1. Introduction
In the era of big data and data-driven research and innovation, the availability of real-world data (RWD) holds immense potential for advancing various domains, particularly healthcare-related data analytics paradigms. In this sense, Living Labs (LLs) have become key for integrating research and innovation processes into real-life environments and digital health applications. As those ecosystems are especially focused on people and their participation in research procedures, the data collected and stored there is naturally generated by real subjects in their daily lives. Thus, Living Labs play an important role in providing valuable insights into human behaviour, physiological patterns, and environmental interactions. However, the inherent sensitivity and privacy concerns related to the General Data Protection Regulation (GDPR) [
1] and associated with the data collected in Living Labs present challenges in terms of sharing and collaborating with external researchers (ERs).
In this context, the H2020 VITALISE project, with the aim of opening Living Lab infrastructures to facilitate and promote research activities in the field of health and well-being across Europe and beyond [
2], is actively working to harmonise health and wellbeing Living Labs research procedures and services, including data capture protocols. A significant component of this project is the development of a
Shareable Data Publishing and Access Service composed of different information and communication technologies (ICT) tools. This innovative service integrates Synthetic Data Generation (SDG) technologies into Living Labs’ data management and sharing processes, ensuring compliance with privacy, ethics, and Intellectual Property Rights (IPR) legislation and policies that apply to each Living Lab infrastructure. The project aims to accelerate the creation and development of innovative and data-driven digital health products and services [
3]. Controlled access to data analysis paradigms for both industrial and scientific communities is being provided, and the computational infrastructure of Living Labs is being used to achieve this. Furthermore, integrating synthetic data generation technologies within the service enhances the capability to offer external researchers shareable (anonymised or synthetically generated) data versions, ensuring robust privacy protection.
The Shareable Data Publishing and Access Service enables Living Lab managers to securely publish collected data in an on-premises node, the VITALISE certified node (VCN). Subsequently, external researchers can access the VITALISE discovery portal (VDP) to download shareable data versions derived from stored data across different Living Labs. Whether anonymised or synthetically generated, these shareable data versions provide realistic and representative data for researchers to develop, test, and debug processing scripts, adhering to legal and ethical data handling practices. Finally, external researchers can request the remote execution of the same algorithms against the real on-premises data in Living Labs, comparing the outcomes with those obtained using shareable data, and register the obtained results with a Research Analysis Identifier (RAI) in the VITALISE RAI Server (VRS). Considering all these steps, the described VITALISE ecosystem was divided into three main components according to their functionalities:
VITALISE certified node (VCN): common information and communication technologies infrastructure tools for Living Labs to upload, capture, store, manage, and securely offer access to shareable data harmonised with the VITALISE data model [
4].
VITALISE discovery portal (VDP): enables external researchers to register on the system, explore metadata of data stored in Living Labs, register new VITALISE-certified nodes within the system, request shareable versions of such data, schedule the remote execution of data analysis scripts, access experiment results, register experiment results to the VITALISE RAI server and verify experiment results registration.
VITALISE RAI server (VRS): cloud server to register and store the remotely executed analysis results with an immutable identifier, the Research Analysis Identifier (RAI).
In this paper, we present the
Shareable Data Publishing and Access Service for Living Labs implemented within the framework of the H2020 VITALISE project. Building upon previous research on incorporating synthetic data generation techniques within a controlled data processing workflow, as presented by Hernandez et al. [
5], this paper presents an extended and complete service architecture and workflows for sharing anonymised and synthetic data from Living Labs. Additionally, the remote execution of experiments and their registration were added to the system. Overall, the described VITALISE ecosystem is intended as a coupling interface between Living Labs and external researchers to accelerate research on data analytics paradigms in digital health, ensuring compliance with data protection laws. However, the service can be extended to other domains, such as industrial processes or business.
The remainder of this paper is organised as follows:
Section 2 provides an overview of the background and related work, presenting basic concepts of anonymisation and synthetic data generation and discussing existing research in sensitive and private data publishing and sharing services;
Section 3 presents the detailed methodology followed for our service, describing the complete architecture, workflows, and techniques employed for data publishing and sharing. Next,
Section 4 offers the methodology used when implementing and validating the service with real-world usage examples and the results obtained on the evaluations, supported by relevant visualisations and analyses. Finally,
Section 5 discusses the obtained results, and concludes the work by summarising key achievements and contributions of our
Shareable Data Publishing and Access Service as a catalyst for innovation in the digital health domain.
2. Background and Related Work
This section provides a comprehensive overview of the background and related research that lays the foundation for our Shareable Data Publishing and Access Service for LLs. Firstly, we present some basic concepts concerning anonymisation and SDG. Then, existing research in sensitive and private data publishing and sharing services is discussed.
2.1. Basis of Anonymisation and Synthetic Data Generation
This subsection gathers basic information about data anonymisation and SDG, along with a brief overview of the state of the art of the most recent and relevant technologies researched for the VITALISE
Shareable Data Publishing and Access Service. Our research into data anonymisation and SDG for this service is motivated by the imperative of LLs to share real data in compliance with the GDPR [
1]. The GDPR provides guidelines to carefully manage and process personal data throughout a research process, considering that traceable personal data cannot be published without explicit participant consent. Given this requirement, which may result in restricted data access for ERs, we propose anonymisation and SDG techniques as possible solutions. These techniques ensure that LL data can be shared with ERs while adhering to legal mandates and mitigating specific access constraints.
2.1.1. Data Anonymisation
Anonymisation is the process of irreversibly altering personal data so it can no longer be directly or indirectly identified [
6]. Among the most used anonymisation techniques, the following four can be found:
Randomisation consists of modifying personal data attributes using predetermined random patterns. This technique maintains variable correlations and is suitable for cases requiring realistic anonymised values and unaltered data models, often seen in software testing.
Permutation involves swapping values between subjects but does not fully address privacy concerns with new external data. While this technique retains some information from the real data, it distorts certain parameters, potentially posing challenges in interpretability and utility.
Generalisation replaces specific datapoints with broader, generic values, thereby providing a less detailed version of the same information. The widely used k-anonymity algorithm clusters data attributes into groups of at least k subjects, establishing a range of values to replace the original datapoints. This technique ensures that accessing any group of subjects requires involving at least k-1 other subjects, enhancing privacy protection.
Pseudonymisation removes personally identifiable information like social security numbers, names, and national identity numbers, replacing them with tokenised IDs, pseudonyms, or hashed values. However, with this technique, attackers can gain substantial information if they gain access to real datasets.
2.1.2. Synthetic Data Generation
SDG technologies have emerged as a robust solution to address many challenges in data privacy, compliance with regulations like the GDPR and limitations inherent in traditional anonymisation methods. The European Data Protection Supervisor defines Synthetic Data (SD) as artificial data derived from original datasets through models trained to replicate the underlying characteristics and structural patterns [
7].
The origins of SDG can be traced back to D. Rubin’s pioneering work in 1993 [
8], which laid the foundation for subsequent advancements, including the seminal creation of Generative Adversarial Networks (GANs) by Goodfellow et al. in 2014 [
9]. Since then, SDG has been widely used to preserve data privacy, ensuring a secure data exchange across diverse domains, encompassing signals, images, and tabular data by different authors [
10,
11,
12,
13,
14,
15,
16,
17,
18]. In all these studies, they demonstrated the potential and improvement of SDG over traditional anonymisation methods. SDG yields highly realistic and representative data and reduces re-identification risks while ensuring superior machine learning performance.
SDG approaches are being extended into biomedical applications, underscoring their relevance in the healthcare sector. Nowadays, SDG is used for the generation of medical images [
18], Electronic Health Records (EHR) synthesis [
13,
14] and the generation of time series data integrated with subject-related metadata [
19]. As highlighted by Hernandez et al., prevalent SDG techniques within the digital health domain encompass classical statistical and probabilistic models, deep learning approaches such as GANs and autoencoders, and composite methods involving multiple sequential steps or modules [
20].
This evolution of SDG methodologies accentuates their increasing adoption in addressing privacy concerns and enabling novel data-driven innovations across diverse domains, especially within the rapidly expanding digital health realm.
2.2. Sensitive and Private Data Publishing and Sharing
This section provides a concise overview of the state of the art on sensitive and private data publishing and sharing, a critical domain that has significantly influenced the development of our Shareable Data Publishing and Access Service for LLs. Our research into this topic for this service is driven by the importance of safeguarding LLs data privacy and security while facilitating access for ERs.
Different approaches can be found in the literature for publishing and sharing sensitive and private data. Many of these heavily rely on privacy-preserving technologies. For instance, Rankin et al. [
13] and Hernandez et al. [
5] have introduced workflows and pipelines incorporating SDG approaches. These methodologies enable the creation of highly realistic shareable data versions suitable for sharing with ERs. Additional studies have proposed conceptual frameworks employing diverse privacy techniques, including data anonymisation [
21], differential privacy [
22], federated learning [
12], blockchain networks [
23], or even a fusion of these technologies [
24]. Some researchers have explored user access control and authorisation mechanisms to facilitate queries for shareable data within healthcare organisations [
25].
So far, research in sensitive and private data publishing and sharing in the context of health and well-being has primarily been limited to creating and obtaining shareable data versions of private data. However, the focus has largely been on research activities rather than providing ER platforms or services for downloading shareable data (anonymised or synthetic) derived from real-world data for executing analysis locally and then executing those analyses remotely and in a controlled environment with real-world data. This represents a notable gap in enabling and expediting research in the digital health domain, particularly in the context of data-driven analysis paradigms.
3. Materials and Methods
Using our Shareable Data Publishing and Access Service, LLs managers can securely host data within a VCN. Then, ERs can access the VDP to obtain shareable data versions, whether anonymised or synthetically generated from various LLs. These shareable datasets offer realistic and representative information, enabling researchers to develop and test processing scripts while adhering to legal and ethical data handling standards. Furthermore, ERs can request remote execution of algorithms on the actual protected datasets hosted in LLs’ nodes, allowing for comparisons with results obtained using shareable data and using the system to register the results.
This section comprehensively describes the methodology employed in designing, developing, and implementing our Shareable Data Publishing and Access Service for LLs developed within the context of the H2020 VITALISE project. In the following subsections, we delve into pivotal aspects of our methodology, presenting the overall architecture of the service, presenting the VITALISE data model, describing the workflows for data publishing and sharing, remote execution and experiments registration, and exploring the techniques and logic applied to the service.
