. Since transformers operate on 1D sequences, the flattened patches are projected into a sequential representation suitable for processing. The transformer encoder’s output is passed through a multilayer perceptron (MLP) head for image classification. ViT demonstrates exceptional computational efficiency and scalability, achieving superior performance on large-scale datasets without degradation or saturation, and can handle up to 100 billion parameters. However, its reliance on extensive pretraining with large datasets remains a significant constraint.
DeiT [
24] is a model proposed by Facebook AI that enhances the efficiency of ViT by significantly reducing data and computational requirements while achieving comparable accuracy. In contrast to ViT, which necessitates pretraining on extensive datasets like JFT-300M, DeiT attains state-of-the-art performance using only the ImageNet dataset, with training completed in three days on a single 8 GPU setup. DeiT leverages hard-label knowledge distillation, transferring informative representations from a CNN teacher model to imbue the transformer with inductive bias, thereby improving generalization and performance. In hard-label distillation, the model minimizes the cross-entropy loss as
where
represents the label predicted by the teacher model, and
is the predicted probability of the student model for class
i. This approach avoids the use of temperature scaling and additional hyperparameters, making it computationally efficient. Furthermore, DeiT incorporates a distillation token, [DIST], analogous to the class token in ViT, which interacts with other embeddings via self-attention. The output of the distillation token is jointly optimized with the ground truth labels through the combined loss function:
where
balances the contributions of the distillation and ground truth losses. This methodology establishes DeiT as a computationally efficient and data-effective transformer-based architecture for image classification tasks, overcoming the heavy reliance on large-scale datasets and high-specification hardware required by ViT.
5. Experiments
In this section, we present empirical evidence regarding the performance of the crop disease recognition network, which is based on the combination of data augmentation methods proposed in this study. Specifically, we report the experimental results, and we analyzed the feature maps of the proposed model. The experiments in this study were conducted on a PC with an Intel® Core™ i9-9900KF CPU @ 3.60 GHz, NVIDIA TITAN RTX, and Windows 10, using the Python 3.8 environment to validate the performance of the DeiT model. The remaining models were evaluated on a PC with an Intel® Xeon® Silver 4208 CPU @ 2.10 GHz, NVIDIA TESLA V100 32 GB, and Ubuntu 18.04.6 LTS, using the Python 3.10 environment to assess the performance of the proposed network.
5.1. Experimental Settings
The crop disease classification model utilized six pretrained deep learning models: VGGNet, ResNet, DenseNet, EfficientNet, ViT, and DeiT. The input sizes of the models varied depending on their size and type. To examine the performance differences of the combined data augmentation methods in the same environment, the input size for all six models was standardized to
. The images, resized to
pixels as described in
Section 5, underwent data preprocessing based on strategies (a) and (b). The test dataset, which was used for model validation, underwent only resizing, tensor conversion, and normalization without data augmentation for image transformation.
The training and validation datasets were divided into five folds per class for cross-validation, with the model trained on the corresponding fold for each class. The crop pest and disease classification model was trained and validated for 100 epochs per fold. Model performance was evaluated using four metrics commonly applied in classification tasks: accuracy, recall, precision, and F1 score. Recall measures the proportion of correctly predicted positive instances among all actual positives, while precision evaluates the proportion of correctly classified positive predictions among all instances predicted as positive. Accuracy represents the ratio of correctly classified instances to the total instances. The F1 score, the harmonic mean of recall and precision, is particularly useful for handling imbalanced datasets. These metrics were calculated at each epoch to assess model performance comprehensively.
Equations (3)–(6) provide the equations for these metrics. True positive (
TP) refers to the number of correctly identified positive cases, false negative (
FN) is the number of positive cases that were incorrectly identified as negative, false positive (
FP) is the number of negative cases that were incorrectly identified as positive, and true negative (
TN) refers to the number of correctly identified negative cases. The experiments employed cross-entropy loss [
37] as the objective function, which was optimized using the Adam optimizer [
38]. The learning rate was also dynamically adjusted using CosineAnnealingLR [
39] to guide the model toward an optimal solution.
5.2. Results on Validation Dataset
The performance evaluation results showed that both strategy (a), which used only geometric transformation data augmentation, and strategy (b), which combined geometric transformation and color space transformation data augmentation, achieved F1 scores of over 95% fpr all six models.
Table 5 presents the performance results of the models using strategies (a) and (b). Both strategies demonstrated high performance in classifying the 24 classes. However, as shown in
Table 5, under the same conditions, strategy (b) showed a maximum F1 score difference of over 3%, and, except for the VGGNet and DeiT models, all models achieved an F1 score of over 98%. As shown in strategy (a), the top three performing models among the six were DenseNet, EfficientNet, and ViT. All three models achieved an F1 score of over 97%, with the ViT model achieving the highest F1 score of 97.68% and a low standard deviation across the five folds.
