Comparative Genomic Analysis of Mycobacterium tuberculosis Isolates Circulating in North Santander, Colombia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Type of Study
2.2. Study Population
2.3. Sample Design
2.4. Bacterial Culture and DNA Extraction
2.5. Data Management
2.5.1. Study of Variables
2.5.2. Analysis of Variables
2.6. Total Sequencing of Mtb Genomes
2.7. Annotation and Identity Analysis of Mtb Genomes
2.8. Phylogenetic Analysis
2.9. Identification of Mtb Variants
2.10. Comparative Genomics Analysis
2.11. Ethical Considerations
3. Results
3.1. Population Characteristics
3.2. Annotation and Identity Analysis of Mtb Genomes
3.3. Phylogenetic Analysis
3.4. Identification of Mtb Variants
3.5. Comparative Genomics Analysis
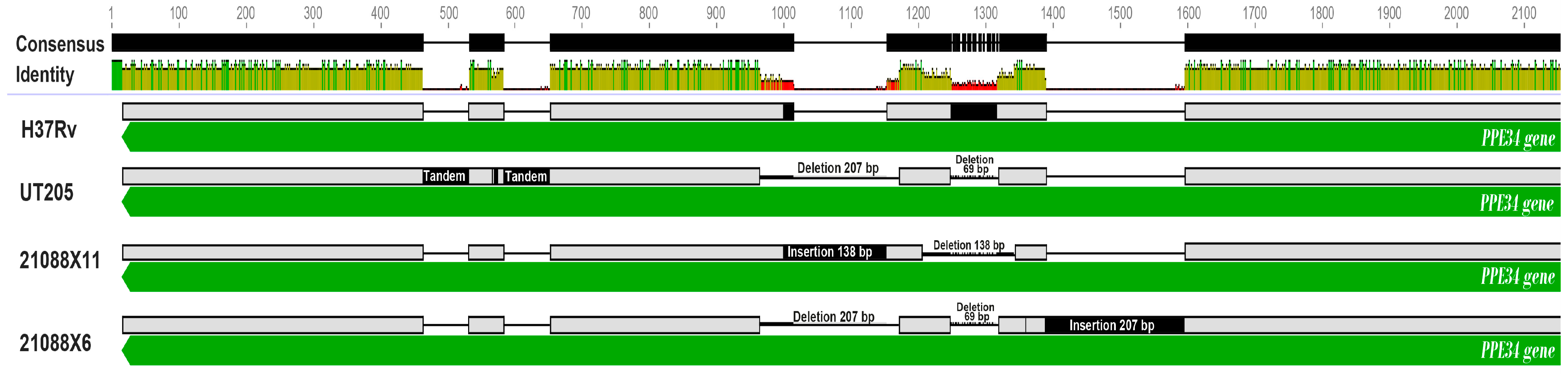
3.5.1. Comparative Genomics Analysis of Mtb Isolates against the H37Rv Genome
3.5.2. Comparative Genomics Analysis of Mtb Isolates against to UT205 Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2023. 2023. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023 (accessed on 7 February 2024).
- WHO. Global Tuberculosis Report 2023. Tuberculosis Data. Interactive Visualizations. Diagnosis Notification and Treatment of Rifampicin-Resistant TB. 2023. Available online: https://www.who.int/teams/global-tuberculosis-programme/data (accessed on 3 March 2024).
- Gagneux, S. Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 2018, 16, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Coscolla, M.; Gagneux, S.; Menardo, F.; Loiseau, C.; Ruiz-Rodriguez, P.; Borrell, S.; Otchere, I.D.; Asante-Poku, A.; Asare, P.; Sánchez-Busó, L.; et al. Phylogenomics of Mycobacterium africanum reveals a new lineage and a complex evolutionary history. Microb. Genom. 2021, 7, 000477. [Google Scholar] [CrossRef] [PubMed]
- Guyeux, C.; Senelle, G.; Meur, A.L.; Supply, P.; Gaudin, C.; Phelan, J.E.; Clark, T.G.; Rigouts, L.; de Jong, B.; Sola, C.; et al. Newly Identified Mycobacterium africanum Lineage 10, Central Africa-Volume 30, Number 3—March 2024-Emerging Infectious Diseases Journal-CDC. Available online: https://wwwnc.cdc.gov/eid/article/30/3/23-1466_article (accessed on 3 March 2024).