3.1. Service Architecture
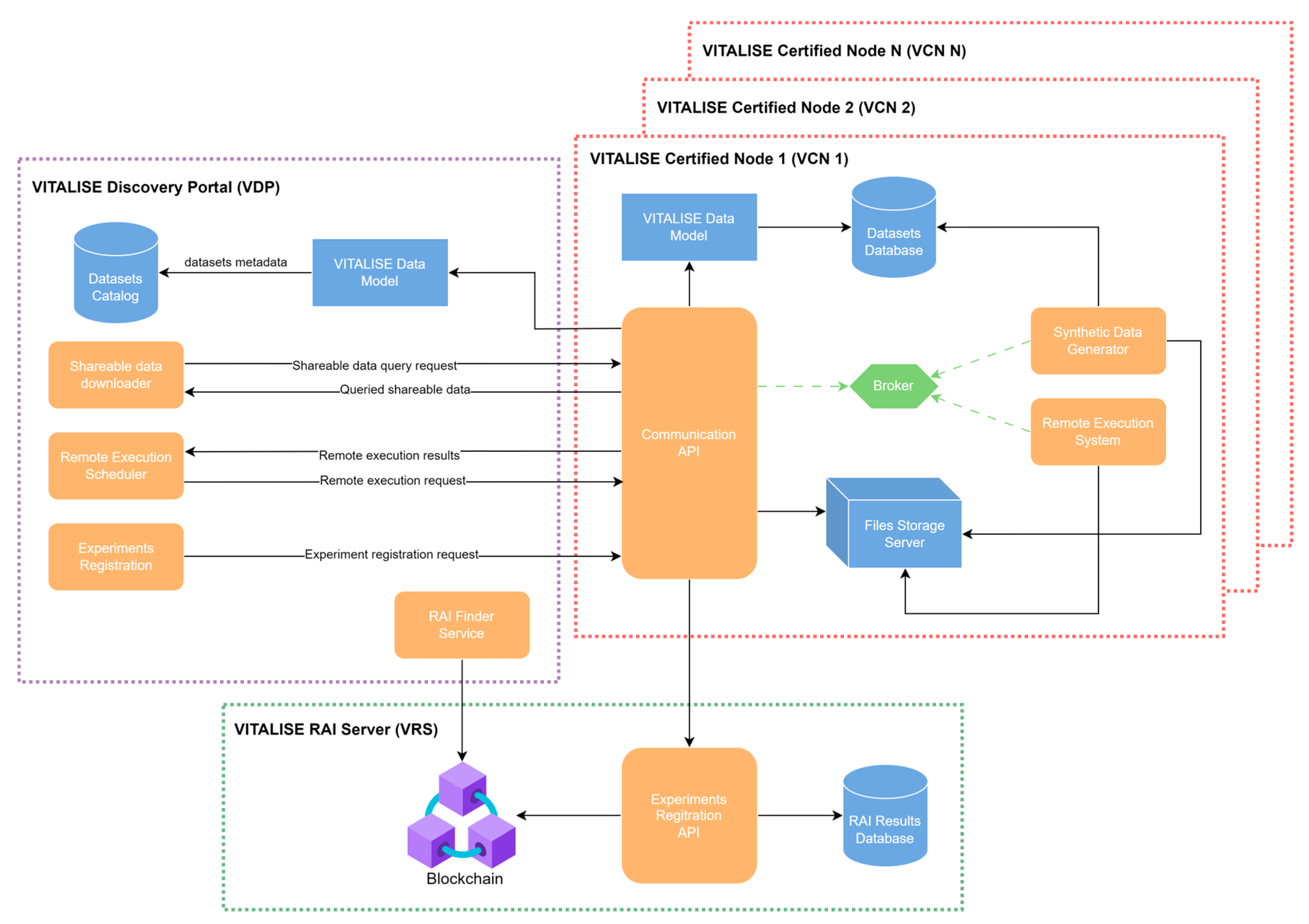
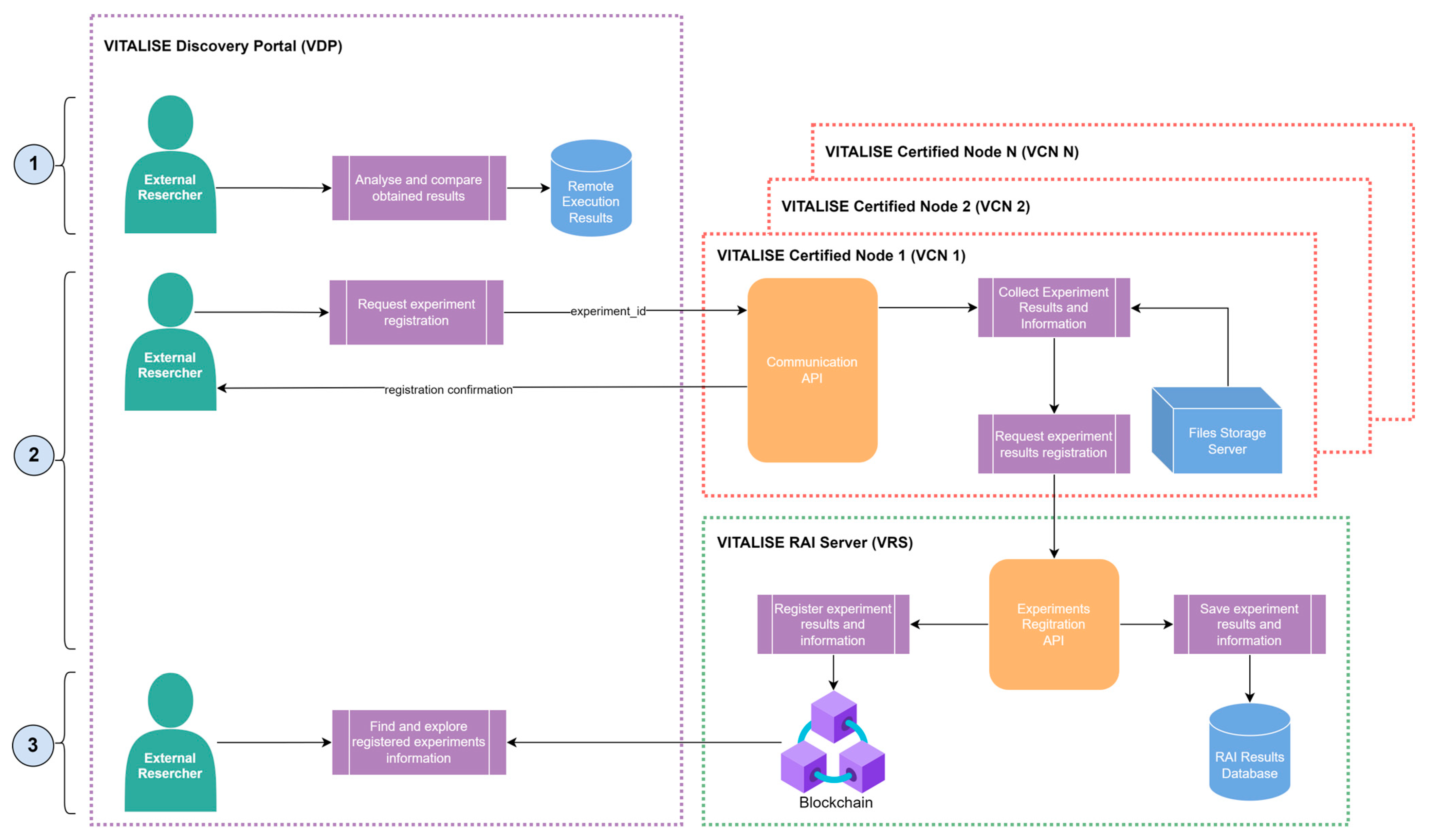
Figure 1 illustrates the architecture of our
Shareable Data Publishing and Access Service, showcasing its various interconnected modules. Both main modules (VDP and VCN) were individually designed, developed, and tested before integration and deployment. The designed architecture supports multiple VCNs, which are individually installed in each LL.
The VDP serves as the central gateway for ERs to interact with the other modules of the service, establishing communication channels with the Communication API of each VCN. This module provides ER access to the metadata and datasets summary catalogue compliant with the VITALISE data model [
4]. Additionally, it offers interfaces for ERs to request shareable data versions stored within the LLs and schedule remote data analysis tasks.
Each VCN is installed and deployed within its respective LL, providing essential ICT tools for data storage, processing, and remote analysis execution. The ER initiates tasks through interactions with the VDP. The correspondent VCN receives these requests, executes tasks in a controlled environment, and sends task status updates to the VDP. The VCN also enables LL managers to register their LL and upload data to their VCN.
In the VRS, all information regarding the registered remotely executed analyses is stored under an immutable identifier (RAI). The module receives the experiment registration request through the Communication API of the VCN, and the information registered is hashed and written into a blockchain network. This way, the ER can make requests using the VDP to query all registered experiments and find information about them.
The following subsections provide detailed technical and architectural explanations of each module, starting with the VDP, continuing with the VCN and finishing with the VRS.
3.1.1. VITALISE Discovery Portal
The VDP is an operational and scalable web application, serving as the interface for ERs to interact with our Shareable Data Publishing and Access Service. The purpose of the VDP is to provide an effective environment, which, on the one hand, acts as an intermediary between the ER and the available VCNs and on the other hand offers the user dataset search and discovery possibilities through the metadata of the LL data.
The VDP offers a user-friendly web interface for ERs, enabling them to initiate procedures available in the VCNs. Functions accessible via the VDP include:
User Registration and Login: The VDP facilitates user registration and login functionalities, enabling individuals to create personalised accounts to access the platform’s features. Once registered, ERs can log in securely to their accounts, ensuring privacy and personalised experiences.
Access to User Profile: Upon logging in, ERs gain access to their individual profiles within the platform, where they can view activity history, such as Data and Experiment Requests and their status. This feature enhances user engagement and enables seamless interaction with the platform’s resources and functionalities.
VCN Registration: The system offers seamless VCN registration, facilitating the integration of additional data sources and expanding the platform’s network of contributors. VCNs can be registered within the platform to share data and be available for experiments, fostering collaboration and enriching the diversity of available metadata. This functionality promotes inclusivity and interoperability, empowering the VDP to serve as a comprehensive ecosystem for metadata discovery across living labs.
Data Discovery: ERs can access LL dataset metadata through the VDP, utilising search bars and filters to find datasets tailored to their needs. Dataset overviews (e.g., title, description, VCN host) are presented, and individual dataset summaries can be accessed for further information, including descriptive statistics and visualisations.
Request Shareable Data: After reviewing dataset metadata, ERs can request shareable versions (anonymised or synthetic) for their queries, which may involve one or multiple datasets of the same type. The VDP’s Application Programming Interface (API) manages these requests and provides the results for the ER to download from the VDP UI.
Run Experiment: ERs can initiate a remote experiment execution in the VCN, utilising data stored within the LLs, by providing experiment code, requirements, and configuration, including the IDs of the real datasets. The VDP API handles the remote execution request to the VCN, and the results can be downloaded via the VDP UI.
Register Experiment: ERs can request the registration of previously executed experiments in the VRS. The information related to the experiment is stored in the RAI Results Database of the VRS and hashed and saved in the blockchain network.
RAI Finder Service: The VDP includes a convenient link to a publicly available finder service (
https://rai-finder.iti.gr/ accessed on 26 February 2024), allowing ERs to locate experiment results registered on the VRS using unique identifiers associated with the Experiment Results. This feature enhances transparency and accessibility, enabling users to easily access and verify experiment outcomes for research, validation, and replication purposes.
These methods, including the user-friendly UI, API communications, diverse functions, and authentication strategies, entail a comprehensive solution for ER, facilitating metadata and LL dataset access, along with various functions and procedures within our Shareable Data Publishing and Access Service.
3.1.2. VITALISE-Certified Node
The VCN is responsible for (1) storing data generated in LLs, (2) providing shareable data (anonymised or synthetic) to ERs, and (3) executing analyses remotely submitted by ERs with real data stored within LL VCNs. This module, independently installed in each LL, follows a microservice architecture composed of six services that communicate with each other.
These services are bundled using containerisation (Docker) and orchestrated with docker-compose for efficient deployment and updates across different LLs. Each service and its communication are described as follows:
Datasets Database: a MongoDB NoSQL database system [
26] is used to securely store the LL data uploaded in the VCN.
Broker: this is a RabbitMQ open-source message broker [
27] used to enable communication among the services of the module and to queue tasks regarding SDG and remote analysis execution.
Files Storage Server: employs MinIO, an object storage server [
28], to store trained SDG models, generated shareable data (anonymised and synthetic) in both CSV and JSON formats, real data used for SDG, files necessary for remote experiment execution, and results produced during remote execution.
Communication API: Developed in Python using the FastAPI framework [
29], this API handles LL managers’ requests, ER interactions through the VDP and the experiment registration schedule in the VRS. It also serves as an intermediary between the Synthetic Data Generator and the Remote Execution System, facilitating task queuing and data access.
Synthetic Data Generator: A Python-based client subscribed to the Broker’s synthetic data topic, responsible for training SDG models and generating synthetic data for ER’s non-anonymised data queries. It queries available data in the datasets database, stores trained SDG models and generates synthetic data in the files storage server. It uses the SDG models provided by the open-source Synthetic Data Vault (SDV) Python package [
30,
31] to generate synthetic, thus shareable versions of non-anonymised datasets.
Remote Execution System: Developed using Celery [
32] and the Python programming language, this distributed system processes and queues tasks related to remote analysis execution with data stored in datasets databases. It accesses the Broker for task queuing and the files storage server for access to real data and required files to perform the execution of analysis and store their results.
3.1.3. VITALISE RAI Server
The VRS is the application module responsible for managing experiment data registration requests, storing them in the internal database and triggering the procedure to register them to the blockchain network. Furthermore, it allows ERs to query information related to the RAI through the VDP. This component features a blockchain network that communicates and is integrated via specific drivers. The motivation for using a blockchain network is to perform an immutable registration of the remotely executed analyses, to guarantee the reproducibility and accreditation of them, reflecting authorship, source dataset, executed algorithm or script and obtained results. It is composed of three main components that interact with each other:
Experiments Registration API: This component is the core logic of the VRS developed in NodeJS leveraging HAPIJS to provide a REST API for interaction from the VCNs and VDP, and storage request data and interaction with the Blockchain Network. Its main task is accepting requests from other services, registering the data of the related experiment to the RAI Results Database if needed, and triggering the blockchain registration process.
RAI Results Database: A MongoDB service [
26] is used for storing experiment registration requests and related data and for log persistence.
Blockchain Network: A distributed network of nodes that records data via block transactions on the distributed ledger. This network leverages blockchain technology to implement a distributed service allowing the registration of remotely executed experiments under an immutable RAI. Each experiment registration in the system generates a unique identifier, allowing subsequent verifications of its integrity via blockchain integration.
3.2. VITALISE Data Model
The
VITALISE Data Model is a versatile, standardised and extensible reference model designed for the presented architecture able to (1) represent information that is made available via the ICT tools, (2) provide additional information including descriptive statistics, (3) provide non-person identifiable user information for analysis, (4) ensure harmonisation of data exchanged among the individual components, and (5) achieve that the provided data model can be further extended with domain knowledge pertinent to the H2020 VITALISE use cases [
5].
This model adheres to the VITALISE modelling and implementation requirements, developed in collaboration with our LLs partners. Based on standards such as OmH [
33], Web Things [
34], Open Connectivity Foundations [
35], and Schema [
36] health data standards, the data model incorporates common schemas that define essential distinctions for various clinical measures (i.e., heart rate or oxygen saturation) that enhance the clinical utility of digital health data. This data model leads to the creation of new JSON schemas designed around proposed properties and relationships, including the
VITALISE Data Point model and the
VITALISE Dataset model.