For strategy (b), except for the VGGNet and DeiT models, the remaining four models all achieved F1 scores of over 98%. Furthermore, excluding the DeiT model, all models achieved accuracies of over 99% and showed lower standard deviations across the five folds compared to the results for strategy (a), indicating a more uniform distribution. This suggests that a combination of geometric transformation and color space transformation data augmentation is effective for both convolution-based and transformer-based models. However, it is worth noting that the training times of strategy (b), which incorporated both data augmentation techniques, were longer than those of strategy (a). An interesting observation was that the DeiT model, which combines the convolution and transformer model architectures, showed minimal improvement with data augmentation. The DeiT model achieved F1 scores of 95.54% (strategy (a)) and 95.90% (strategy (b)). Despite being a distillation model designed to transfer knowledge from a teacher model to a student network for optimal performance, the combination of convolution and transformer model structures did not align well with the agricultural dataset and had the least impact on the effectiveness of the data augmentation techniques.
5.3. Results on Test Dataset
The test performance of the models revealed that strategy (b), which combined geometric and color space transformation data augmentation, allowed the models to recognize agricultural disease patterns more prominently than strategy (a), which only used geometric transformation data augmentation.
Table 6 presents the test results obtained using the models with the highest performances. As observed in
Table 6, when strategy (a) was used, the performance was similar to or slightly lower than the cross-validation results. However, when strategy (b) was used, the performance improved compared to the cross-validation results, and all six models achieved an F1 score of 98%. Therefore, when constructing a crop disease classification network, it is important to analyze the disease patterns, which can vary depending on the type of disease, and consider the corresponding color distribution to enhance the model’s performance.
6. Visualization Feature Maps
To better understand the six models and crop diseases, we loaded the weights of the model that achieved the highest F1 score according to
Table 5 and visualized the feature maps. The feature maps represent the process of extracting patterns as the models pass through the layers and capture the characteristics of crop diseases. By examining the feature maps, we saw how the models perceived the features of agricultural pests and diseases. The red bounding box highlights regions within the feature map where the disease object is prominently represented, providing an analytical visualization of its salient characteristics.
Figure 7 shows the images from which the feature maps were extracted using the VGGNet and ResNet models. The VGGNet model appears to focus on the edges of the leaves as it progresses through the convolution layers. Additionally, since VGGNet employs only 16 layers, its feature maps maintain the shape of the original image even after passing through the convolutions, unlike those of the other models. In contrast, the ResNet model emphasizes the bottom of the leaves to locate the disease. The first right row in
Figure 7 shows images from the higher layers of the ResNet model. In all three images, the disease is observable in the exact location. Although the VGGNet and ResNet models identified the disease in different locations, both accurately recognized the objects associated with the disease.
