1. Introduction
The size of biomedical data, as well as the rate at which it is being produced, is increasing dramatically. The biomedical data is also being collected from many different sources, such as hospitals (clinical Big Data), laboratories (genomic and proteomic Big Data), and the internet (online Big Data). There is a growing need for statistically predictive causal discovery algorithms that incorporate the biological knowledge gained from modern statistical, machine learning, and informatics approaches used in the learning of causal relationships from biomedical Big Data comprised of clinical, omics (genomic and proteomic), and environmental components.
While earlier available studies focus on statistical methods to infer causality [
1,
2,
3,
4], recent statistical machine learning methods have been introduced which aim at analyzing big datasets [
5,
6,
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18]. However, given many different types of clinical, genomic, and environmental data, it is rather uncommon to see statistical machine learning methods that utilize prior knowledge relevant to the mechanisms behind the phenomena which generates those different data types. The statistical machine learning methods that recognize that there are many variables which are not collected in the data, but are still related to the mechanisms which produced the data (hidden variables), are also limited. Furthermore, there is a lack of statistical methods that evaluate how well the methods perform at inferring causality when hidden confounded variables are present.
There are many aspects of causality, from its representation (syntax) to its semantics and many different related concepts to causality, e.g., theory of inferred causation, counterfactual analyses, incomplete interventions, confounding effect, etc. [
1,
9]. However, in learning mechanisms from a phenomenon with collected data, the goal is to infer cause and effect relationships among complicated knitted random variables in the dataset with reasonable confidence.
Thus, the focus in this study is on the learning of causal relationships among random variables in the collected data, particularly when using causal Bayesian networks (CBNs). CBNs are directed acyclic graphs in which each arc is interpreted as a direct causal influence between a parent node and a child node relative to the other nodes in the network [
19]. CBNs consist of a structure (such as an example in
Figure 1) and a set of probabilities that parameterize said structure (not shown). In general, for each variable there is a conditional probability of that variable given the states of its direct causes. Thus, the probability associated with
Gliomas Grade is
P (
Gliomas Grade|
PTNP1,
LPL,
EGFR). That is, we provide the probability distribution over the values of the
Gliomas Grade conditioned on each of the possible expression levels of the genes
PTNP1,
LPL, and
EGFR. For variables that have no direct causes in the network, a prior probability is specified. The causal Markov condition [
9] specifies the conditional independence relationships which are represented by a causal network: Let X and Y be variables. Suppose that Y is neither a direct nor an indirect effect of X. Then X is independent of Y, conditioned on any state of the direct causes of X. The causal Markov condition permits the joint distribution of the n variables in a CBN to be factored as follows [
19]:
where
xi denotes a state of variable
Xi,
πi denotes a joint state of the parents of
Xi, and
K denotes background knowledge (prior probability). Since the initial research for a general Bayesian formulation for learning causal structure (including latent variables) and parameters from observational data using CBN [
20,
21], Bayesian causal discovery has become an active field of research in which numerous advances have been made [
1,
7,
8,
10,
22,
23].
CBNs have been suitable in analyzing Big Data sets consisting of different types of large data including clinical, genomic, and environmental data [
8,
12,
23,
24,
25,
26,
27,
28,
29]. Such causal statistical models help to provide a more comprehensive understanding of human physiology and disease. More importantly, CBNs have been used as a natural way to express “causal” knowledge as a graph using nodes (representing random variables) and arcs (representing “causal” relationships). Indeed, there are many causal models made from existing causal knowledge—from simple and intuitive causal models (e.g., a model to predict whether neighbor is out [
30], a sprinkler model [
1], etc.), to expert causal models (e.g., a multiple diseases model [
31], an ALARM monitoring system [
32], etc.). The learning of causal relationships from data has been discussed in different articles [
1,
9,
33], and this especially holds true for cases where researchers have used Bayesian Networks for learning structures [
29,
34,
35,
36,
37]. Also, other algorithms, such as PC [
9], K2 [
5], and more recently the Bayesian Inference for Directed Acyclic Graphs (BiDAG) [
12], have been used to learn causal relationships from data.
Earlier structure learning methods concentrated on model selection, where we select a model
M* from
or
where we assume we have
p number of mutually exclusive models,
[
38]. Later methods incorporated model averaging [
29], where we summarize how likely a feature
F that is found in a subset of the models and is defined by a set of indices,
where
f includes those indices of the models where
F is observed. Thus, in model averaging, we calculate the probability of a feature
F as the following:
or
However, most of the structure learning methods do not address hidden variables. Since we cannot observe all relevant variables in a natural phenomenon, to better learn the underlying mechanistic process from Big Data, we need to address and evaluate the learning of causal relationships with hidden variables.
In this paper, we show that searching through the order (we describe further about what we mean by “order” in the method section) of variables in CBNs can help provide a better understanding of the underlying mechanistic process that generated the data even in the presence of hidden variables. In addition, we propose a novel algorithm in searching through the order (we call it the PrePrior algorithm) which evidences a promising performance when attempting to learn the underlying mechanistic process from data containing hidden variables. The algorithm utilizes model averaging techniques such as searching through a relative order (e.g., if gene A is regulating gene B, then we can say that gene A is in a higher order than gene B) and incorporates relevant prior mechanistic knowledge to guide the Markov chain Monte Carlo (MCMC) search through the order.
2. Methods
Given a CBN structure
S and a dataset
D, the Bayesian scoring method that assesses how well the structure fits the given data can be calculated using a closed form [
39]:
In the above scoring method, Dirichlet uniform parameter priors are used and parameter independence is assumed [
40];
n represents the number of variables in the structure;
qi represents the number of configurations of the parents for a given variable
Xi; and
ri represents the total amount of states for a variable
Xi. For example, if
Xi is a binary random variable and it has two binary random variables as direct causes (parents), then
ri is equivalent to two and
qi is equivalent to four.
Nijk represents the counts for a given variable
Xi under a given parent configuration (indexed by
j) and a given state (indexed by
k) for variable
Xi.
N’
ijk represents the Dirichlet uniform prior, which in this case may be calculated as the following:
The number of possible structures increases exponentially with the number of variables, and so the above formula is sufficient for determining the best BN when the number of variables in the CBN is small. However, when the number of variables is large, it becomes impossible to determine the best structure in this manner. The problem of finding the best CBN is NP-hard [
41], and thus it is not always possible to find the best CBN that fits the data. This is the key limitation of model selection methods [
38] when used as a means of extending our current mechanistic understanding through the learning of causal relationships from data.