Specifically, the
VITALISE Data Point model consists of two models: the Data Person and Data Point models. The
Data Person model encompasses the metadata of the subject to whom the measurements in the LLs are recorded, including birthdate and gender. The second one, the
Data Point model, represents the clinical measurements (heart rate, steps count, oxygen saturation, etc.) gathered in LLs. Each datapoint must be assigned to a dataset, but it may not necessarily be assigned to a specific person since it can represent environmental measurement. This model fully aligns with the LL requirements specified by Petsani et al. [
37].
On the other hand, the VITALISE Dataset model contains metadata of a VITALISE dataset (e.g., name, description, author) and descriptive statistics about the subjects and datapoints within the dataset. Overall, it provides essential structure and standardisation for publishing and sharing data collected in LLs. It facilitates data processing steps for LL managers and enables data processing and access for ERs, ensuring data consistency and accelerating digital health research.
3.3. Shareable Data Publishing and Access Flows
The Shareable Data Publishing and Access Service presented in this paper enables LL managers to securely publish collected data in the VCN to then make them shareable and available for the ER through the VDP. Three data flow modalities can be found in the service to provide different options for publishing and sharing data: (1) VITALISE data model-compliant anonymised data, (2) VITALISE data model-compliant non-anonymised data, and (3) non-VITALISE data model-compliant (NDM) anonymised data. Different workflows were developed for publishing and accessing each data modality to ensure that highly realistic and representative data is given to ERs with no privacy risks. In the following subsections, each data flow is described in more detail.
3.3.1. VITALISE Data Model-Compliant Anonymised Data
For publishing VITALISE data model-compliant anonymised data, LL managers must ensure prior anonymisation of their data using their preferred technique (randomisation, permutation, generalisation, or pseudonymisation).
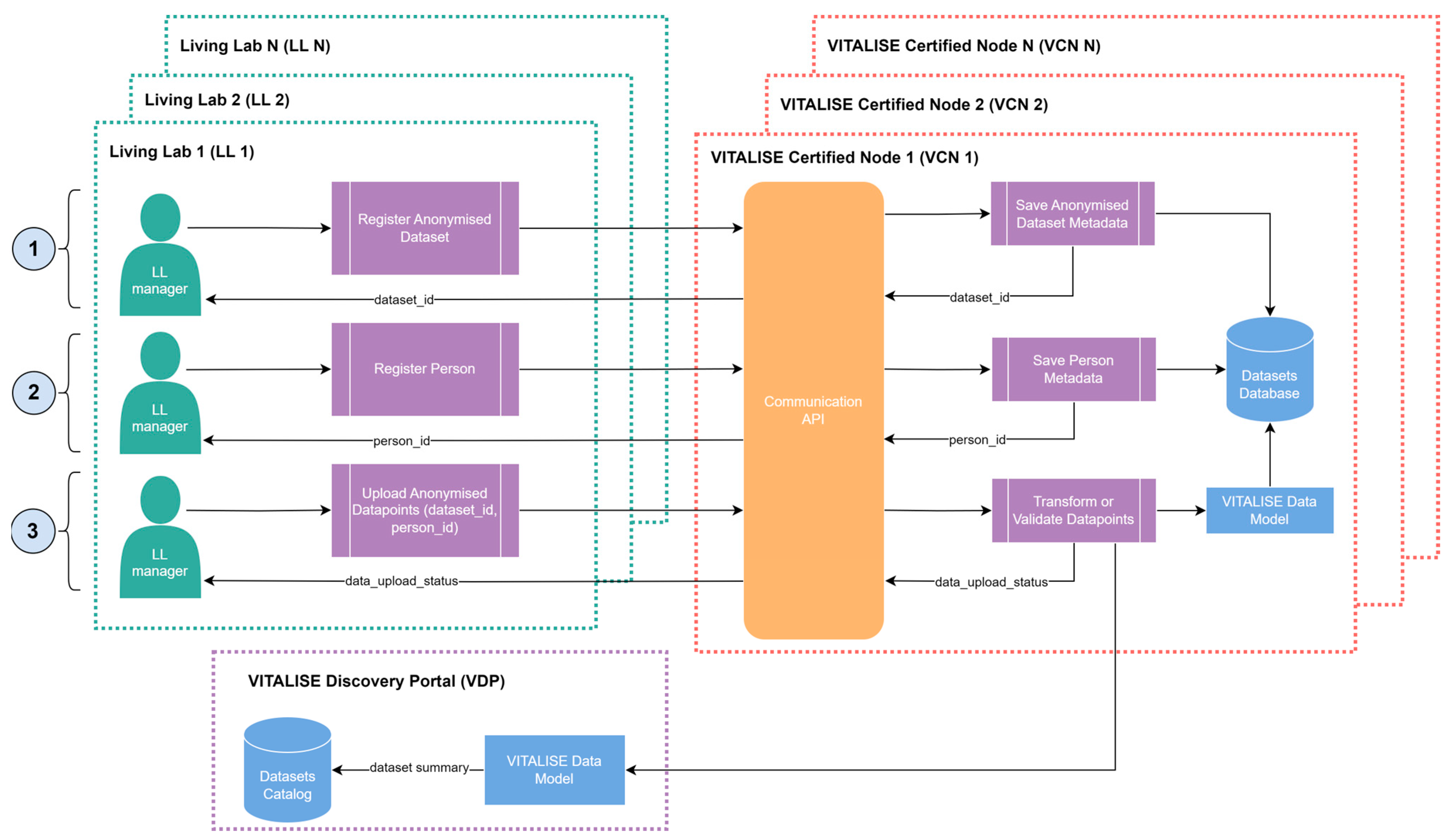
Figure 2 illustrates how LL managers can publish previously anonymised VITALISE data model-compliant data in the VCN of their LL.
LL managers initiate the process by registering an anonymous dataset using the Communication API of the VCN. This registration includes essential information such as the dataset’s title, description, and the author’s name. Upon registration, a unique identifier is assigned to the dataset, and its metadata is stored in the Datasets Database.
Next, LL managers register individual data of persons included in the dataset. This involves specifying each person’s gender and date of birth individually through the communication API. Each person added through this process receives a unique identifier, and their data is securely stored in the Datasets Database.
Finally, LL managers upload the datapoints generated by their sensors using the Communication API. These datapoints are associated with a dataset identifier and can optionally include an identifier to map datapoints to individuals previously registered. LL managers have the flexibility to choose between manually transforming the datapoints into the VITALISE data model format or uploading them in their original sensor-collected format if transformation logic is available. In the former case, the VCN validates that the uploaded datapoints comply with the required VITALISE data model format, while in the latter case, the VCN performs the necessary transformations to align the datapoints with the VITALISE data model format.
Once the datapoints are successfully uploaded to the datasets database, the VDP receives a summary of the updated dataset that contains metadata and descriptive statistics, and it is used to update the datasets catalogue of VITALISE data model-compliant anonymised datasets. After the VITALISE data model-compliant anonymised datasets catalogue is updated, the VDP ER can access and download the mentioned data through the workflow illustrated in
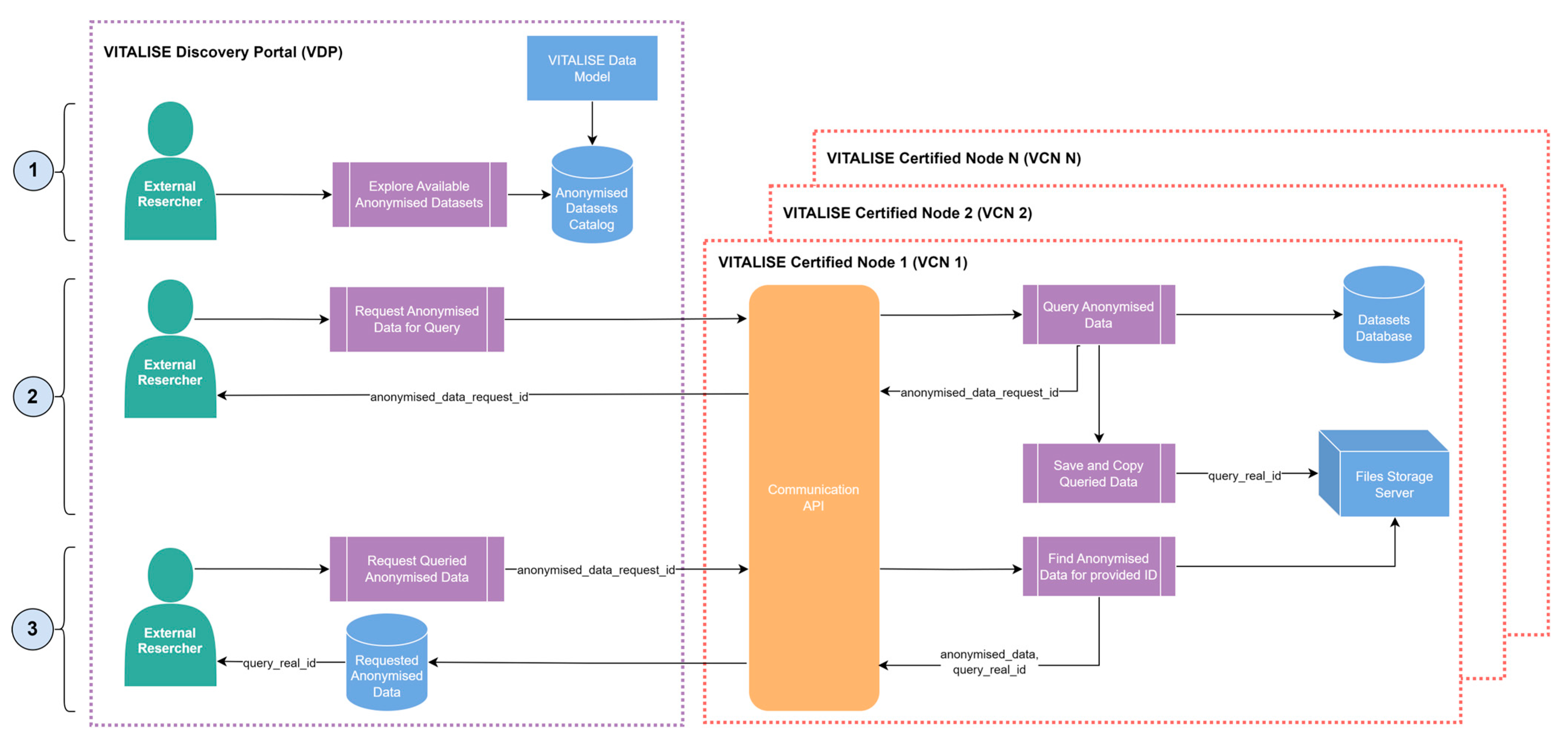
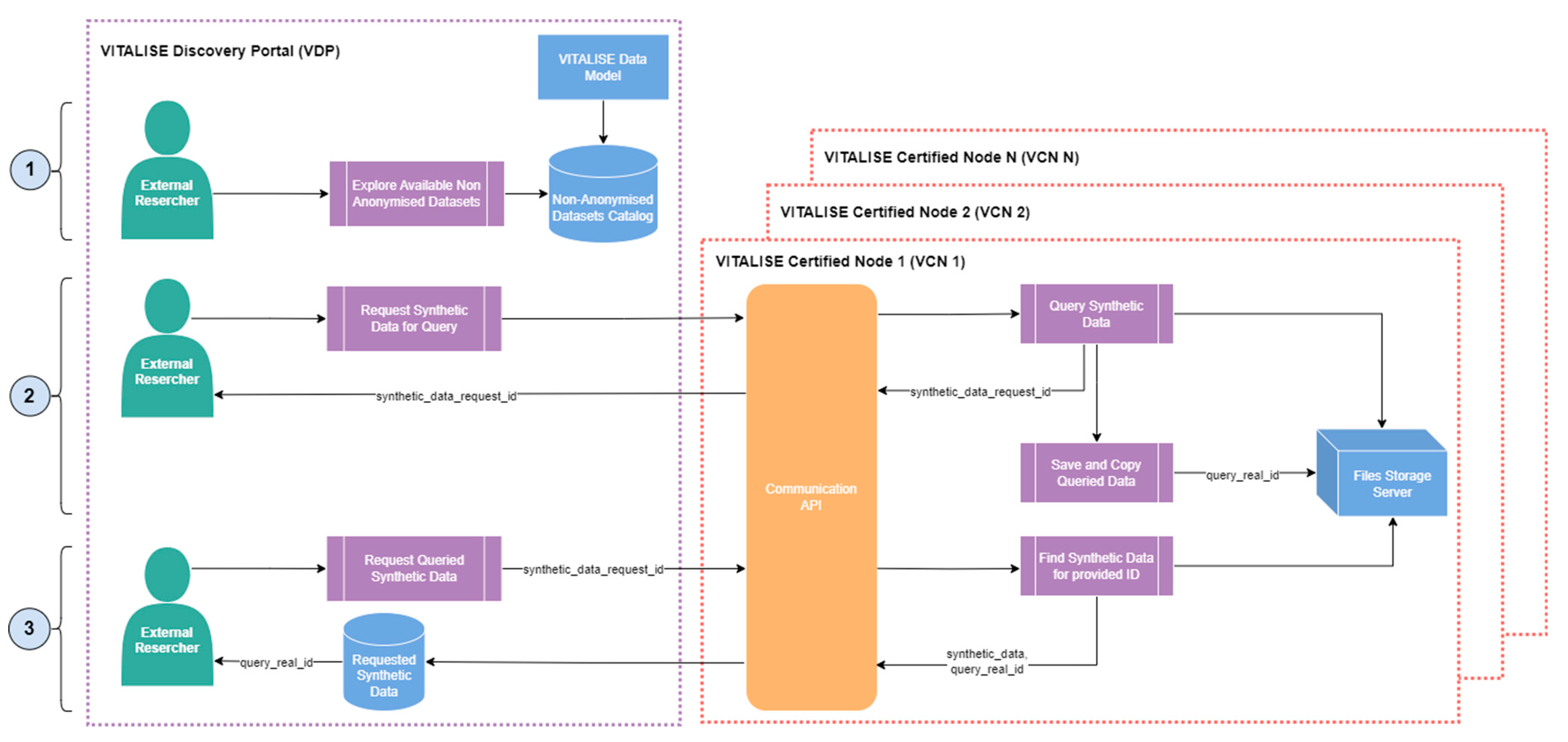
Figure 3.