- Napier, G.; Campino, S.; Merid, Y.; Abebe, M.; Woldeamanuel, Y.; Aseffa, A.; Hibberd, M.L.; Phelan, J.; Clark, T.G. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020, 12, 114. [Google Scholar] [CrossRef] [PubMed]
- MinSalud; INS. Informe Evento Tuberculosis PEXII 2023 [Internet]. 2023. Available online: https://www.ins.gov.co/buscador-eventos/Informesdeevento/TUBERCULOSIS%20PE%20XII%202023.pdf (accessed on 12 July 2024).
- DANE. Estadísticas Vitales (EEVV). 2022. Available online: https://www.dane.gov.co/files/investigaciones/poblacion/pre_estadisticasvitales_IIItrim_2022pr.pdf (accessed on 7 March 2024).
- DANE. Estadísticas Vitales. Cifra de Defunciones. 2023. Available online: https://www.dane.gov.co/files/operaciones/EEVV/pres-EEVV-Defunciones-IIItrim2023.pdf (accessed on 7 March 2024).
- Plata-Casas, L.; Gutierrez-Lesmes, O.; Cala-Vitery, F. Tuberculosis Disability Adjusted Life Years, Colombia 2010–2018. Trop. Med. Infect. Dis. 2022, 7, 250. [Google Scholar] [CrossRef]
- Jia, X.; Yang, L.; Dong, M.; Chen, S.; Lv, L.; Cao, D.; Fu, J.; Yang, T.; Zhang, J.; Zhang, X.; et al. The Bioinformatics Analysis of Comparative Genomics of Mycobacterium tuberculosis Complex (MTBC) Provides Insight into Dissimilarities between Intraspecific Groups Differing in Host Association, Virulence, and Epitope Diversity. Front. Cell Infect. Microbiol. 2017, 7, 88. [Google Scholar] [CrossRef]
- Brites, D.; Gagneux, S. The Nature and Evolution of Genomic Diversity in the Mycobacterium tuberculosis Complex. In Strain Variation in the Mycobacterium tuberculosis Complex: Its Role in Biology, Epidemiology and Control; Gagneux, S., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–26. [Google Scholar] [CrossRef]
- Gagneux, S. Host–pathogen coevolution in human tuberculosis. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 850–859. [Google Scholar] [CrossRef]
- McHenry, M.L.; Bartlett, J.; Igo, R.P.; Wampande, E.M.; Benchek, P.; Mayanja-Kizza, H.; Fluegge, K.; Hall, N.B.; Gagneux, S.; Tishkoff, S.A.; et al. Interaction between host genes and Mycobacterium tuberculosis lineage can affect tuberculosis severity: Evidence for coevolution? PLoS Genet. 2020, 16, e1008728. [Google Scholar] [CrossRef]
- Zwyer, M.; Rutaihwa, L.K.; Windels, E.; Hella, J.; Menardo, F.; Sasamalo, M.; Sommer, G.; Schmülling, L.; Borrell, S.; Reinhard, M.; et al. Back-to-Africa introductions of Mycobacterium tuberculosis as the main cause of tuberculosis in Dar es Salaam, Tanzania. PLoS Pathog. 2023, 19, e1010893. [Google Scholar] [CrossRef]
- Osei-Wusu, S.; Otchere, I.D.; Morgan, P.; Musah, A.B.; Siam, I.M.; Asandem, D.; Afum, T.; Asare, P.; Asante-Poku, A.; Kusi, K.A.; et al. Genotypic and phenotypic diversity of Mycobacterium tuberculosis complex genotypes prevalent in West Africa. PLoS ONE 2021, 16, e0255433. [Google Scholar] [CrossRef]
- Keating, L.A.; Wheeler, P.R.; Mansoor, H.; Inwald, J.K.; Dale, J.; Hewinson, R.G.; Gordon, S.V. The pyruvate requirement of some members of the Mycobacterium tuberculosis complex is due to an inactive pyruvate kinase: Implications for in vivo growth. Mol. Microbiol. 2005, 56, 163–174. [Google Scholar] [CrossRef]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; Sabio y García, J.; Morbidoni, H.R.; Santangelo, M.d.l.P.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed]