The algorithm we introduce in this paper utilizes model averaging techniques, such as searching through a relative
order [
29] (e.g., cause is in a higher
order than effect) and incorporating prior mechanistic knowledge to guide the MCMC (Markov Chain Monte Carlo) search through the
order. An
order describes the relationships between variables based on describing whether a variable can be a direct cause (parent) for another variable.
Definition 1. (Order ): iff .
With the above definition of the
order, we are stating that
Xi is considered to be of a higher order than
Xj if, and only if,
Xj cannot be found in the group of direct causes (parents) of
Xi. A potential ordering for a list of three variables is <
X1,
X2,
X3>. This order implies that
X1 can be a direct cause (parent) of
X2 and/or
X3, but
X2 and
X3 cannot be direct causes (parents) of
X1. Similarly,
X2 can be a direct cause (parent) of
X3, but
X3 cannot be a direct cause (parent) of
X2. Note that any given order of random variables can better summarize mechanistic (causal) relationships than just one structure. For example, an order <
X1,
X2,
X3> includes the following three structures (
Figure 2):
Orders are useful because, in a manner similar to structures, they can be scored. Since an order represents a set of structures, it may be scored by summing over all structures consistent with the given order. This method for scoring an order is not efficient because it would require that we have a score for all structures that meet a given order. With that being the case, we consider an alternative method for scoring orders presented by Friedman and Koller [
29], which uses the direct cause (parent) sets of variables. The equation for this scoring procedure is:
The above equation is an expansion of Bayesian scoring presented by Heckerman [
33]. Here,
O represents an ordering,
Ui,o represents the possible parent-sets for a given variable under a given ordering, and
qi,U represents the possible configurations of the parents for a variable
i within a parent-set
U. All other parameters in the equation are represented in the same manner as in Equation (6).
The benefit in scoring orders over scoring structures is that in the case where one is dealing with two or more variables, there are more structures than orders. For example, when the number of variables equals four, there are 543 structures but only 24 different orders.