ERs can explore the metadata and descriptive statistics available in the data model-compliant anonymised datasets catalogue of the VDP. Upon identifying datasets of interest, they can request anonymous data for specific queries from the selected datasets. This request can include the selection of multiple datasets, various measurement types, and data filtering based on person metadata such as age and gender. The Communication API of the VCN, in which the selected datasets are stored, performs the requested query within the internal Datasets Database. Subsequently, it generates a copy of the query result in both CSV and JSON formats, storing them on the file storage server and assigning a unique query identifier to the copied data.
ERs are then given the identifier for their anonymous data request through the VDP. ER can download the requested data in CSV or JSON format using this identifier. The Communication API of the VCN retrieves the queried data from the file storage server and delivers the anonymised data to the ERs through the VDP in their requested format.
3.3.2. VITALISE Data Model-Compliant Non-Anonymised Data
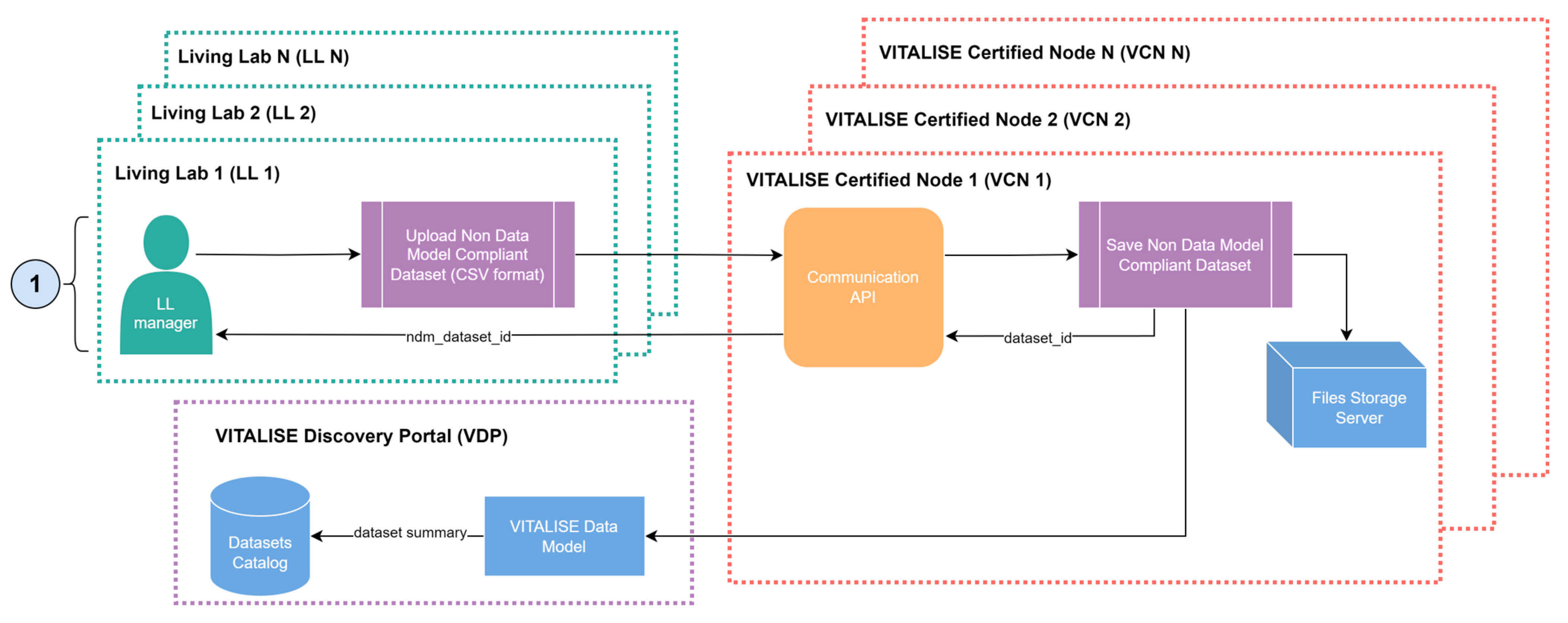
The workflow for publishing VITALISE data model-compliant non-anonymised data closely resembles the previous process, with a key difference being that LL managers can upload non-anonymised data, knowing that a synthetic data version will be shared with the ER.
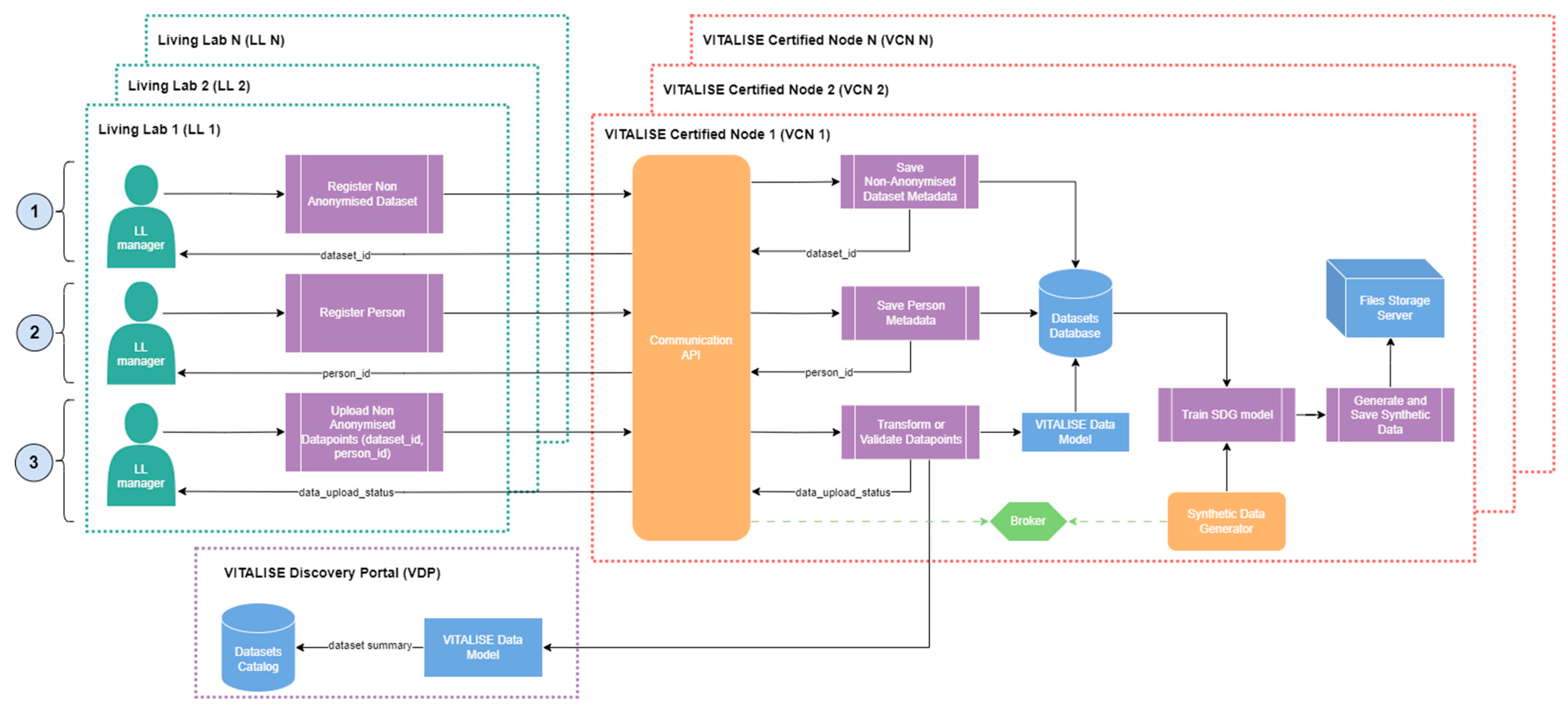
Figure 4 illustrates how LL managers can publish non-anonymised VITALISE data model-compliant datasets in their LL, with a subsequent synthetic version generated within the VCN.
As with VITALISE data model-compliant anonymised data, LL managers start by registering the dataset and the persons included in the dataset using the Communication API of the VCN. LL managers then upload and associate with a dataset identifier and person identifier the desired non-anonymised datapoints, which may have been previously transformed or uploaded in the source format. Once the datapoints are successfully uploaded to the dataset database, the Communication API dispatches a task to the Synthetic Data Generator through the Broker of the VCN.
Upon receiving the task, the Synthetic Data Generator trains an SDG model with the data related to the dataset for which new VITALISE data model-compliant non-anonymised datapoints were uploaded. This training only happens if more than one day has passed since the last training session. This model is then used to generate a synthetic version of the mentioned dataset, which is saved as a shareable dataset version in the Files Storage Server of the VCN. Currently, the service prototype employs SDG models that combine several probabilistic graphical modelling and Deep Learning (DL)-based techniques provided by the open-source SDV Python package [
30,
31]. These models were widely used in the literature and served as a baseline for comparing the different data types and scenarios [
38,
39,
40,
41]. Future enhancements to the service will incorporate the SDG approaches proposed by Isasa et al. [
19], which consider subject metadata for related time series generation.
While the SDG process is in progress, the VDP receives an updated dataset summary containing metadata and descriptive statistics of the dataset, which is then used to update the dataset catalogue of VITALISE data model-compliant non-anonymised datasets. As in the VITALISE data model-compliant anonymised data, once the datasets catalogue of the VDP is updated, the ER can access and download the synthetic and highly representative shareable data through the workflow illustrated in
Figure 5.
ERs can explore the metadata and descriptive statistics available in the data model-compliant non-anonymised datasets catalogue of the VDP. After identifying datasets of interest, they can request synthetic data for specific queries from the selected datasets. This request may involve multiple datasets, various measurement types, and data filtering based on person metadata such as age and gender. The Communication API of the VCN, in which the selected datasets and synthetic versions of those are stored, processes the requested query within the synthetic datasets versions stored in the Files Storage Server. A copy of the query result containing the non-anonymised original data is generated in both CSV and JSON formats and assigned a unique query identifier.
ERs are then provided with the identifier for their synthetic data request via the VDP. Using this identifier, they can download the requested data in CSV or JSON format. The Communication API retrieves the queried synthetic data from the File Storage Server and delivers it to the ERs through the VDP in their preferred format.