- Sobkowiak, B.; Banda, L.; Mzembe, T.; Crampin, A.C.; Glynn, J.R.; Clark, T.G. Bayesian reconstruction of Mycobacterium tuberculosis transmission networks in a high incidence area over two decades in Malawi reveals associated risk factors and genomic variants. Microb. Genom. 2020, 6, e000361. [Google Scholar] [CrossRef] [PubMed]
- Faksri, K.; Xia, E.; Ong, R.T.-H.; Tan, J.H.; Nonghanphithak, D.; Makhao, N.; Thamnongdee, N.; Thanormchat, A.; Phurattanakornkul, A.; Rattanarangsee, S.; et al. Comparative whole-genome sequence analysis of Mycobacterium tuberculosis isolated from tuberculous meningitis and pulmonary tuberculosis patients. Sci. Rep. 2018, 8, 4910. [Google Scholar] [CrossRef]
- Hernández Pando, R.; Aguilar, D.; Cohen, I.; Guerrero, M.; Ribon, W.; Acosta, P.; Orozco, H.; Marquina, B.; Salinas, C.; Rembao, D.; et al. Specific bacterial genotypes of Mycobacterium tuberculosis cause extensive dissemination and brain infection in an experimental model. Tuberculosis 2010, 90, 268–277. [Google Scholar] [CrossRef]
- Bohada-Lizarazo, D.P. Variabilidad Genómica de Mycobacterium tuberculosis como Elemento Explicativo del alto Riesgo para Tuberculosis del Departamento de Norte de Santander. Doctoral Thesis, Universidad de Antioquia, Medellín, Colombia, 2024. [Google Scholar]
- INS. Tuberculosis. Manual de Procedimientos. Segunda edición. Instituto Nacional de Salud. 1987. Available online: https://www.minsalud.gov.co/sites/rid/Lists/BibliotecaDigital/RIDE/IA/INS/tuberculosis-manual-procedimientos.pdf (accessed on 10 November 2023).
- van Soolingen, D. Molecular epidemiology of tuberculosis and other mycobacterial infections: Main methodologies and achievements. J. Intern. Med. 2001, 249, 1–26. [Google Scholar] [CrossRef]
- Votintseva, A.A.; Pankhurst, L.J.; Anson, L.W.; Morgan, M.R.; Gascoyne-Binzi, D.; Walker, T.M.; Quan, T.P.; Wyllie, D.H.; Del Ojo Elias, C.; Wilcox, M.; et al. Mycobacterial DNA Extraction for Whole-Genome Sequencing from Early Positive Liquid (MGIT) Cultures. J. Clin. Microbiol. 2015, 53, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Prodan, A.; Tremaroli, V.; Brolin, H.; Zwinderman, A.H.; Nieuwdorp, M.; Levin, E. Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PLoS ONE 2020, 15, e0227434. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Gundabolu, S.C.; Vijaykumar, T.N.; Thottethodi, M. FastZ: Accelerating gapped whole genome alignment on GPUs. In Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, St. Louis, MI, USA, 14–19 November 2021; ACM: New York, NY, USA, 2021; pp. 1–13. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- WMA-The World Medical Association-WMA Declaration of Helsinki–Ethical Principles for Medical Research Involving Human Subject. 2022. Available online: https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects/ (accessed on 7 June 2024).
- Narvaiz de Kantor, I.; Kim, S.J.; Frieden, T.R.; Laszlo, A.; Luelmo, F.; Norval, P.Y.; Rieder, H.; Valenzuela, P.; Weyer, K. Laboratory Services in Tuberculosis Control. 1998. Available online: https://iris.who.int/handle/10665/65942 (accessed on 17 July 2024).