An MCMC search is used to search through the orders. At any given MCMC search process, we have a current order (denote it as o) and a proposed order (denote it as o′), and we decide whether the proposed order will take the place of the current order with a probability that is returned by a decision function . A proposed order is generated by either applying a local perturbation (i.e., swapping two variables in an order: for example, <X1, X2…Xi…Xj…Xn> to <X1, X2…Xj…Xi…Xn>), or a global perturbation (i.e., aka a cutting the deck, swapping groups of variables in an order: for example, <X1, X2…Xi, Xi+1…Xn> to < Xi+1…Xn, X1, X2…Xi >). Initially, a random order is generated.
Friedman and Koller [
29] propose the following two algorithms for MCMC search with different
:
- o
Random Algorithm
- ▪
Uses
- o
Prior Algorithm
- ▪
Uses
where o, o′, and D represent the current order that we are considering: a proposed order and a dataset, respectively.
We further propose a new algorithm called the PrePrior Algorithm with the following MCMC search with the same as the Prior algorithm with an additional step:
- o
PrePrior Algorithm
- ▪
Uses based on user defined prior to sample o′
- ▪
Uses
Note that PrePrior algorithm generates proposed orders based on the prior, and that the user provides.
User’s Prior of an Order. To specify a prior of mechanistic causal knowledge in terms of an order o (if X is known to cause Y, we say X has a higher order than Y, i.e., ) or , we assume the following:
- i.
If no prior is provided, a uniform prior of any given order is assumed. For example, for a pairwise order of X and Y, if no prior is provided then . In general, for n variables a uniform prior for any order o is .
- ii.
The prior of an order is specified as the probability of how likely it is compared to the uniform prior. For example, if prior publications show gene Y is regulating gene X, a user might specify and if there have been studies suggesting that gene Z is regulating gene W, a user might specify .
For mechanism discovery, the correct discovery of the generating structure is the most important aspect of the algorithm. Datasets consisting of 50 and 1000 simulated observational cases from the ALARM Bayesian network were generated [
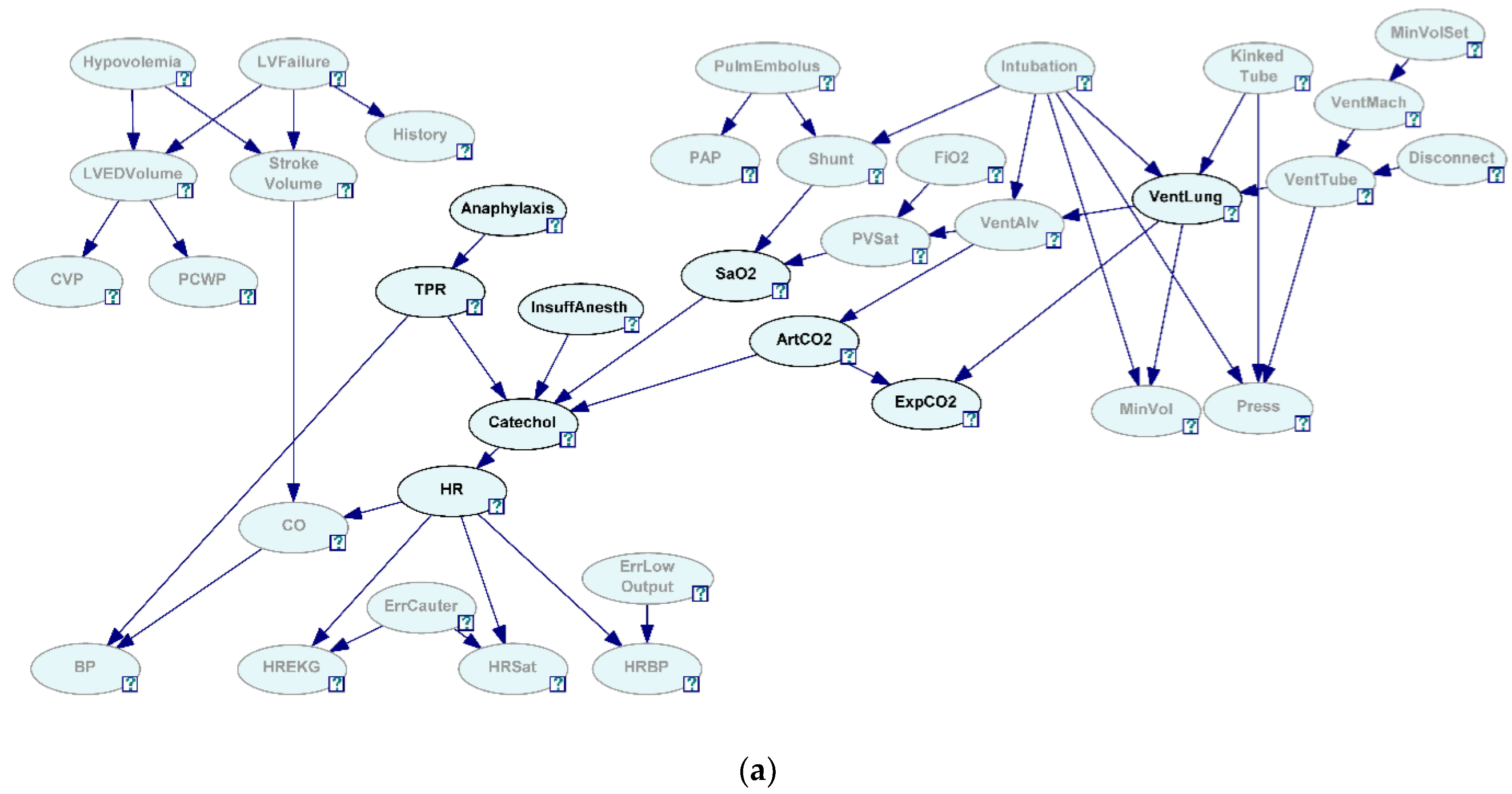
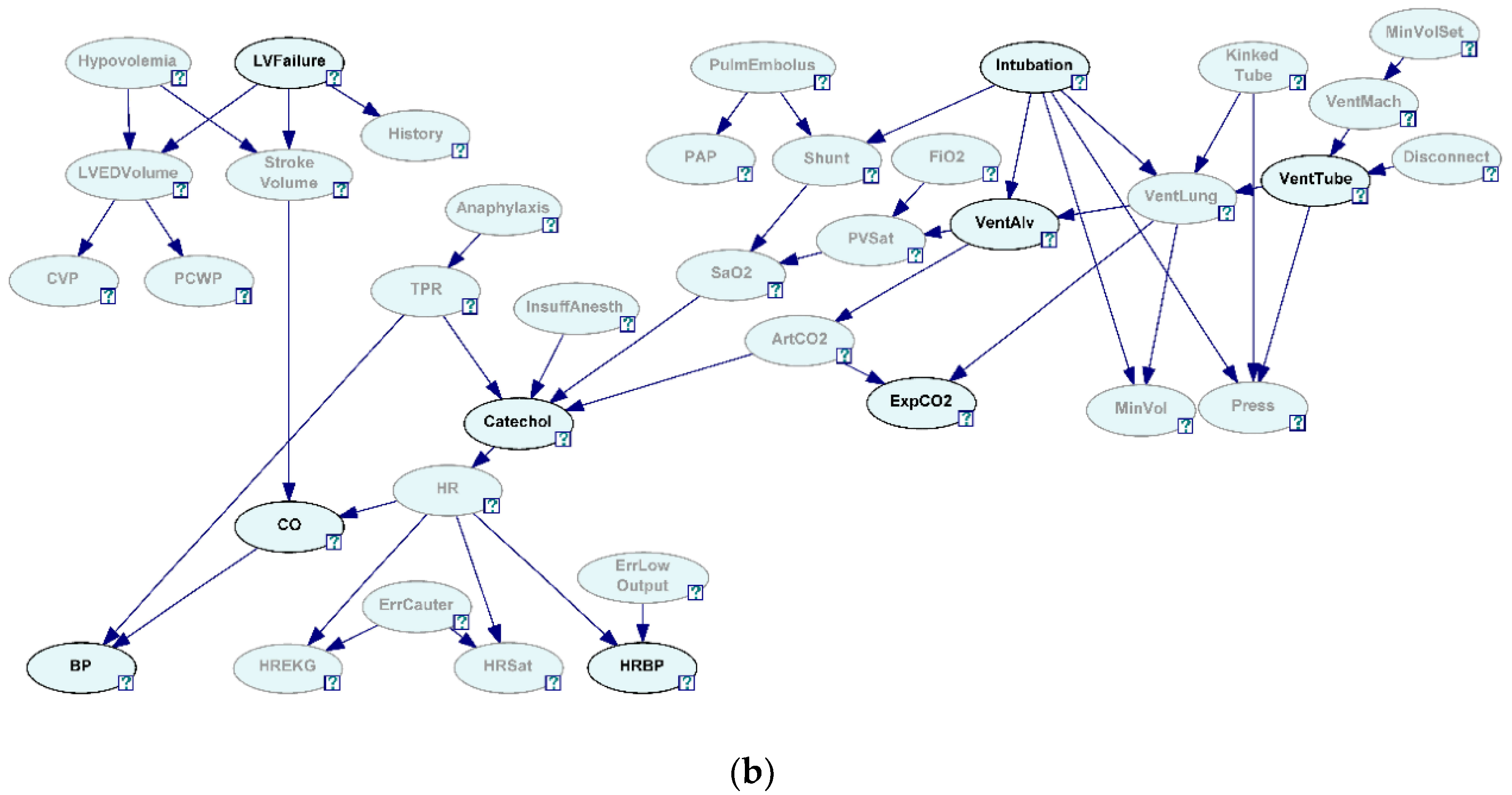
27]. To see how well the algorithm correctly discovered the generating structure in the presence of hidden variables, we have selected two sets of nine variables each selected from 37 variables in the network. The first variable set is referred to as Close 9 variables (C9) and was created by selecting variables that were closely situated in the network (
Figure 3a, all the grayed-out variables are hidden and not selected). The second variable set is referred to as Sparse 9 variables (S9) and was created by selecting variables that were relatively situated further in the network (
Figure 3b, all the grayed-out variables are hidden and not selected).