3.3.3. Non-VITALISE Data Model-Compliant Data
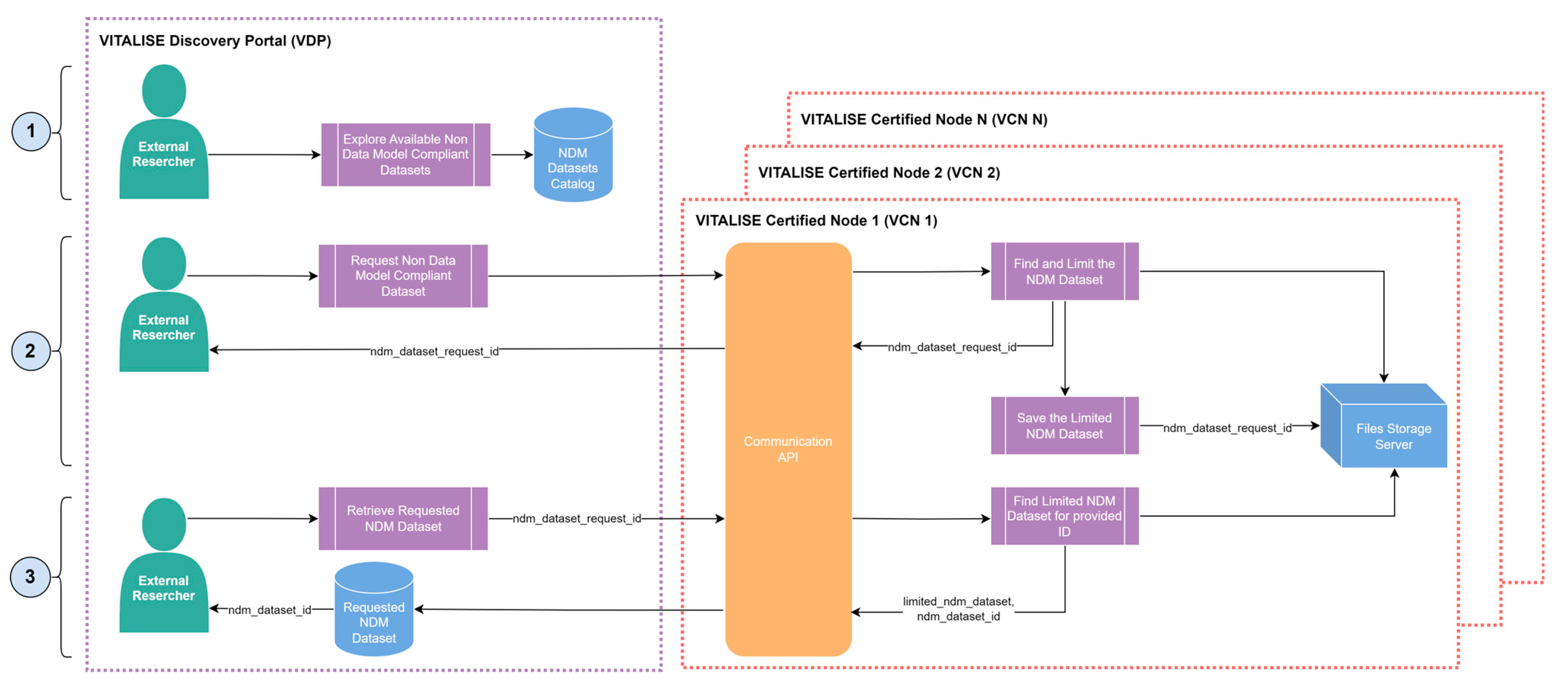
The workflows for publishing and accessing the NDM anonymised datasets are simpler than those for VITALISE data model-compliant anonymised and non-anonymised data. An NDM anonymised dataset is defined as a valid CSV data file containing any data collected in LLs, provided it adheres to a valid tabular format. These datasets do not conform to the defined VITALISE data format and must be anonymised before being uploaded to the system.
Figure 6 shows the workflow for publishing an NDM anonymised dataset in the service. In this scenario, LL managers use the Communication API of the VCN to upload the NDM dataset in CSV format while providing dataset information details such as name, author, description, license, and others. The uploaded NDM anonymised dataset is stored in the Files Storage Server, identified by a generated NDM dataset identifier.
Similar to previous data types, when a new NDM anonymised dataset is uploaded to the VCN, the VDP is notified. The dataset information, including name, author, type of license, description, and descriptive statistics of the variables, is added to the data catalogue. ER can then access and download the desired NDM anonymised dataset through the workflow illustrated in
Figure 7.
ERs can request the specific NDM anonymised dataset they require through the dataset catalogue provided by the VDP. This request is limited to a single NDM anonymised dataset. Upon receiving the request, the Communication API of the VCN, where the selected NDM anonymised dataset resides, generates a limited version of the dataset (sampled version of 1000 rows) in CSV format, which is then stored in the Files Storage Server. ERs are provided with a request identifier to download this limited version of the NDM anonymised dataset in CSV format. The Communication API retrieves the dataset version from the File Storage Server and delivers it to ERs via the VDP.
3.4. Remote Execution and Experiment Registration Flows
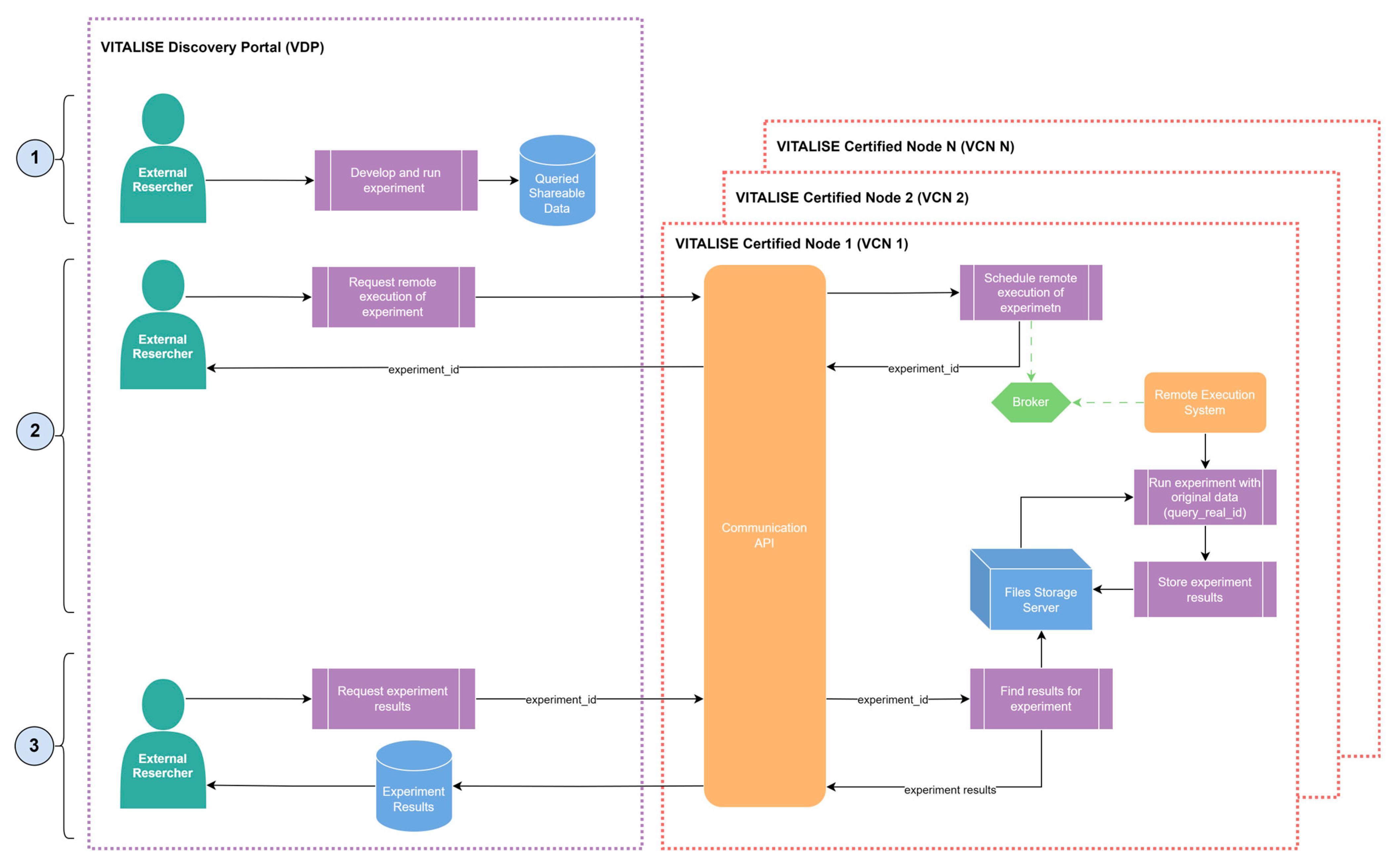
After using the data flows introduced in the section above to obtain a shareable version of the protected datasets hosted in the VCNs through the VDP, our Shareable Data Publishing and Access Service can also be used to remotely execute experiments previously developed by the ER. These experiments are initially developed locally on their computers by ERs using shareable data versions obtained from the data catalogue of the VDP and can consist of any data analytics paradigm experiment (e.g., prediction, clustering, recommendation). ERs can then use the service to register the obtained results in the VRS under the immutable RAI. Only experiments developed in Python 3.8 are supported in the current version of the system.
Figure 8 illustrates the workflow for the remote execution of experiments. Once the ERs have prepared their experiment with the queried shareable data obtained from the VDP, they can request the remote execution of the same experiment, but this time using the data stored in the VCN. For this purpose, an ER makes the remote execution request through the VDP, providing the following items:
The unique query identifier of the real data was obtained when shareable data was requested.
The data format in which the experiment must be performed: CSV or JSON for VITALISE data model-compliant data (anonymised or synthetic), and NDM for non-VITALISE data model-compliant data.
A requirements file indicating all dependencies needed to execute the experiment.
The source code of the experiment adhering to the formatting guidelines provided in the VDP.
Upon receiving the remote execution request in the VCN, the Communication API creates and stores the necessary files for the remote execution in the Files Storage Server, schedules the remote execution task through the Broker, and generates and returns an experiment identifier to the ER.
Since the Remote Execution System is connected to the Broker, when it receives the task for the remote execution of experiments, it (1) reads the real data and the files for the execution from the Files Storage Server and (2) executes the experiment within a dedicated and secure virtual environment. This environment is created individually for each experiment through containerisation technologies, ensuring data security and privacy. When the execution is completed, the results are stored in the Files Storage Server, and the created environment is deleted from the VCN.
After the remote execution of the experiment is finalised, ERs are notified through the VDP, and they can use the experiment identifier to retrieve the obtained results. If, for any reason, the experiment fails to produce results, ERs will receive a file containing the experiment logs. At this stage, ERs can analyse and compare the results obtained from their local analysis with those generated from the remotely executed analysis. This enables them to make informed decisions about whether further analyses are necessary or if the obtained results are suitable to proceed with the registration of the experiment.
Once the ERs have obtained the results of the remotely executed analysis and they want to register the experiment results, the workflow shown in
Figure 9 is executed. First, ERs make the experiment registration request through the VDP, indicating the identifier of the experiment they want to register. Upon receiving the remote execution request in the VCN, the Communication API collects the experiment results and information from the Files Storage Server and sends the request for experiment results registration to the Experiment Registration API of the VRS.
When the VRS processes this request, the results and information of the experiment are registered in the blockchain network and in an internal RAI results database to keep a copy in the VRS. Once the experiment is registered, ERs can query and explore results and information of all registered experiments within the unique and immutable RAI at an additional service of the VITALISE architecture available at
https://rai-finder.iti.gr (accessed on 26 February 2024).
4. Results
In this section, the results obtained when using our
Shareable Data Publishing and Access Service for the different data flows with real-world usage examples are presented to validate the presented service. This validation is based on the demonstration of the usefulness, applicability and good performance of the service. It was performed using a VCN deployed with docker technology in a virtual machine configured with an 8-core CPU running at 2.30 GHz, 32 GB of SSD storage, and 128 GB RAM, and the VDP is available online at
https://vitalise-portal.iti.gr/ (accessed on 26 February 2024).
In the next sections, the obtained results for each validated data flow are described. VITALISE data model-compliant anonymised data and VITALISE data model-compliant non-anonymised data flows were validated to illustrate the capabilities of the service to effectively support various research needs and scenarios. The data flow for non-VITALISE data model-compliant data has not been validated since the data are processed in the same way as for the VITALISE data model-compliant anonymised data, and similar results would be obtained.
4.1. VITALISE Data Model-Compliant Anonymised Data
In this section, the used data, the steps executed and the obtained results for the VITALISE data model-compliant anonymised data are described.
4.1.1. Data Description
The data used to evaluate the VITALISE data model-compliant anonymised data flow is a dataset composed of anonymised heart rate measurements from Fitbit smart wristbands taken over three days for a fictitious person registered in the VCN. The dataset contains a total of 1040 measurements corresponding to three days and sampled every 5 min. These measurements were uploaded to the VCN in the original Fitbit data format since the available version of the service is able to convert this format to the VITALISE Data Model. The data files in the original Fitbit format for this data flow in JSON format are accessible in a Zenodo repository at
https://doi.org/10.5281/zenodo.10777370 (accessed on 20 February 2024).
4.1.2. Data Flow Execution
The following steps were conducted for the VITALISE data model-compliant anonymised data flow execution with the previously described data:
By simulating the role of an LL manager, the VCN was used to upload the described data, following the flow described in
Section 3.3.1.
By simulating the role of an ER, the datasets catalogue of the VDP was inspected, and an anonymised data request was made for heart rate measurements. At this point, a limited version of 1000 samples of the anonymised heart rate data was created for the request made.
Through the VDP, the ER was able to download the anonymised heart rate data in the desired format, either JSON or CSV (flow described in
Section 3.3.1). A zip file was downloaded containing a file for the heart rate measurements and another one with the information of the anonymised data request (request identifier, hash of the real data and the date on which the anonymised data request was performed).
Using the downloaded anonymised data files in CSV format, the listed two analyses were performed locally for the heart rate measurements (with a limited data version of 1000 samples):
Descriptive statistics computation: computation of central tendency statistics (mean, mode and median), dispersion statistics (minimum value, maximum value, first quartile, second quartile and third quartile) and histogram (values and occurrence frequencies).
Forecasting task: The seasonal AutoRegressive Integrated Moving Average with eXogenous regressors (SARIMAX) forecasting model [
42] was trained with 80% of the measurements and tested, making predictions for the other 20% of the measurements. To evaluate the model performance and compare the real values with the predicted values, the Mean Forecasting Error (MFE), Mean Absolute Error (MAE), Mean Squared Error (MSE), and the Root Mean Squared Error (RMSE) were used. The lower these metrics are, the better the ML model is.