- Sánchez-Corrales, L.; Tovar-Aguirre, O.L.; Galeano-Vanegas, N.F.; Jiménez, P.A.C.; Martínez-Vega, R.A.; Maldonado-Londoño, C.E.; Hernández-Botero, J.S.; Siller-López, F. Phylogenomic analysis and Mycobacterium tuberculosis antibiotic resistance prediction by whole-genome sequencing from clinical isolates of Caldas, Colombia. PLoS ONE 2021, 16, e0258402. [Google Scholar] [CrossRef]
- Baena, A.; Cabarcas, F.; Ocampo, J.C.; Barrera, L.F.; Alzate, J.F. Large genomic deletions delineate Mycobacterium tuberculosis L4 sublineages in South American countries. PLoS ONE 2023, 18, e0285417. [Google Scholar] [CrossRef]
- Puerto, G.; Erazo, L.; Wintaco, M.; Castro, C.; Ribón, W.; Guerrero, M.I. Mycobacterium tuberculosis Genotypes Determined by Spoligotyping to Be Circulating in Colombia between 1999 and 2012 and Their Possible Associations with Transmission and Susceptibility to First-Line Drugs. PLoS ONE 2015, 10, e0124308. [Google Scholar] [CrossRef]
- Realpe, T.; Correa, N.; Rozo, J.C.; Ferro, B.E.; Gómez, V.; Zapata, E.; Ribon, W.; Puerto, G.; Castro, C.; Nieto, L.M.; et al. Population structure among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients in Colombia. PLoS ONE 2014, 9, e93848. [Google Scholar] [CrossRef]
- Ritacco, V.; Iglesias, M.-J.; Ferrazoli, L.; Monteserin, J.; Dalla Costa, E.R.; Cebollada, A.; Morcillo, N.; Robledo, J.; de Waard, J.H.; Araya, P.; et al. Conspicuous multidrug-resistant Mycobacterium tuberculosis cluster strains do not trespass country borders in Latin America and Spain. Infect. Genet. Evol. 2012, 12, 711–717. [Google Scholar] [CrossRef]
- Asadi, L.; Croxen, M.; Heffernan, C.; Dhillon, M.; Paulsen, C.; Egedahl, M.L.; Tyrrell, G.; Doroshenko, A.; Long, R. How much do smear-negative patients really contribute to tuberculosis transmissions? Re-examining an old question with new tools. eClinicalMedicine 2022, 43, 101250. [Google Scholar] [CrossRef] [PubMed]
- Ruíz-Roa, S.L.; Rojas, S.M.M.; Galeano, I.A.F.S. Tuberculosis en la frontera colombo-venezolana: Distribución geoespacial. Rev. Peru Med. Exp. Salud Pública 2022, 39, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Shastry, B.S. SNPs: Impact on Gene Function and Phenotype. In Single Nucleotide Polymorphisms: Methods and Protocols; Komar, A.A., Ed.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 3–22. [Google Scholar] [CrossRef]
- Tischler, A.D.; Leistikow, R.L.; Kirksey, M.A.; Voskuil, M.I.; McKinney, J.D. Mycobacterium tuberculosis Requires Phosphate-Responsive Gene Regulation To Resist Host Immunity. Infect. Immun. 2013, 81, 317–328. [Google Scholar] [CrossRef]
- White, D.W.; Elliott, S.R.; Odean, E.; Bemis, L.T.; Tischler, A.D. Mycobacterium tuberculosis Pst/SenX3-RegX3 Regulates Membrane Vesicle Production Independently of ESX-5 Activity. mBio 2018, 9, e00778-18. [Google Scholar] [CrossRef]
- Tischler, A.D.; Leistikow, R.L.; Ramakrishnan, P.; Voskuil, M.I.; McKinney, J.D. Mycobacterium tuberculosis Phosphate Uptake System Component PstA2 Is Not Required for Gene Regulation or Virulence. PLoS ONE 2016, 11, e0161467. [Google Scholar] [CrossRef] [PubMed]