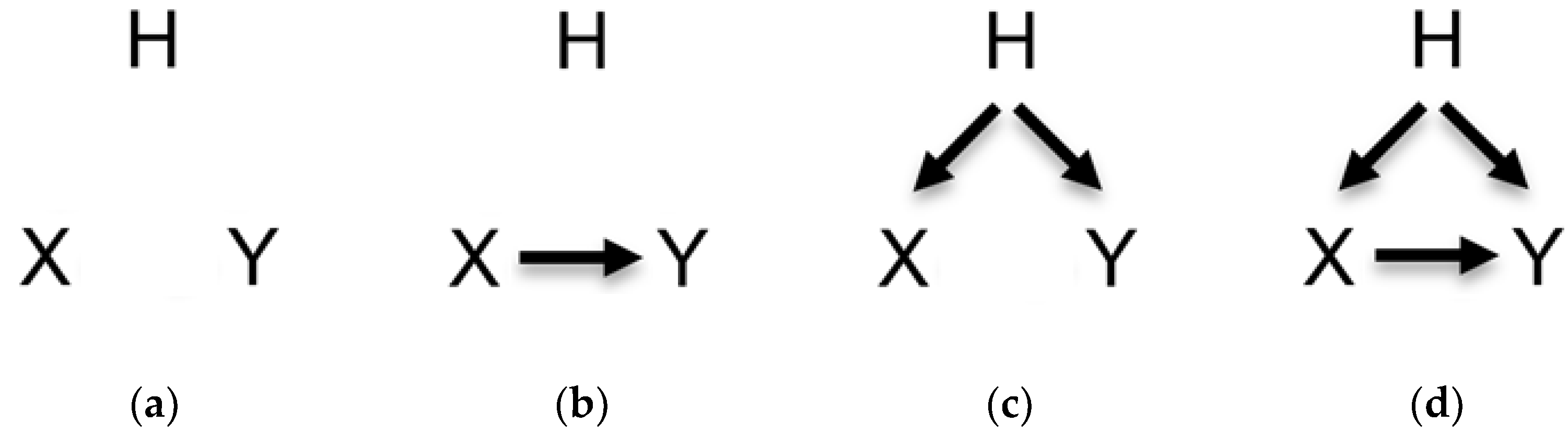
Another reason we have selected these nine variables was to see how well the causal discovery algorithms were predicting the four pairwise relationships shown in
Figure 4. Distinguishing these four pairwise relationships is the first step in better understanding the mechanistic process involved in generating these datasets.
Different numbers of pairwise causal relationships are found in the Close 9 variables (C9) and Sparse 9 variables (S9) (
Table 1). For example, in C9, TPR and VentLung are not confounded nor causally related (denoted as Ø
X Y in
Figure 4a), and TPR and HR are not confounded and causally related (denoted as Ø
X→Y in
Figure 4b). In S9, ExpCO2 and Catechol are confounded but not causally related (denoted as H
X Y in
Figure 4c, ArtCO2 being a variable as H), and ArtCO2 and VentAlv are confounded and causally related (denoted as H
X→Y in
Figure 4d where VentLung takes the role of H).
Two datasets were generated from each of the two sets of variables. Two of the datasets had 50 observational cases each and were named D50C9 and D50S9 because they were generated by the C9 and S9 sets of variables, respectively. The other two datasets had 1000 observational cases each and were named D1KC9 and D1KS9 because they were generated by the C9 and S9 sets of variables, respectively. Many biological mechanistic networks are not completely connected, i.e., each variable has limited (e.g., less than five) causes. As a result, we have limited the number of possible parents to five and scored all the possible orders using Equation (8). It took roughly one month to score all of the possible orders for the four datasets. The dataset of results is referred to as
Dataset Global BDe Best Order.
Dataset Global BDe Best Order contains information on all of the scores for all of the possible orders, and therefore we know which is the best order (and the best Bayesian networks structure) that will be identified if the BDe metric [
5] (similar to Equation (8)) is used given the dataset.
The Random, Prior, and PrePrior algorithms were independently ran three times on D50C9 and D50S9 for 1 h, 2 h, and 4 h; and on D1KC9 and D1KS9 for 2 h, 4 h, and 16 h. We have used five Linux machines to run in parallel of 522 total h (over 21 equivalent days) of runs.
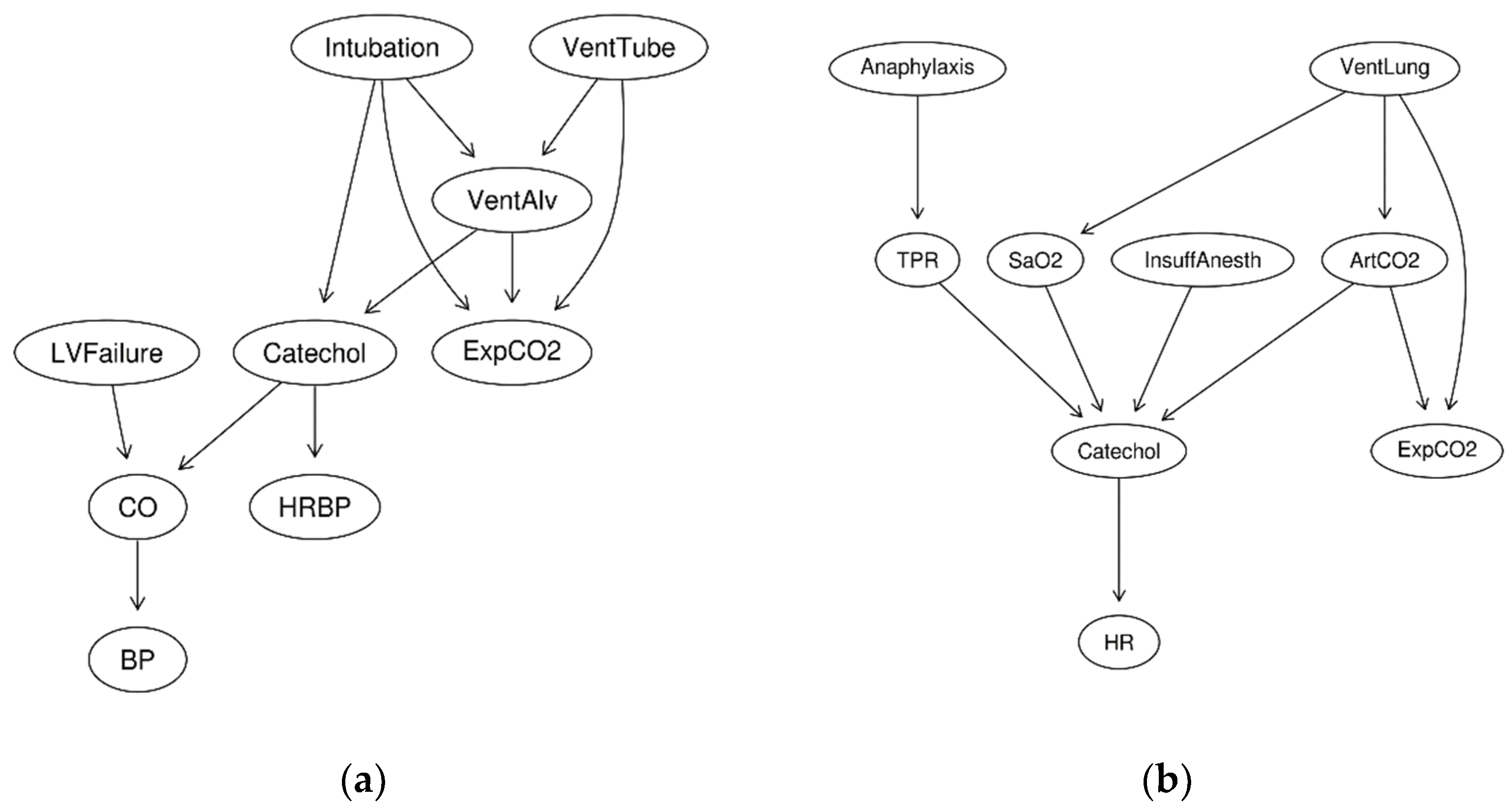
The predictive performance is calculated as a pairwise causal distance from either generating the structure (denoted it as
SG and shown in
Figure 5) or the Dataset Global BDe Order. For each variable pair of
X and
Y, let the underlying relationship between
X and
Y be denoted as
RX,Y where
. Let the likelihood score of
RX,Y assessed from either the generating structure and Dataset Global BDe Order as
PG(
RX,Y) and
PG(
D|
RX,Y) respectively, where
Note that we calculate.