The remote execution of the locally developed analyses was requested by the VDP to be executed with the whole set of anonymised heart rate data hosted in the VCN.
The results obtained from the remotely executed analyses (with the whole set of samples) are compared against the results obtained from the locally executed analyses with the anonymised data limited to 1000 samples.
The remotely executed experiments were registered to the system, and a unique RAI was generated for each one to make the results of the remotely executed experiments available.
The downloaded anonymised data files, the code for the local analyses, the files for requesting the remote execution of the analysis, and the obtained results can be accessed as
Supplementary Materials at
https://doi.org/10.5281/zenodo.10777757 (accessed on 4 March 2024). In the next subsections, the results of the descriptive statistics and forecasting local and remote analyses are presented, which were performed with the downloaded anonymised data and locally stored real data in CSV format. Additionally, the assigned RAI for each remotely executed experiment is presented in
Appendix A.
4.1.3. Local and Remote Analyses Results
Figure 10 shows the obtained results for the descriptive statistics analysis executed remotely (for the whole set of heart rate measurements) and locally (with a limited version of heart rate measurements) for the anonymised data flow. As shown in the figure, similar statistical values were obtained for both analyses. Thus, a representative sample was obtained to perform analysis locally with limited data versions.
Table 1 shows the results obtained for the forecasting analyses executed remotely (for the whole set of heart rate measurements) and locally (with a limited version of heart rate measurements) for the anonymised data flow. In this table, the metric values for the best forecasting model are typed in bold and italic. The forecasting analyses developed locally with the limited version of heart rate measurements gave similar MFE, MAE, MSE and RMSE values to the values obtained with the analysis executed remotely with the whole set of heart rate measurements. Even though the results from the forecasting executed remotely are lower, the obtained anonymised limited version of data is representative enough to perform ML tasks.
4.2. VITALISE Data Model-Compliant Non-Anonymised Data
In this section, the used data, the steps executed and the obtained results for the VITALISE data model-compliant non-anonymised data flow are described.
4.2.1. Data Description
The dataset used to evaluate the VITALISE data model-compliant non-anonymised data flow is a dataset composed of anonymised physiological measurements taken by Withings devices during a period of six months for a fictitious person registered in the VCN. The physiological measurements that were used were Body Weight (kg), Fat Ratio (%), Hydration (kg), Muscle Mass (kg), Fat-Free Mass (kg) and Fat Mass Weight (kg). In total, there are 69 measurements for each physiological measurement listed above. These measurements were transformed into the VITALISE Data Model before being uploaded to the VCN. The data files in the transformed VITALISE Data Model for this data flow in JSON format are accessible in a Zenodo repository at
https://doi.org/10.5281/zenodo.10777370 (accessed on 8 March 2024).
4.2.2. Data Flow Execution
The next steps were involved for the VITALISE data model-compliant non-anonymised data flow execution with the previously described data:
By simulating the role of an LL manager, the VCN was used to upload the described data, following the flow described in
Section 3.3.2. At this moment, the Synthetic Data Generator trained the SDG model and generated the synthetic data that is saved in the Files Storage Server of the VCN.
By simulating the role of an ER, the datasets catalogue of the VDP was inspected, and six synthetic data requests were performed, one per each type of measurement (body weight, fat ratio, hydration, muscle mass, fat-free mass and fat mass weight).
Through the VDP and for each synthetic data request, the ER was able to download the synthetic data in the desired format, either JSON or CSV (flow described in
Section 3.3.2). For each request, a zip file was downloaded with one file per each requested synthetic measurement and the other one with the information of synthetic data (request identifier, hash of the real data and the date on which the data request was performed).
Using the downloaded synthetic data files in CSV format, two analyses were performed locally for each request:
Descriptive statistics computation: computation of central tendency statistics (mean, mode and median), dispersion statistics (minimum value, maximum value, first quartile, second quartile and third quartile) and histogram (values and occurrence frequencies).
Forecasting task: The SARIMAX forecasting model [
42] was trained with 80% of the measurements and tested, making predictions for the other 20% of the measurements. To evaluate the model performance and compare the real values with the predicted values, the MFE, MAE, MSE, and RMSE were used. The lower these metrics are, the better the ML model is.
The remote execution of the locally developed analyses was requested by the VDP to be executed with the real data hosted in the VCN.
The results obtained from the remotely executed analyses (with real data) are compared against the results obtained from the locally executed analyses with synthetic data.
The remotely executed experiments were registered to the system, and a unique RAI was generated for each one to make the results of the remotely executed experiments available.
The downloaded synthetic data files, the code for the local analyses, the files for requesting the remote execution of the analysis, and the obtained results can be accessed as
Supplementary Materials at
https://doi.org/10.5281/zenodo.10777757 (accessed on 8 March 2024). In the next subsections, the results of the descriptive statistics and forecasting for the local and remote analyses are presented, which were performed with the downloaded synthetic data and locally stored real data in CSV format. Additionally, the assigned RAI for each remotely executed experiment is presented in
Appendix A.
4.2.3. Local and Remote Analyses Results
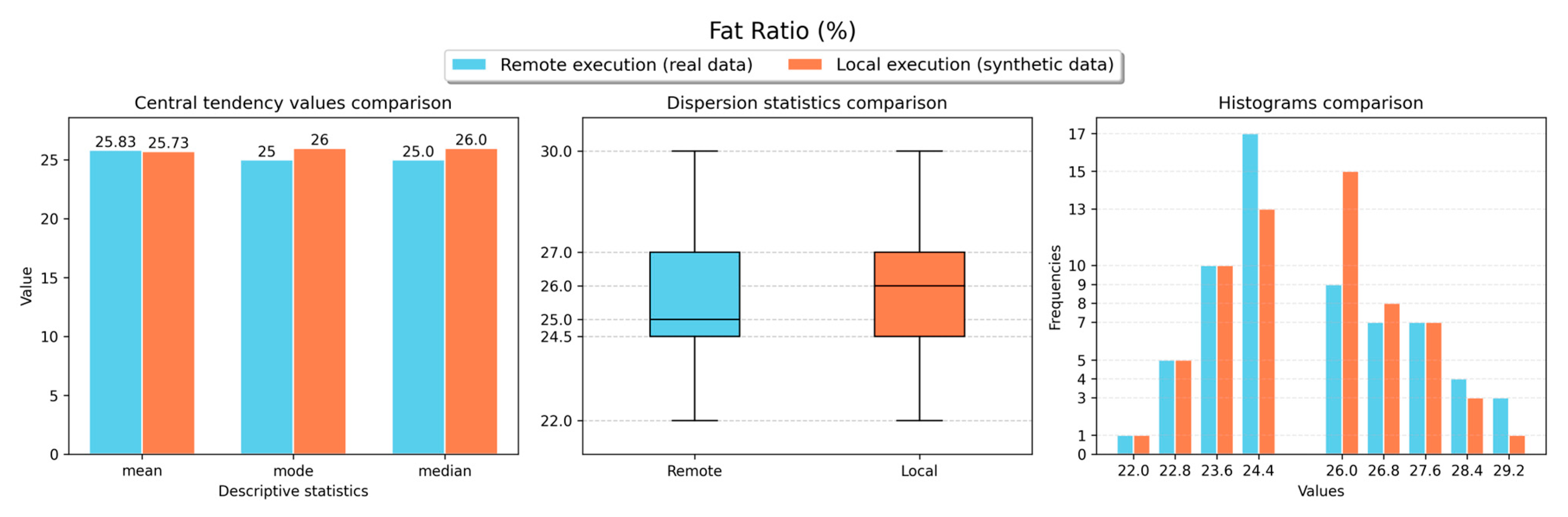
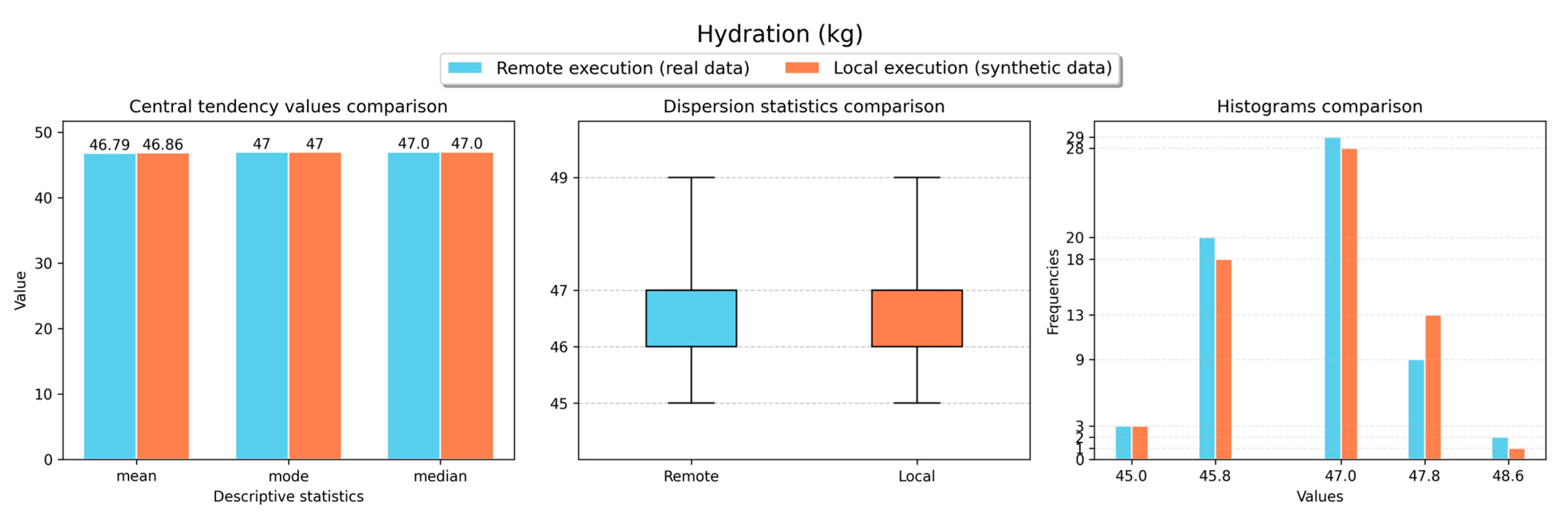
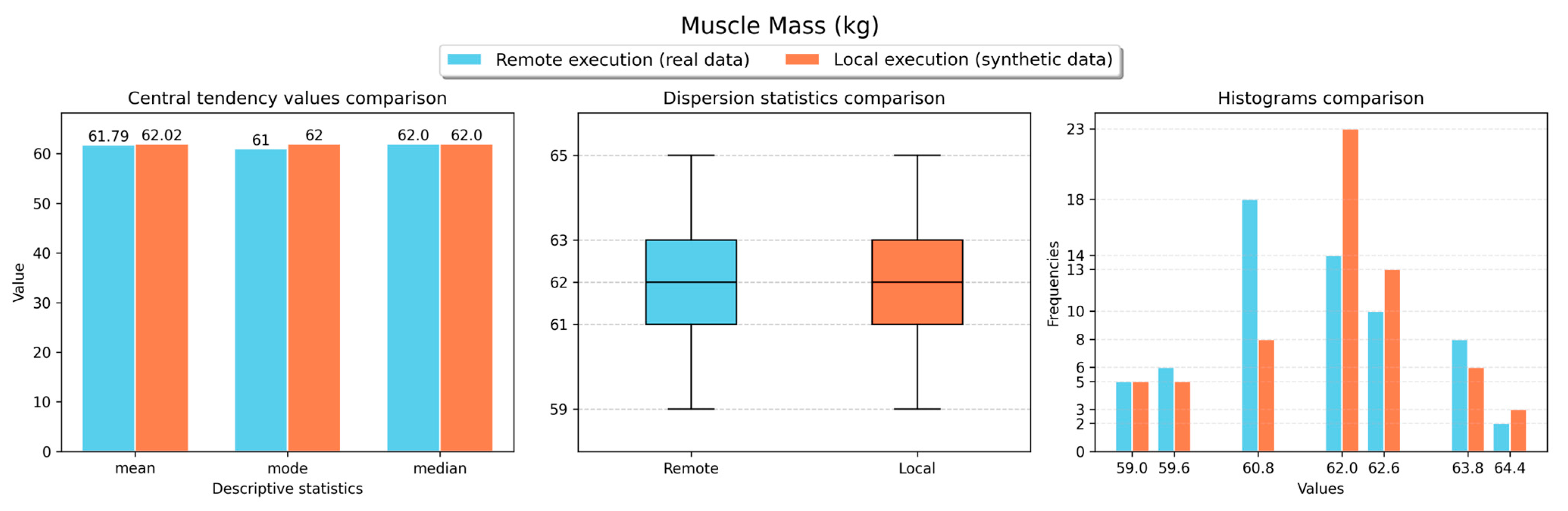
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15 and
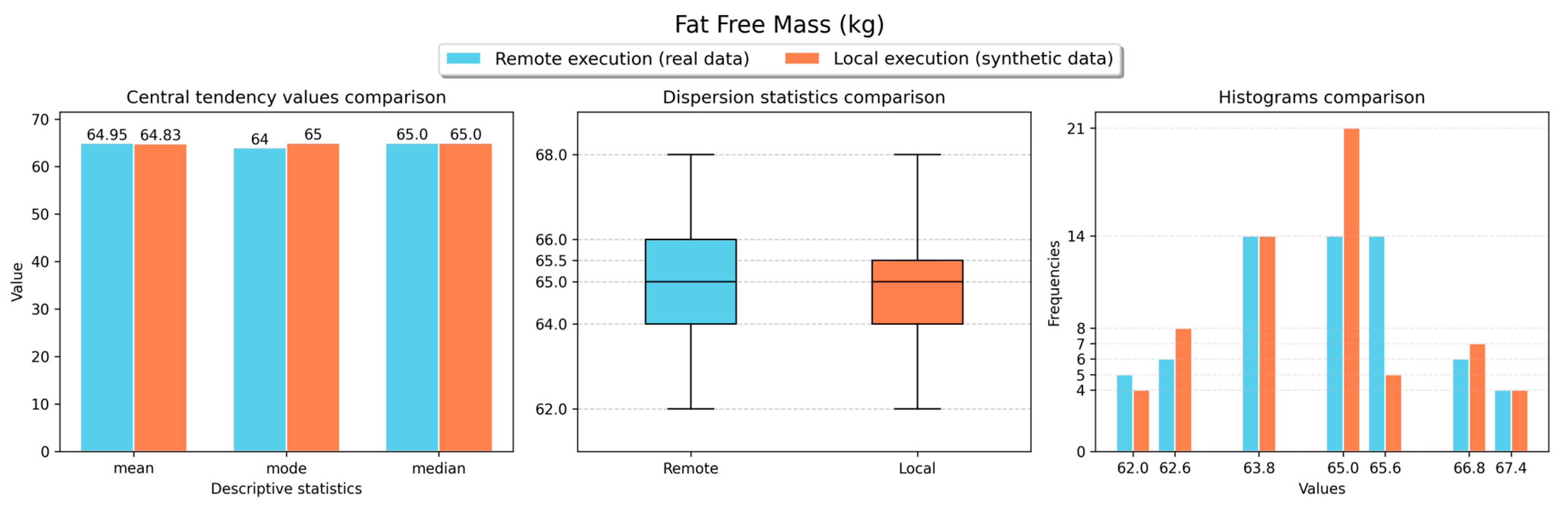
Figure 16 show the obtained results for the descriptive statistics analysis executed remotely (with real data) and locally (with the retrieved synthetic data) for the body weight, fat ratio, hydration, muscle mass, fat-free mass and fat mass weight measurements, respectively. As shown in the figures, similar statistical values were obtained for both local and remote execution and for all measurement types, resulting in comparable values. Thus, a representative sample was obtained to perform analyses locally with synthetic data versions of non-anonymised data stored in the VCN.