- Argyrou, A.; Vetting, M.W.; Blanchard, J.S. Characterization of a New Member of the Flavoprotein Disulfide Reductase Family of Enzymes from Mycobacterium tuberculosis. J. Biol. Chem. 2004, 279, 52694–52702. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.S.S.; Siu, G.K.H.; Tam, K.K.G.; To, S.W.C.; Rajwani, R.; Ho, P.L.; Wong, S.S.-Y.; Zhao, W.W.; Ma, O.C.-K.; Yam, W.-C. Comparative Genomic Analysis of Two Clonally Related Multidrug Resistant Mycobacterium tuberculosis by Single Molecule Real Time Sequencing. Front. Cell. Infect. Microbiol. 2017, 7, 478. [Google Scholar] [CrossRef]
- Jiang, Y.; Wei, J.; Liu, H.; Li, G.; Guo, Q.; Qiu, Y.; Zhao, L.; Li, M.; Zhao, X.; Dou, X.; et al. Polymorphisms in the PE35 and PPE68 antigens in Mycobacterium tuberculosis strains may affect strain virulence and reflect ongoing immune evasion. Mol. Med. Rep. 2016, 13, 947–954. [Google Scholar] [CrossRef]
- D’Souza, C.; Kishore, U.; Tsolaki, A.G. La familia PE-PPE de Mycobacterium tuberculosis: Proteínas camufladas. Immunobiology 2023, 228, 152321. [Google Scholar] [CrossRef]
- Bansal, K.; Sinha, A.Y.; Ghorpade, D.S.; Togarsimalemath, S.K.; Patil, S.A.; Kaveri, S.V.; Balaji, K.N.; Bayry, J. Src Homology 3-interacting Domain of Rv1917c of Mycobacterium tuberculosis Induces Selective Maturation of Human Dendritic Cells by Regulating PI3K-MAPK-NF-κB Signaling and Drives Th2 Immune Responses. J. Biol. Chem. 2010, 285, 36511–36522. [Google Scholar] [CrossRef]
- Gómez-González, P.J.; Grabowska, A.D.; Tientcheu, L.D.; Tsolaki, A.G.; Hibberd, M.L.; Campino, S.; Phelan, J.E.; Clark, T.G. Functional genetic variation in pe/ppe genes contributes to diversity in Mycobacterium tuberculosis lineages and potential interactions with the human host. Front. Microbiol. 2023, 14, 1244319. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, C.R.E.; Cloete, R.; Müller, B.; Schürch, A.C.; van Helden, P.D.; Gagneux, S.; Warren, R.M.; van Pittius, N.C.G. Comparative Analysis of Mycobacterium tuberculosis pe and ppe Genes Reveals High Sequence Variation and an Apparent Absence of Selective Constraints. PLoS ONE 2012, 7, e30593. [Google Scholar] [CrossRef] [PubMed]
- Guilhot, C.; Chalut, C.; Daffé, M. Biosynthesis and Roles of Phenolic Glycolipids and Related Molecules in Mycobacterium tuberculosis. In The Mycobacterial Cell Envelope; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2008; pp. 271–289. [Google Scholar] [CrossRef]
- Pérez, E.; Constant, P.; Laval, F.; Lemassu, A.; Lanéelle, M.-A.; Daffé, M.; Guilhot, C. Molecular dissection of the role of two methyltransferases in the biosynthesis of phenolglycolipids and phthiocerol dimycoserosate in the Mycobacterium tuberculosis complex. J. Biol. Chem. 2004, 279, 42584–42592. [Google Scholar] [CrossRef] [PubMed]
- Tundup, S.; Mohareer, K.; Hasnain, S.E. Mycobacterium tuberculosis PE25/PPE41 protein complex induces necrosis in macrophages: Role in virulence and disease reactivation? FEBS Open Bio 2014, 4, 822–828. [Google Scholar] [CrossRef]
- Stucki, D.; Brites, D.; Jeljeli, L.; Coscolla, M.; Liu, Q.; Trauner, A.; Fenner, L.; Rutaihwa, L.; Borrell, S.; Luo, T.; et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat. Genet. 2016, 48, 1535–1543. [Google Scholar] [CrossRef]
- Noda, G.S.; Barrios, M.F.; Díaz, Y.R. Evasión del sistema inmune por Mycobacterium tuberculosis: Mecanismos moleculares. Rev. Cuba Tecnol. Salud 2018, 9, 191–204. [Google Scholar]