and
where
O is the set of orders that satisfy
and
So is the set of structures that satisfy an order
and
for all possible orders (denote them as
) and all possible structures that satisfies an order
(denote them as
).
Additionally, we calculate
PSG (
D|
RX,Y).
S is the set of structures that satisfies for all possible structures (denote them as ) from all possible orders.
We use PSG(D|RX,Y) and PG(D|RX,Y) for all X and Y to generate a consensus causal structure by drawing arcs between X and Y with the thickest arc when PSG(D|RX,Y) or PG(D|RX,Y) are above 0.9999, and with the thinnest arc when PSG(D|RX,Y) or PG (D|RX,Y) are close to 0.0001. If PSG(D|X→Y) and PSG(D|Y→X) both are less than 0.0001, then no arcs are drawn between X and Y.
We first compare generating causal structure and
Dataset Global BDe Best Order by calculating the following:
These results will show us how the BDe metric approximates the generated causal structure given the generated datasets. In addition to comparing the predictive ability of these algorithms, we compared the causal structure predictive ability of the algorithms that use BDe metric with the Dataset Global BDe Best Order.
We report each Dataset Global BDe Best Order prediction using a Markov blanket of a variable (Catechol) appearing both from Close 9 variables (C9) and Sparse 9 variables (S9) and compared that with the Markov blanket of the Catechol from the generating structure.
Denote the probability of
RX,Y predicted from an algorithm as
PA(
D|
RX,Y) and P
SA(
D|
RX,Y). Note that P
A(
D|
RX,Y) is calculated the same way we calculated
PG(
D|
RX,Y) described above. We report the distance from the generating structure as
and the distance from the
Dataset Global BDe Order as
Note here we consider indirect causation to assess RX,Y, i.e., we check whether X appears as an ancestor of Y (i.e., repeatedly applying parent-of(Y) function–parent-of(parent-of(Y)), parent-of(parent-of(parent-of (Y)))…), or whether Y appears as an ancestor of X in the overall network.
We report how well algorithms predict the Markov blanket of each variable in
Close 9 variables (C9) and
Sparse 9 variables (S9) (denote all Markov Blankets as
AM) and compare with the Markov blanket of the variable from the
Dataset Global BDe Best Order (denote all Markov Blankets as
GM) by calculating the following distance:
Note that and can be calculated by incorporating the order weight (as we calculated or by multiplying ) and and can be calculated by not incorporating the order weight (as we calculated or by not multiplying ).
We also report all algorithms’ predicted performance, as how well they predict four causal pairwise relationships–Ø
X Y, Ø
X→Y, H
X Y, and H
X→Y–introduced in
Table 1 by comparing the algorithm’s prediction of
RX,Y ∈ {
X→Y, X←Y, X(none)Y} with the true underlying relationships
TX,Y ∈ {Ø
X Y, Ø
X→Y, H
X Y, H
X→Y}. In addition to the predictive performance, we also report the following for each
RX,Y and for each
TX,Y:
where
is the number of underlying true relationships (i.e., counts in
Table 1). Finally, we report the percentage of the algorithm’s most probable prediction of
RX,Y given the true underlying true relationships
TX,Y by calculating the following:
We have also run other causal discovery algorithms, such as PC [
9], K2 [
5], and BiDAG [
12] on the same datasets, i.e., 50 and 1000 cases for Sparse 9 variables (in D50S9 and D1KS9); and 50 and 1000 cases for Close 9 variables (in D50C9 and D1KC9). Since BiDAG could only incorporate binary random variables for learning, we converted all the variables in the datasets as continuous variables. This was done by adding normal noise with
to each measurement of discrete data. The reason we have used these parameters for noise was that they have given the most consistent conditional independencies among the variables when we compared the original discrete data and converted continuous data.
3. Results
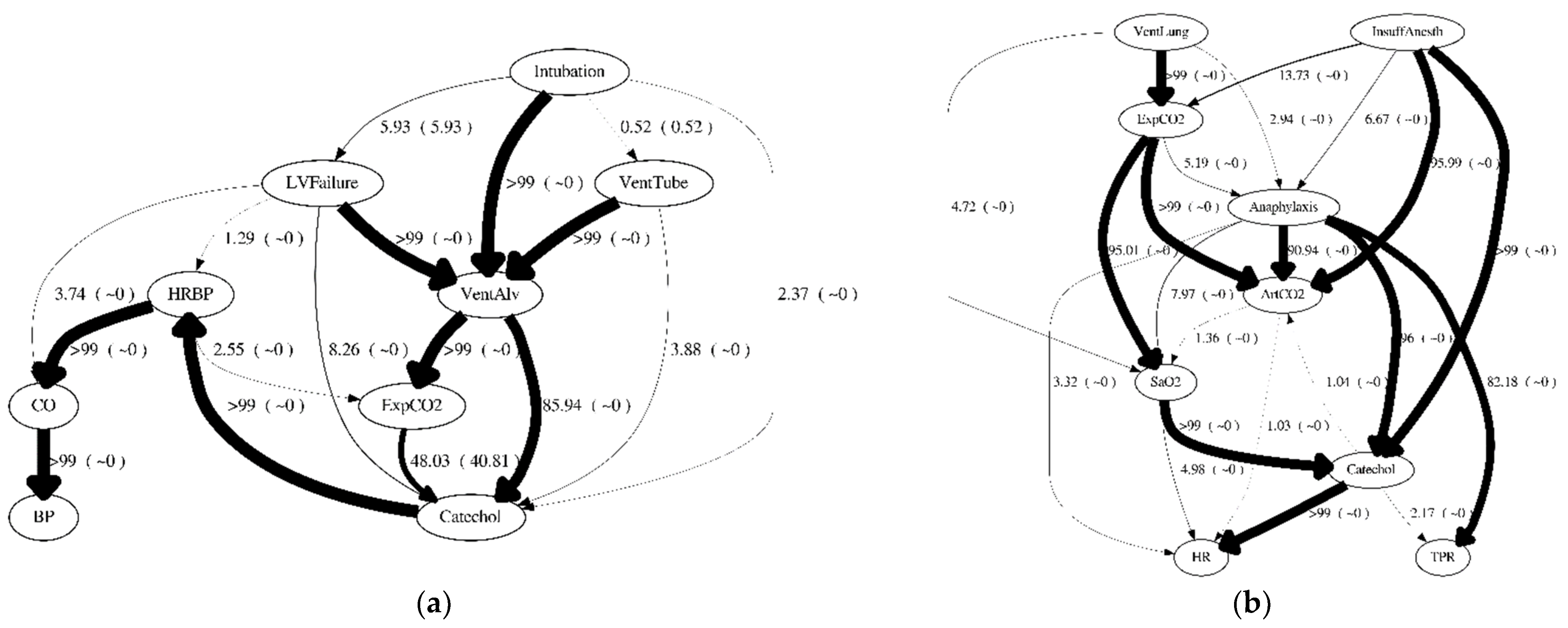
Figure 6 reports the highest scored structure reported by BDe scores for each dataset. It is interesting to note that even with a large number of samples and a significantly more likely Global BDe Structure, i.e., for 1000 cases (D1KS9) and its BDe percentage structure score of >99%, it predicts incorrect mechanisms, e.g., HRBP is predicted as a cause of CO and CO is predicted as a cause of LVFailure (
Figure 6c). However, the generating structure shows that HRBP is not a cause of CO (they are confounded by Catechol), and LVFailure is a cause of CO (
Figure 5a). Another interesting result to notice is that even with many cases (i.e., 1000 cases), the highest BDe scored structure may obtain a mere 4% of the total BDe structure score.