Table 2 presents the results obtained for the forecasting analyses executed remotely (with synthetic data versions) and locally (with the real data) for all requested measurements. In this table, the metric values for the best forecasting model for each measurement type are typed in bold and italic. For fat ratio and muscle mass measurements, the forecasting analyses executed locally (with synthetic data) resulted in better results than the analyses executed remotely (with real data). However, for both measurement types, metric differences lower than 1 were observed in most cases. On the other hand, for the other measurement types (body weight, hydration, fat-free mass and fat mass weight), the forecasting analyses executed remotely (with real data) delivered better results than the analyses executed locally (with synthetic data). Even though, for most cases, metric differences lower than 1 were observed. These results demonstrate that the obtained synthetic data can be effectively used for performing ML tasks with comparable results to using real data.
5. Discussion
The real-world usage examples of the VITALISE Data Model Anonymised Data and VITALISE Data Model Non-Anonymised data flow execution were successfully performed. Firstly, the data were correctly stored in the VCN, and then, the shareable data versions (limited version of the anonymised data measurements and the synthetic data version of the non-anonymised data measurements) were requested and obtained for different physiological monitoring variables (heart rate, body weight, fat ratio, hydration, muscle mass, fat-free mass and fat mass weight) in two data formats (JSON and CSV). Finally, local analysis was performed with each obtained shareable data version, and the remote execution of the same analyses with the complete set of measurements of real internal data was requested.
Regarding the performed descriptive statistics description and forecasting analyses, similar results were obtained when performing the experiments locally with shareable data versions and when performing them remotely with the real data stored internally in the VCN. In the descriptive statistics analyses, it was proven that although statistics from the shareable data versions are not equal to the statistics of the complete set of real internal data, in all cases, basic characteristics and trends are preserved. On the other hand, in the forecasting analyses, similar prediction errors were obtained for the shareable data version and the real internal data. For most of the cases, lower errors were obtained for the remotely executed forecasting analyses. Since the aim of this paper is to present the Shareable Data Publishing and Access Service and verify its usefulness, an accurate resemblance of the shareable data versions is not critical. However, solid results were obtained; shareable data versions have resembled most of the distributions of the real internal data while preserving privacy. Furthermore, it must be considered that the kind of analyses that can be remotely executed is up to the ER and can be extended to decision-making optimisation as far as enough data are available. In conclusion, with the presented results, it can be assured that the proposed service can be used for AI and ML model development without having access to the real data stored internally in the VCN, thus ensuring compliance with personal data protection laws and adhering to legal and ethical data handling practices, while maintaining the usefulness of the datasets, allowing them to be available to the Research Community. However, the service must be deployed and validated in terms of usability in a real-world environment to analyse the final potential it has.
With the validation of the service with real-world examples of usages, the efficiency of the presented
Shareable Data Publishing and Access Service was demonstrated along with how it can be used as an intermediary between LLs and ERs for secure data exchange and to accelerate research on data analytics paradigms in digital health, ensuring compliance with data protection laws. Furthermore, the developed service extends the controlled data processing workflow presented by Hernandez et al. [
5], providing a more complete service architecture compatible with more data modalities and generating an RAI accessible to the Research Community and interested stakeholders for each remotely executed experiment. This feature allows us to verify the integrity of the obtained results for executed experiments via blockchain integration. Despite the service having been developed for LL infrastructures, it can be applied and adapted to other types of entities or infrastructures that work on different application domains in the future. Furthermore, future work includes making our approach compatible with Data Space initiatives and implementing components that they are standardising such as data connectors, identity providers or metadata catalogues. The proposed system should not be limited to the health and wellbeing domain but could also be extended to other domains where privacy concerns can arise, such as demographics, industrial processes, business, etc.
Supplementary Materials
The following supporting information can be downloaded at:
https://doi.org/10.5281/zenodo.10777757, Data S1. Original uploaded data; Data S2. Retrieved shareable data; Results S1. Local execution analyses; Results S2. Remote execution analyses.
Author Contributions
Conceptualisation, M.H., E.K., G.E., F.L., D.P., M.T., V.F., L.M., C.M.-N. and I.M.; Data curation, M.H., D.P. and I.M.; Funding acquisition, E.K., G.E. and P.D.B.; Methodology, M.H., E.K., G.E., F.L., M.T., V.F., L.M. and C.M.-N.; Project administration, E.K., G.E. and P.D.B.; Resources, G.E.; Software, M.H., F.L., V.F., L.M. and C.M.-N.; Supervision, E.K., G.E. and P.D.B.; Validation, M.H., F.L. and V.F.; Visualisation, M.H.; Writing—original draft, M.H.; Writing—review and editing, M.H., E.K., G.E., F.L., D.P., V.F., L.M., C.M.-N. and I.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was partly funded by the VITALISE (Virtual Health and Wellbeing Living Lab Infrastructure) project, funded by the Horizon 2020 Framework Program of the European Union for Research Innovation (grant agreement 101007990).
Informed Consent Statement
Not applicable. The data used are fictitious or the data were collected during previous studies, for which the retainment period expired and have, therefore, been fully anonymised.
Data Availability Statement
Acknowledgments
VITALISE Consortium partners participating in the design and implementation of the VITALISE ecosystem and external persons participating in requirements shaping open sessions. Edwin Dokla, Stefanos Vrochidis and Andoni Beristain participated in the conceptualisation of the service. Edwin Dokla participated in the development of the VITALISE RAI Server. Spiros Nikolopoulos and Ioannis Kompatsiaris supervised the development and provided resources for the VITALISE Discovery Portal. Imanol Isasa and Naiara Aginako participated in revising and technical editing of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A
Table A1 and
Table A2 show the generated and assigned RAI immutable identifiers for the experiments executed remotely for validating the data flows for our
Shareable Data Publishing and Access Service. More information about the assigned metadata for each identifier can be found by typing the RAI of the experiment in the service available at
https://rai-finder.iti.gr (accessed on 8 March 2024).
Table A1.
Generated RAI immutable identifiers for the experiments executed remotely for validating the VITALISE data model-compliant anonymised flow.
Table A1.
Generated RAI immutable identifiers for the experiments executed remotely for validating the VITALISE data model-compliant anonymised flow.
| Measurement | Analysis | Generated RAI by Registration of the Experiment |
|---|
| Heart Rate (beats/min) | Descriptive statistics | cd53f3c29ba74d3383cc2129ccd6c223f4a6f0bff8faa07a03e782b42e8eddfd |
| Forecasting task | d5fa13187d2aeb27f7f4734460b01cb715666af64c0223cde491f1586ede4ace |
Table A2.
Generated RAI immutable identifiers for the experiments executed remotely for validating the VITALISE data model-compliant non-anonymised flow.
Table A2.
Generated RAI immutable identifiers for the experiments executed remotely for validating the VITALISE data model-compliant non-anonymised flow.
| Measurement | Analysis | Generated RAI by Registration of the Experiment |
|---|
Body Weight
(kg) | Descriptive statistics | d6294ce2df588423b64d1e168e89cb2d696b99b3fd571adf818a5195e556cd7a |
| Forecasting task | da8ebe82dc55baec0e3eb68d0acd9d1d265f3e733163ec5225008d53b49185a1 |
Fat Ratio
(%) | Descriptive statistics | 1904de27915e0d632a8b7a6e3b38abfb4b482ea1dd0b9372aa7b149f84275ef2 |
| Forecasting task | a749c934071edc5b6e9661195a665b496eb9c477ab2c6725d8ee3171ada3f490 |
Hydration
(kg) | Descriptive statistics | 0e773b1eb64074b78845f8d42dc604954cd1f1e011f9e47877a332a03de8a019 |
| Forecasting task | 9c96d686bae2e81665b2a0d529353426b0a2253ed715e29967343aa39e48c3b5 |
Muscle Mass
(kg) | Descriptive statistics | 67b0af4df6c0dba5c5fa24ad744f3ef5da659bd0e44b107b3dbbc3507e5a0a14 |
| Forecasting task | cf18585c97203bcc9dc881c1105bdb9f6855e26e7018201c28f2228ed0cb421b |
Fat-Free Mass
(kg) | Descriptive statistics | 7aa9c93fcbfaaf0dfa2bddced0b3a70e8faf16612f1c831c937a75005778704b |
| Forecasting task | c3ee2a62be5f48b4a0cadcda6f9f2c5c8d154f782e8143c0669f4205794a2118 |
| Fat Mass Weight (kg) | Descriptive statistics | 311ad8c6bfd249d0f4bcac4fe0356c061f5fd4415534b887d585a701ed7a4e23 |
| Forecasting task | 09f2939b5d0bc758adf4123610ecb9141f39b9f48a7010e3570e37d49b5109ff |
References
- General Data Protection Regulation (GDPR)—Official Legal Text. Available online: https://gdpr-info.eu/ (accessed on 20 June 2023).
- VITALISE Project—Home. Available online: https://vitalise-project.eu/ (accessed on 9 August 2023).
- VITALISE Project—Why VITALISE. Available online: https://vitalise-project.eu/why-vitalise/ (accessed on 9 August 2023).
- Maga-Nteve, C.; Epelde, G.; Hernandez, M.; Tsolakis, N.; Konstantinidis, E.; Meditskos, G.; Bamidis, P.; Vrochidis, S. Standardized and Extensible Reference Data Model for Clinical Research in Living Labs. Procedia Comput. Sci. 2022, 210, 165–172. [Google Scholar] [CrossRef]
- Hernandez, M.; Epelde, G.; Beristain, A.; Álvarez, R.; Molina, C.; Larrea, X.; Alberdi, A.; Timoleon, M.; Bamidis, P.; Konstantinidis, E. Incorporation of Synthetic Data Generation Techniques within a Controlled Data Processing Workflow in the Health and Wellbeing Domain. Electronics 2022, 11, 812. [Google Scholar] [CrossRef]
- Emam, K.E. Guide to the De-Identification of Personal Health Information; CRC Press: Boca Raton, FL, USA, 2013; ISBN 978-1-4665-7906-4. [Google Scholar]
- Synthetic Data|European Data Protection Supervisor. Available online: https://edps.europa.eu/press-publications/publications/techsonar/synthetic-data (accessed on 20 June 2023).