- Fishbein, S.; van Wyk, N.; Warren, R.M.; Sampson, S.L. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol. Microbiol. 2015, 96, 901–916. [Google Scholar] [CrossRef]
- Baena, A.; Cabarcas, F.; Alvarez-Eraso, K.L.F.; Isaza, J.P.; Alzate, J.F.; Barrera, L.F. Differential determinants of virulence in two Mycobacterium tuberculosis Colombian clinical isolates of the LAM09 family. Virulence 2019, 10, 695–710. [Google Scholar] [CrossRef]
- Isaza, J.P.; Duque, C.; Gomez, V.; Robledo, J.; Barrera, L.F.; Alzate, J.F. Whole genome shotgun sequencing of one Colombian clinical isolate of Mycobacterium tuberculosis reveals DosR regulon gene deletions. FEMS Microbiol. Lett. 2012, 330, 113–120. [Google Scholar] [CrossRef]
- Baruzzo, G.; Serafini, A.; Finotello, F.; Sanavia, T.; Cioetto-Mazzabò, L.; Boldrin, F.; Lavezzo, E.; Barzon, L.; Toppo, S.; Provvedi, R.; et al. Role of the Extracytoplasmic Function Sigma Factor SigE in the Stringent Response of Mycobacterium tuberculosis. Microbiol. Spectr. 2023, 11, e0294422. [Google Scholar] [CrossRef]
- Sachdeva, P.; Misra, R.; Tyagi, A.K.; Singh, Y. The sigma factors of Mycobacterium tuberculosis: Regulation of the regulators. FEBS J. 2010, 277, 605–626. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Pooja Borah, K. Mycobacterium tuberculosis carbon and nitrogen metabolic fluxes. Biosci. Rep. 2022, 42, BSR20211215. [Google Scholar] [CrossRef]
- Xu, J.Y.; Zhao, L.; Xu, Y.; Li, B.; Zhai, L.; Tan, M.; Ye, B.C. Dynamic Characterization of Protein and Posttranslational Modification Levels in Mycobacterial Cholesterol Catabolism. mSystems 2020, 5, e00424-19. [Google Scholar] [CrossRef] [PubMed]
- Abou-Fadel, J.S. Steroid-CoA Ligases, FadD17A1 and FadD19A1, and Their Role in Cholesterol Side-Chain Degradation in Mycobacterium tuberculosis. ETD Collect Univ Tex El Paso. 2018, pp. 1–129. Available online: https://scholarworks.utep.edu/dissertations/AAI10814786 (accessed on 6 July 2024).
- López, M.; Quitian, L.V.; Calderón, M.N.; Soto, C.Y. The P-type ATPase CtpG preferentially transports Cd2+ across the Mycobacterium tuberculosis plasma membrane. Arch. Microbiol. 2018, 200, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Botella, H.; Peyron, P.; Levillain, F.; Poincloux, R.; Poquet, Y.; Brandli, I.; Wang, C.; Tailleux, L.; Tilleul, S.; Charrière, G.M.; et al. Mycobacterial P1-Type ATPases Mediate Resistance to Zinc Poisoning in Human Macrophages. Cell Host Microbe 2011, 10, 248–259. [Google Scholar] [CrossRef]
- Chaiyachat, P.; Chaiprasert, A.; Nonghanphithak, D.; Smithtikarn, S.; Kamolwat, P.; Pungrassami, P.; Reechaipichitkul, W.; Ong, R.T.-H.; Teo, Y.-Y.; Faksri, K. Whole-genome analysis of drug-resistant Mycobacterium tuberculosis reveals novel mutations associated with fluoroquinolone resistance. Int. J. Antimicrob. Agents 2021, 58, 106385. [Google Scholar] [CrossRef]





| Sample | Sex | Age | Municipality of Origen | Commune/Township of Cúcuta | Overcrowding | Homeless | Type of TB | Comorbidity | HIV Result | BK Result * | Drug Susceptibility |
|---|---|---|---|---|---|---|---|---|---|---|---|
| col180 | M | 42 | Cúcuta | 6 | No | HL | Pulmonary | HIV | Positive | 2+ | Sensitive |