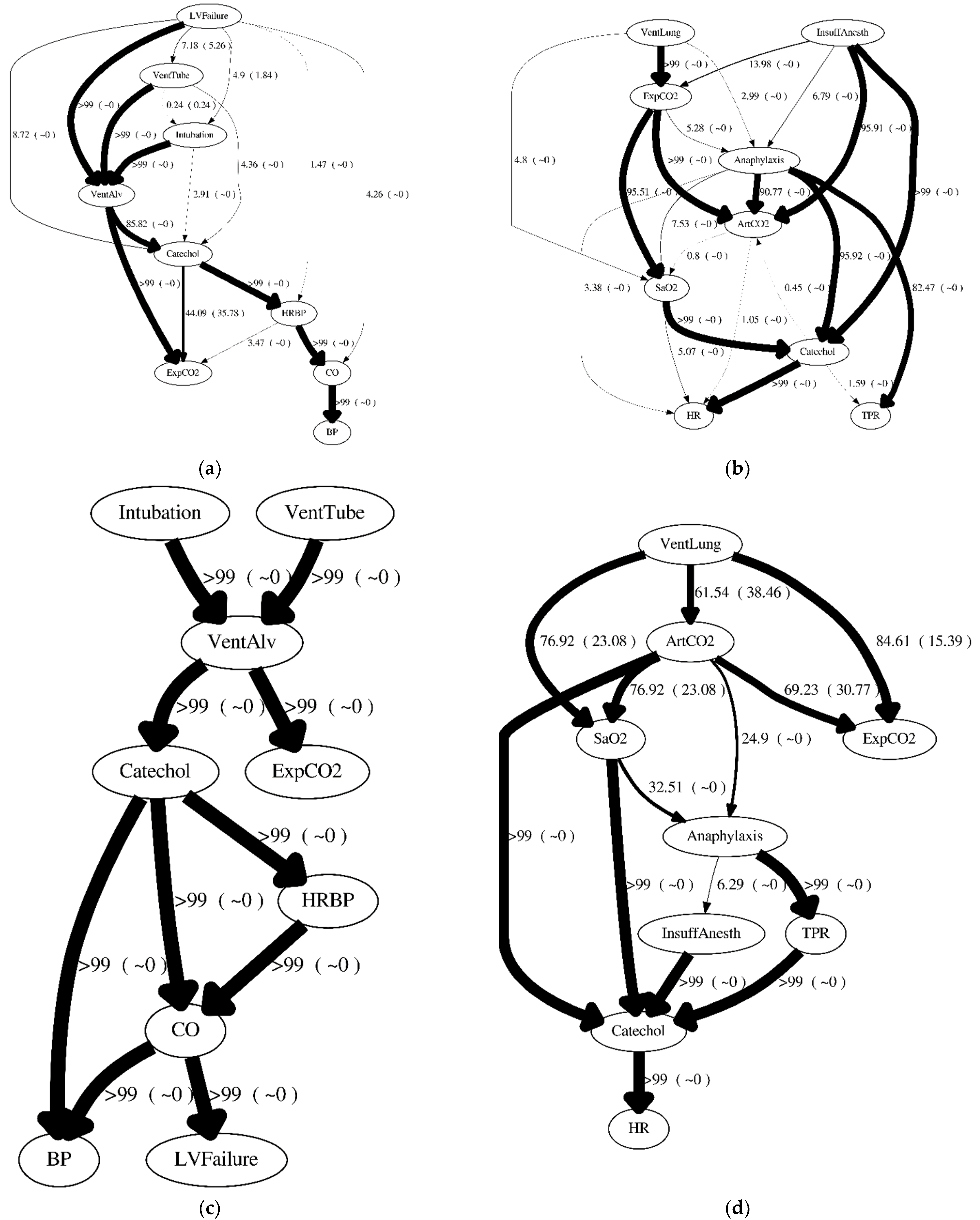
Figure 7 shows consensus structures using
PSG(
D|
RX,Y) (without incorporating the order weight) for D50S9, D50C9, D1KS9, and D1KC9. The arcs thicknesses are based on
PSG(
D|
X→Y) or P
SG(
D|
Y→X). If
PSG(
D|
X→Y) is displayed as a percentage, then
PSG(
D|
Y→X) is also displayed as a percentage in the parentheses. If
PSG(
D|
X→Y) and
PSG(
D|
Y→X) both are less than 0.0001, then no arcs are drawn between
X and
Y. >99 or ~0 indicates where the pairwise causal relationship probability is greater than 0.9999 or less than 0.0001, respectively. Similarly,
Figure 8 shows consensus structures using
PG(
D|
RX,Y) (with incorporating the order weight) for D50S9, D50C9, D1KS9, and D1KC9.