Figure 8 shows the images from which the feature maps were extracted using the DenseNet model. Unlike the previous two models, the DenseNet model detects the disease in the center of the leaves. It can be observed that the DenseNet model consistently maintains the recognition of disease patterns as it passes through the dense blocks without losing them. Similar to DenseNet, the EfficientNet model recognizes the disease in the exact location. The EfficientNet model appears to have uniform intensity in the images and detects the brightness of the background more rapidly than the previous three models.
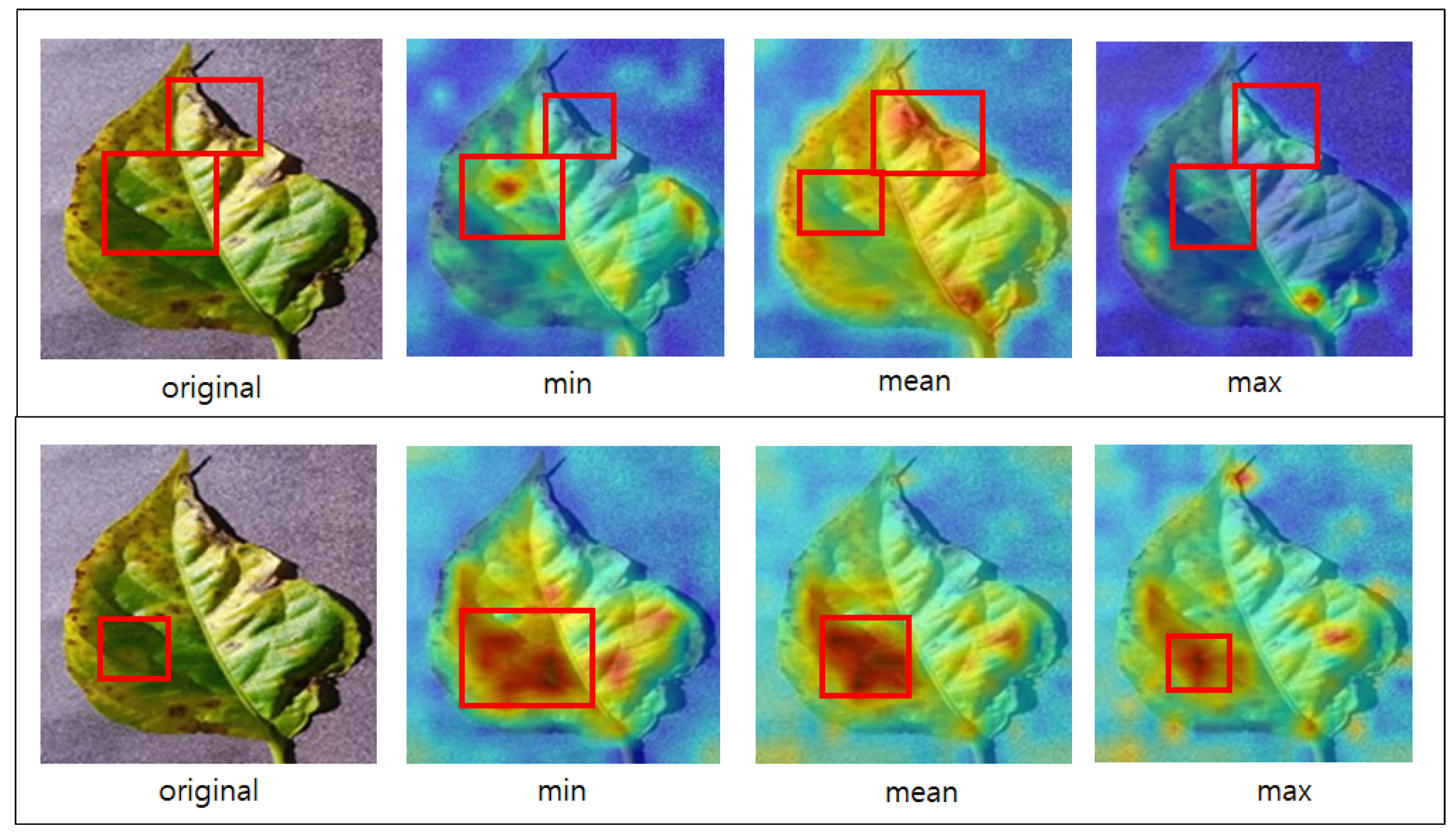
Figure 9 shows an image from which the attention map was extracted using the ViT model and Deit, specifically the multihead attention’s minimum (min), mean, and maximum (max) values. The above attention map corresponds to the ViT model, and the one below corresponds to the DeiT model. When the mean value is emphasized, each head focuses on a different position, allowing the model to recognize diseases at the edges of the image. However, the minimum and maximum values are concentrated in the localized areas of the image. The attention map of DeiT exhibits a pattern different from that of ViT. ViT’s attention map shows a wide distribution when emphasizing the mean value but that of DeiT shows variations in distribution based on the minimum, mean, and maximum values but still focuses on common areas. By visually examining the image, the model may seem to focus on normal leaf regions rather than diseased parts. However, upon closer examination of the attention maps for mean and maximum values, it becomes apparent that the model recognizes the diseases.
7. Discussion and Conclusions
In this section, we compare the proposed approach with prior methods using the same crop disease data from the PlantVillage dataset employed in our experiments.
Table 7 presents a comparative analysis between our method and prior methods leveraging the same dataset. As shown in
Table 7, ML-based models exhibited a notable decline in performance on identical crop data, while DL-based models demonstrated performance comparable to or marginally superior to our results. However, our study employed a unified model capable of addressing multiple crop types simultaneously, unlike prior studies that optimized distinct models for individual crops. Naturally, such crop-specific models achieved higher performance. Moreover, while the PlantVillage dataset includes only leaf images, our study incorporated both leaf and fruit images, which introduced additional complexity but enhanced the model’s generalizability. If separate classification tasks had been conducted for leaf and fruit images, our model’s performance would likely have improved further. Despite these challenges, our model achieved nearly 99% accuracy, underscoring its effectiveness across diverse data types.
This study analyzed data augmentation techniques to develop a method that enables deep learning models to perform disease diagnosis efficiently. A novel dataset comprising 24 classes significantly enhanced the model’s generalization capability across diverse crop types. The integration of geometric transformations and color space modifications resulted in deep learning architectures, including VGGNet, ResNet, DenseNet, EfficientNet, ViT, and DeiT, achieving F1 scores exceeding 98%. Furthermore, our approach emphasizes the potential for reducing energy consumption and carbon emissions by employing a single model for multiple crop types, contributing to sustainable agriculture through scalable disease detection methods. However, reliance on image data alone imposes limitations on broader applicability. To address this, future research will explore integrating image and text data to develop multimodal classification systems, further enhancing robustness and versatility.