- Rubin, D.B. Statistical Disclosure Limitation. J. Off. Stat. 1993, 9, 461–468. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Volume 27. [Google Scholar]
- Piacentino, E.; Guarner, A.; Angulo, C. Generating Synthetic ECGs Using GANs for Anonymizing Healthcare Data. Electronics 2021, 10, 389. [Google Scholar] [CrossRef]
- Hazra, D.; Byun, Y.-C. SynSigGAN: Generative Adversarial Networks for Synthetic Biomedical Signal Generation. Biology 2020, 9, 441. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, X.; Qiu, X.; Zhang, X.; Brusic, V.; Zhao, J. A Privacy Preserving Framework for Federated Learning in Smart Healthcare Systems. Inf. Process. Manag. 2023, 60, 103167. [Google Scholar] [CrossRef]
- Rankin, D.; Black, M.; Bond, R.; Wallace, J.; Mulvenna, M.; Epelde, G. Reliability of Supervised Machine Learning Using Synthetic Data in Health Care: Model to Preserve Privacy for Data Sharing. JMIR Med. Inform. 2020, 8, e18910. [Google Scholar] [CrossRef] [PubMed]
- Yale, A.; Dash, S.; Dutta, R.; Guyon, I.; Pavao, A.; Bennett, K.P. Generation and Evaluation of Privacy Preserving Synthetic Health Data. Neurocomputing 2020, 416, 244–255. [Google Scholar] [CrossRef]
- Rashidian, S.; Wang, F.; Moffitt, R.; Garcia, V.; Dutt, A.; Chang, W.; Pandya, V.; Hajagos, J.; Saltz, M.; Saltz, J. SMOOTH-GAN: Towards Sharp and Smooth Synthetic EHR Data Generation. In Proceedings of the Artificial Intelligence in Medicine, Minneapolis, MN, USA, 25–28 August 2020; Michalowski, M., Moskovitch, R., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 37–48. [Google Scholar]
- Yoon, J.; Drumright, L.N.; van der Schaar, M. Anonymization Through Data Synthesis Using Generative Adversarial Networks (ADS-GAN). IEEE J. Biomed. Health Inform. 2020, 24, 2378–2388. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, A.; Ray, P.; Soper, B.; Stevens, J.; Coyle, L.; Sales, A.P. Generation and Evaluation of Synthetic Patient Data. BMC Med. Res. Methodol. 2020, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Pinaya, W.H.L.; Tudosiu, P.-D.; Dafflon, J.; Da Costa, P.F.; Fernandez, V.; Nachev, P.; Ourselin, S.; Cardoso, M.J. Brain Imaging Generation with Latent Diffusion Models. In Proceedings of the Deep Generative Models, Singapore, 22 September 2022; Mukhopadhyay, A., Oksuz, I., Engelhardt, S., Zhu, D., Yuan, Y., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 117–126. [Google Scholar]
- Isasa, I.; Hernandez, M.; Epelde, G.; Londoño, F.; Beristain, A.; Larrea, X.; Alberdi, A.; Bamidis, P.; Konstantinidis, E. Comparative Assessment of Synthetic Time Series Generation Approaches in Healthcare: Leveraging Patient Metadata for Accurate Data Synthesis. BMC Med. Inform. Decis. Mak. 2024, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.; Epelde, G.; Alberdi, A.; Cilla, R.; Rankin, D. Synthetic Data Generation for Tabular Health Records: A Systematic Review. Neurocomputing 2022, 493, 28–45. [Google Scholar] [CrossRef]
- Victor, N.; Lopez, D. A Conceptual Framework for Sensitive Big Data Publishing. In Proceedings of the International Conference on Communication and Computational Technologies, Jaipur, India, 30–31 August 2019; Purohit, S.D., Singh Jat, D., Poonia, R.C., Kumar, S., Hiranwal, S., Eds.; Springer: Singapore, 2021; pp. 523–533. [Google Scholar]
- Ficek, J.; Wang, W.; Chen, H.; Dagne, G.; Daley, E. Differential Privacy in Health Research: A Scoping Review. J. Am. Med. Inform. Assoc. 2021, 28, 2269–2276. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Namasudra, S.; Chilamkurti, N.; Kim, B.-G.; Gonzalez Crespo, R. Blockchain-Based Privacy Preservation for IoT-Enabled Healthcare System. ACM Trans. Sens. Netw. 2023, 19, 1–17. [Google Scholar] [CrossRef]
- Javed, L.; Anjum, A.; Yakubu, B.M.; Iqbal, M.; Moqurrab, S.A.; Srivastava, G. ShareChain: Blockchain-Enabled Model for Sharing Patient Data Using Federated Learning and Differential Privacy. Expert Syst. 2023, 40, e13131. [Google Scholar] [CrossRef]
- Gao, S. Advanced Health Information Sharing with Web-Based GIS. Ph.D. Thesis, Department of Geodesy and Geomatics Engineering, Technical Report No. 272, University of New Brunswick, Frederiction, NB, Canada, March 2010; 188p. [Google Scholar]
- MongoDB: The Data Platform for Applications. Available online: https://www.mongodb.com (accessed on 7 September 2023).
- RabbitMQ: Easy to Use, Flexible Messaging and Streaming—RabbitMQ. Available online: https://www.rabbitmq.com/ (accessed on 7 September 2023).
- MinIO|High Performance, Kubernetes Native Object Storage. Available online: https://min.io (accessed on 7 September 2023).
- FastAPI. Available online: https://fastapi.tiangolo.com/ (accessed on 7 September 2023).
- Patki, N.; Wedge, R.; Veeramachaneni, K. The Synthetic Data Vault. In Proceedings of the 2016 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Montreal, QC, Canada, 17–19 October 2016; pp. 399–410. [Google Scholar]
- The Synthetic Data Vault. Put Synthetic Data to Work! Available online: https://sdv.dev/ (accessed on 13 June 2023).
- Celery-Distributed Task Queue—Celery 5.3.4 Documentation. Available online: https://docs.celeryq.dev/en/stable/ (accessed on 7 September 2023).
- mHealth Data Interoperability. Available online: https://www.openmhealth.org/ (accessed on 11 September 2023).
- WebThings. Available online: https://webthings.io (accessed on 11 September 2023).
- Open Connectivity Foundation (OCF). Available online: https://openconnectivity.org/ (accessed on 11 September 2023).
- Schema.Org. Available online: https://schema.org/ (accessed on 11 September 2023).
- Petsani, D.; Santonen, T.; Merino-Barbancho, B.; Epelde, G.; Bamidis, P.; Konstantinidis, E. Categorizing Digital Data Collection and Intervention Tools in Health and Wellbeing Living Lab Settings: A Modified Delphi Study. Int. J. Med. Inform. 2024, 185, 105408. [Google Scholar] [CrossRef] [PubMed]
- Hittmeir, M.; Mayer, R.; Ekelhart, A. A Baseline for Attribute Disclosure Risk in Synthetic Data. In Proceedings of the Tenth ACM Conference on Data and Application Security and Privacy, New Orleans, LA, USA, 16–18 March 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 133–143. [Google Scholar]
- Mayer, R.; Hittmeir, M.; Ekelhart, A. Privacy-Preserving Anomaly Detection Using Synthetic Data. In Data and Applications Security and Privacy XXXIV; Singhal, A., Vaidya, J., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; Volume 12122, pp. 195–207. ISBN 978-3-030-49668-5. [Google Scholar]
- Hittmeir, M.; Ekelhart, A.; Mayer, R. Utility and Privacy Assessments of Synthetic Data for Regression Tasks. In Proceedings of the 2019 IEEE International Conference on Big Data (Big Data), Los Angeles, CA, USA, 9–12 December 2019; pp. 5763–5772. [Google Scholar]
- Hittmeir, M.; Ekelhart, A.; Mayer, R. On the Utility of Synthetic Data: An Empirical Evaluation on Machine Learning Tasks. In Proceedings of the 14th International Conference on Availability, Reliability and Security, Canterbury, UK, 26–29 August 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 1–6. [Google Scholar]
- Seabold, S.; Josef, P. Statsmodels: Econometric and statistical modeling with python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
Figure 1.
Architecture of the Shareable Data Publishing and Access Service.
Figure 1.
Architecture of the Shareable Data Publishing and Access Service.
Figure 2.
Data publishing workflow for VITALISE data model-compliant anonymised data.
Figure 2.
Data publishing workflow for VITALISE data model-compliant anonymised data.
Figure 3.
Data accessing workflow for VITALISE data model-compliant anonymised data.
Figure 3.
Data accessing workflow for VITALISE data model-compliant anonymised data.
Figure 4.
Data publishing workflow for VITALISE data model-compliant non-anonymised data.
Figure 4.
Data publishing workflow for VITALISE data model-compliant non-anonymised data.
Figure 5.
Data accessing workflow for VITALISE data model-compliant non-anonymised data.
Figure 5.
Data accessing workflow for VITALISE data model-compliant non-anonymised data.
Figure 6.
Data publishing workflow for NDM anonymised data.
Figure 6.
Data publishing workflow for NDM anonymised data.
Figure 7.
Data accessing workflow for NDM anonymised data.
Figure 7.
Data accessing workflow for NDM anonymised data.
Figure 8.
Remote execution of experiments with original data stored in the VCNs.
Figure 8.
Remote execution of experiments with original data stored in the VCNs.
Figure 9.
Registration of experiments in the VRS.
Figure 9.
Registration of experiments in the VRS.
Figure 10.
Results of the descriptive statistics computation analyses performed for heart rate measurements.
Figure 10.
Results of the descriptive statistics computation analyses performed for heart rate measurements.
Figure 11.
Results of the descriptive statistics computation analyses performed for body weight measurements.
Figure 11.
Results of the descriptive statistics computation analyses performed for body weight measurements.
Figure 12.
Results of the descriptive statistics computation analyses performed for fat ratio measurements.
Figure 12.
Results of the descriptive statistics computation analyses performed for fat ratio measurements.
Figure 13.
Results of the descriptive statistics computation analyses performed for hydration measurements.
Figure 13.
Results of the descriptive statistics computation analyses performed for hydration measurements.
Figure 14.
Results of the descriptive statistics computation analyses performed for muscle mass measurements.
Figure 14.
Results of the descriptive statistics computation analyses performed for muscle mass measurements.
Figure 15.
Results of the descriptive statistics computation analyses performed for fat-free mass measurements.
Figure 15.
Results of the descriptive statistics computation analyses performed for fat-free mass measurements.
Figure 16.
Results of the descriptive statistics computation analyses performed for fat mass weight measurements.
Figure 16.
Results of the descriptive statistics computation analyses performed for fat mass weight measurements.
Table 1.
Results of the forecasting analyses performed for the validation of VITALISE data model-compliant anonymised data flow. The best results between each local and remote experiments pair are marked in bold and italic.
Table 1.
Results of the forecasting analyses performed for the validation of VITALISE data model-compliant anonymised data flow. The best results between each local and remote experiments pair are marked in bold and italic.
| Measurement | Execution | Train Size | Test Size | MFE | MAE | MSE | RMSE |
|---|
| Heart Rate (beats/min) | Local execution | 800 | 200 | 4.2250 | 10.4950 | 142.8650 | 11.9526 |
| Remote execution | 830 | 210 | 4.2644 | 8.1971 | 120.6875 | 10.9857 |
Table 2.
Results of the forecasting analyses performed for the validation of VITALISE data model-compliant non-anonymised data flow. The best results between each local and remote experiments pair are marked in bold and italic.
Table 2.
Results of the forecasting analyses performed for the validation of VITALISE data model-compliant non-anonymised data flow. The best results between each local and remote experiments pair are marked in bold and italic.
| Measurement | Execution | Train Size | Test Size | MFE | MAE | MSE | RMSE |
|---|
| Body Weight (kg) | Local execution | 55 | 14 | 1.5000 | 1.9285 | 5.6428 | 2.3754 |
| Remote execution | 55 | 14 | 1.0714 | 1.6428 | 5.5000 | 2.3452 |
| Fat Ratio (%) | Local execution | 55 | 14 | 0.8462 | 1.3076 | 2.6923 | 3.1865 |
| Remote execution | 55 | 14 | 2.9230 | 2.9230 | 10.1538 | 1.6400 |
| Hydration (kg) | Local execution | 55 | 14 | 1.2307 | 1.2307 | 1.8462 | 1.3580 |
| Remote execution | 55 | 14 | 0.9231 | 0.9231 | 1.0769 | 1.0300 |
| Muscle Mass (kg) | Local execution | 55 | 14 | 1.8462 | 2.0000 | 5.3846 | 2.3204 |
| Remote execution | 55 | 14 | 2.3076 | 2.3076 | 6.6154 | 2.5720 |
| Fat-Free Mass (kg) | Local execution | 55 | 14 | 1.6923 | 1.8462 | 5.0769 | 2.2530 |
| Remote execution | 55 | 14 | 0.3076 | 1.3846 | 2.7692 | 1.6000 |
| Fat Mass Weight (kg) | Local execution | 55 | 14 | 3.3846 | 3.3846 | 12.6153 | 3.5518 |
| Remote execution | 55 | 14 | 0.0769 | 2.0769 | 6.3846 | 2.5267 |
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).