The Global BDe structure using D50S9 was marginally better (maximum likelihood of 0.1423) than other structures. All of the models incorrectly identified causal effects from
LVFailure to
VentAlv; from
Catechol to
ExpCO2; and from
HRBP to
CO when compared to the generating structure (
Figure 5a). In D50S9, the consensus structures generated with the order weight (
Figure 8a) and without the order weight (
Figure 7a) were different than the Global BDe structure (
Figure 6a). A significant difference between the consensus structures generated with the order weight (
Figure 8a), and without the order weight (
Figure 7a), was a causal relationship between
Catechol to
ExpCO2. The consensus structure generated with the order weight predicted P
G(
D|
ExpCO2 → Catechol) = 0.4803 as the most probable relationship; however, the consensus structure generated without the order weight predicted P
G(
D|
Catechol → ExpCO2) = 0.4409 as the most probable relationship, as the generating structure (
Figure 3a) showed that
Catechol and
ExpCO2 had no direct causal influence between each other. It is also noteworthy that one of their common causes,
VentAlv, was correctly predicted to be a common cause in both consensus structures. This showed that, to some extent, we can use the disagreement between the consensus structures generated with and without the order weight to identify confounded relationships without any direct causal relationship.
The Global BDe structure using D50C9 was marginally better (maximum likelihood of 0.1571) than other structures. In D50C9, the consensus structures generated with the order weight (
Figure 8b) or without the order weight (
Figure 7b) were slightly different than the Global BDe structure (
Figure 6b). All models incorrectly identified causal effects from
Anaphylaxis to
ArtCO2; from
InsuffAnesth to
ArtCO2; and predicted a reversed causal direction of
ArtCO2 and
ExpCO2 compared to the generating structure (
Figure 5b). Compared to the same 50 cases, D50S9, no significant differences were observed between the consensus structures generated with the order weight (
Figure 8b) and without the order weight (
Figure 7b).
Only in D1KS9, both consensus structures generated with (
Figure 8c) or without the order weight (
Figure 7c) agreed with the Global BDe structure (
Figure 6c). This is not surprising because the Global BDe structure was significantly better (>0.9999) than any other structures. However, all models incorrectly predicted the following three causal relationships: between
CO and
LVFailure (reversed causal prediction); between
Intubation and
ExpCO2 (missing causal prediction); and added between
Catechol and
BP (unnecessary causal prediction) compared to the generating structure (
Figure 5a).
The Global BDe structure using D1KC9 was marginally better (maximum likelihood of 0.0403). Among the four datasets, it resulted in the lowest maximum likelihood, making D1KC9 the most difficult dataset to learn causal relationships from. All models incorrectly identified a causal effect from
ArtCO2 to
SaO2 (
Figure 5b). In D1KC9, the consensus structures generated with the order weight (
Figure 8d) and without the order weight (
Figure 7d) were different than the Global BDe structure (
Figure 6d). A significant difference between the consensus structures generated with the order weight (
Figure 8d) and without the order weight (
Figure 7d) was the prediction of a causal relationship between
VentLung and
ArtCO2. The consensus structure generated with the order weight predicted P
G(
D|
ArtCO2 → VentLung) = 0.5556 as being the most probable relationship; however, the consensus structure generated without the order weight predicted P
G(
D|
VentLung → ArtCO2) = 0.6154 as being the most probable relationship. As the generating structure (
Figure 3b) shows
VentLung and
ArtCO2 have a direct causal influence between each other and their common cause,
Intubation is hidden in the dataset. This shows how difficult it is to learn reliable causal relationships among the upstream variables in which most of the confounded causes are hidden in the dataset.
We believe all these results are due to the omission of 28 variables and random sampling effects. Also, as the later results will show, with 50 cases, it is more difficult to learn the generating structure of C9, and with 1000 cases it is more difficult to learn the generating structure of S9.
Table 2 shows all the orders (from the total of 9! = 362,880 orders) that received a combined percentage score of >99%. Interestingly, the means were all 7.1429%. However, depending on the dataset, the standard deviation of the scores were different. The data sampled from S9 tended to show tighter percentage scores among the orders than the data sampled from C9. This means that order scores from S9 had less impact than those from C9.
Table 3 summarizes our claim that incorporating the ordering results can help us gain mechanistic knowledge. According to the distances, the BDe score had difficulties in learning the true underlying mechanisms from the generating structure with 50 cases of C9. However, by adding more samples, i.e., with 1000 cases of C9, we improved the ability to learn the true underlying mechanisms from the generating structure.
Overall, the results shown in
Table 3 illustrate that order weight improves in learning the true underlying mechanisms from the generating structure. In the 1000 cases of S9 (D1KS9), as it was mentioned earlier (shown in
Figure 6c), there was only one structure that was significant in terms of BDe score (i.e., >99% of the total BDe structure score). Because of this fact, all orders that were compliant with the dominating structure had a very similar score with a very tight margin, resulting in almost all the same order score (
Table 2). Therefore, in this situation we can see why the order score will not improve in learning the true underlying mechanisms from the generating structure.
Table 4 and
Table 5 compare the structure distances between (1) the algorithm’s predicted structures and the generated structures (Generated δ), and (2) the algorithm’s predicted structures and the best BDe structure scores (Global BDe δ). In some sense, Generated δ measures how well the algorithm learns the underlying mechanism from a phenomenon, and Global BDe δ measures how well the algorithm estimates the best BDe (or BGe) score from the sample.
In 50 cases spanning
Table 4a and
Table 5a, it is clear that all the MCMC ordering algorithms (Random, Prior, and PrePrior) outperformed constrained variant algorithms (BiDAG, K2, and PC) in terms of Generated δ and Global BDe δ with datasets D50S9 and D50C9. Also, in general, algorithms with the order weight predicted better in generating structures (i.e., lower Generated δ and Global BDe δ,) with a higher confidence (i.e., lower variance.)
With the maximum hours (4 h) run, Random and PrePrior converged on their predictions; however, Prior showed some variance in performance. We note that with a lesser number of hours (1 and 2 h), PrePrior showed better performances (better predictions with confidence, i.e., less variance) than Random in D50S9 and comparable predictions in D50C9 (in 1 h run, Random Generated δ was 22.31 with variance of 0.302, and PrePrior Weak Correct achieved Generated δ 22.65 with a very low variance, 0.001 (
Table 4a)).
The structure distances of 1000 cases are shown in
Table 4b and
Table 5b. K2 showed the best Generated δ and Global BDe δ in D1KS9; however, its performance was the lowest among all the algorithms in D1KC9. We believe this was because, in D1KS9, as it was mentioned earlier (shown in
Figure 6c), there was only one structure that was significant in terms of its BDe score (>99% of the total BDe structure scores).
The BiDAG performance in Global BDe δ in D1KS9 was the second best (next to K2′s); however, Generated δ in D1KS9 was either comparable or worse than the MCMC ordering algorithms (Random, Prior, and PrePrior). It seems MCMC ordering algorithms need more than 16 h to converge, although structure distances were generally decreasing in D1KC9, however, that trend is questionable in D1KS9.
We could not find a general pattern as we saw in 50 cases that better predicted the generating structures (lower Generated δ and Global BDe δ) with a higher confidence, i.e., a lower variance with order weight in 1000 cases. We believe this fact has to do with the results that we mentioned earlier, i.e., that MCMC ordering algorithms needs more than 16 h to converge.
With the outstanding performance of K2 in D1KS9 reported earlier, however, we must also mention the outstanding performance of the Prior algorithm with the Strong Correct prior, which achieved a better performance that was statistically significant in a mere 2h run in D1KC9. In D1KC9, all algorithms showed larger than ten for Generated δ, except for Prior. Prior achieved lower than ten for Generated δ with a high confidence (variance of 8.136; significantly lower than the second lowest variance of 18.0 from BiDAG).
Table 6 and
Table 7 compare the Markov blanket distances between the algorithm’s predicted Markov blanket of each variable in the structures (for short, we refer it to MB) and MB in the generated structure (Generated δ), as well as the distance of the algorithm’s predicted MB and the MB of the best BDe structure scores (Global BDe δ).
In 50 cases from
Table 6a and
Table 7a, it is clear that all the MCMC ordering algorithms (Random, Prior, and PrePrior) outperformed the constrained variant algorithms (BiDAG, K2, and PC) in Generated δ and Global BDe δ with datasets D50S9. In dataset D50C9, BiDAG was slightly better (16.0 vs. 16.19) in Generated δ; however, it was significantly worse in Global BDe δ. Also, in general, Generated δ and Global BDe δ of the algorithms with the order weight did not change much because the MB distances were low to begin with (Generated δ ranged from 16.00 to 16.53, and with the order weight it ranged from 16.00 to 16.50; Global BDe δ ranged from 0.00 to 8.93, and with the order weight it ranged from 0.00 to 5.03). We note that with the order weight, the 1h runs in D50C9 showed lower Global BDe δ with a higher confidence, i.e., a lower variance.
With the maximum hour (4 h) run, Random and PrePrior predictions converged; however, Prior showed some variance in its performance. We note that with a smaller number of hours (1 and 2 h) runs, PrePrior showed better performances (better predictions with higher confidence (i.e., lower variance) than Random in D50S9, and comparable performances in D50C9 (in 1 h run, Random Generated δ was 16.17 with variance of 0.0, PrePrior Weak Correct achieved Generated δ 16.16 with a very low variance, 0.0 (
Table 6a).
MB distances of 1000 cases are shown in
Table 6b and
Table 7b. In D1KS9, PrePrior with Strong and Weak Prior achieved the best Generated δ (16.00) with a variance of 12.0. K2 showed the best Global BDe δ (0.0) in D1KS9. Also, in general, Generated δ and Global BDe δ of the algorithms with the order weight did not change much because the MB distances were low to begin with (Generated δ ranged from 7.99 to 18.0, and with the order weight it ranged from 7.81 to 18.0; Global BDe δ ranged from 4.66 (excluding 0.0 from K2) to 13.83 (excluding 18.0 from BiDAG), and with the order weight it ranged from 4.80 (excluding 0.0 from K2) to 13.75 (excluding 18.0 from BiDAG)).
In D1KC9, most of the MCMC ordering algorithms (Random, Prior, and PrePrior) outperformed the constrained variant algorithms (BiDAG, K2, and PC) in Generated δ and Global BDe δ. In 2 h runs, Prior with Weak Correct prior achieved the best Generated δ (7.99; the runner-up was PrePrior Weak Correct prior with 9.15) and Global BDe δ (5.82; the runner-up was PrePrior Weak Correct prior with 8.16); however, the most confident prediction came from PrePrior Weak Correct prior in Generated δ (0.912; the runner-up was Prior Weak Correct prior with 0.938).
Also, in D1KC9 with 4 h runs, PrePrior with Strong Correct prior achieved the best Generated δ (10.27; the runner-up was Random with 10.31) and Random achieved the best Global BDe δ (8.07; the runner-up was PrePrior Strong Correct prior with 9.97). In 16 h runs, Random achieved the best Generated δ (8.38; the runner-up was PrePrior Weak Correct prior with 9.43) and Global BDe δ (4.66; the runner-up was PrePrior Strong Correct prior with 7.26).
Table 8 and
Table 9 show algorithm’s predicted probabilities of four causal pairwise relationships shown in
Figure 4. In all four datasets, all the MCMC ordering algorithms (Random, Prior, and PrePrior) outperformed the constrained variant algorithms (BiDAG, and K2) in the confounded relationships H
X Y (no causal relationship) or H
X→Y (causal relationship). K2 and BiDAG incorrectly predicted (with probability of 0.0) the true underlying confounded relationships: for example, with 1000 cases, using D1KS9, BiDAG predicted all the three true H
X→Y relationships with a probability of 0.0, and using D1KC9, BiDAG, and K2 predicted all of the four true H
X Y relationships with probability of 0.0. Typically, algorithms with the order weight tended to perform better in correctly predicting true causally independent relationships (Ø
X Y and H
X Y) and performed worse in correctly predicting true causal predictions (Ø
X→Y and H
X→Y).
Table 10 and
Table 11 show the algorithm’s most probable prediction rates of four causal pairwise relationships shown in
Figure 4. As it was noticed earlier in
Table 8 and
Table 9, in all four datasets, all the MCMC ordering algorithms (Random, Prior, and PrePrior) outperformed the constrained variant algorithms (BiDAG, and K2) in confounded relationships H
X Y (no causal relationship) or H
X→Y (causal relationship). Algorithms with the order weight changed most the probable prediction rates of the confounded and causally independent predictions (H
X Y) of MCMC ordering algorithms except PrePrior with Weak Correct prior in D50S9 (one relationship prediction of Y→X was changed to X→Y). Another change by weighing order was noticed in D1KC9. There, algorithms with the order weight changed most the probable prediction rates of the confounded and causally independent predictions (H
X Y), and the confounded causal predictions (H
X→Y) of PrePrior with Weak Correct prior. For H
X Y, five relationships prediction of X→Y were correctly changed to the true underlying relationship, X Y; and for H
X→Y, one relationship prediction of Y→X was correctly changed to the true underlying relationship, X→Y.