| col179 | M | 42 | Cúcuta | 6 | No | HL | Pulmonary | HIV | Positive | 3+ | Sensitive |
| col119 | F | 45 | Cúcuta | 4 | No | No | Pulmonary | None | Negative | Negative | Sensitive |
| col41 | M | 68 | Cúcuta | Aguaclara | No | No | Pulmonary | None | Negative | 2+ | Sensitive |
| col207 | F | 56 | Cúcuta | 6 | No | No | Pulmonary | None | Negative | Negative | Sensitive |
| col178 | M | 32 | Cúcuta | 5 | No | No | Pulmonary | None | Negative | 2+ | Sensitive |
| col137 | M | 71 | Cúcuta | 3 | No | No | Pulmonary | DM | Negative | 1+ | Sensitive |
| col177 | M | 73 | Chinácota | Without commune | No | No | Pulmonary | None | Negative | 1+ | Sensitive |
| col117 | M | 26 | Cúcuta | 9 | No | No | Pulmonary | None | Negative | Negative | Sensitive |
| col213 | F | 17 | El Zulia | Without commune | No | No | Pulmonary | None | Negative | 3+ | Sensitive |
| col201 | M | 72 | Tibú | Without commune | No | No | Pulmonary | None | Negative | 1+ | Sensitive |
| col174 | F | 28 | Cúcuta | Buena Esperanza | No | No | Pulmonary | Malnutrition | Negative | 1+ | Sensitive |
| col173 | M | 51 | Cúcuta | 7 | No | No | Pulmonary | Malnutrition | Negative | 1+ | Sensitive |
| col132 | M | 71 | Cúcuta | 9 | No | HL | Pulmonary | Malnutrition | Negative | 3+ | Sensitive |
| col124 | F | 33 | Cúcuta | 3 | No | No | Pulmonary | None | Negative | Negative | Sensitive |
| col123 | F | 33 | Cúcuta | 3 | No | No | Pulmonary | None | Negative | Negative | Sensitive |
| col210 | M | 32 | Cúcuta | 2 | No | No | Extrapulmonary | None | Negative | Without Data | RR |
| col121 | F | 22 | Cúcuta | 1 | No | No | Pulmonary | None | Negative | 3+ | Sensitive |
| Gene Involved | Protein Coding | Type of Modification | Variant Frequency | Isolates Involved | Protein Effect | Biological Function |
|---|---|---|---|---|---|---|
| rv2958c | PGL/p-HBAD biosynthesis glycosyltransferase | Substitutions | 11.10% | 21088X12, X15 | Protein spread | PGL/p-HBAD Biosynthesis |
| rv2962c | PGL/p-HBAD biosynthesis rhamnosyltransferase | SNP Transition | 22% | 21088X4, X5, X12, X13 | Truncation | |
| rv2959c | Rhamnosyl O-methyltransferase | SNP Transition | 38.9% | 21088X1, X2, X8, X11, X14, X15, X17 | Truncation | |
| rv0279c | PE PGRS4 | Substitutions | 33.3% | 21088X1, X6, X7, X9, X10, X11, X12, X14, X17 | Protein spread | Virulence and evasion of the host immune response |
| rv0978c | PE PGRS17 | SNP transversion | 41.90% | 21088X1, X2, X3, X4, X5, X6, X7, X8, X9, X11, X13, X16, X18 | Protein spread | |
| rv0980c | PE PGRS18 | SNP transversion | 21.70% | 21088X10, X12, X14, X15, X17 | Truncation | |
| rv3344c | PE PGRS49 | Substitutions | 35.70% | 21088X1, X2, X4, X5, X7, X9, X10, X12, X13, X14 | Loss of start codon | |
| rv3508 | PE PGRS54 | Substitutions | 5.3% | 21088X5 | Protein spread | |
| 23.50% | 21088X1, X2, X3, X6, X12, X13, X15, X18 | Truncation | ||||
| rv0304c | PPE5 | Substitutions | 50% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18 | Loss of start codon | |
| rv0305c | PPE6 | Substitutions | 2.9% | 21088X11 | Truncation | |
| 45.70% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X12, X13, X14, X15, X16, X17, X18 | Protein spread | ||||
| rv0354c | PPE7 | SNP transversion | 47.10% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X12, X13, X14, X15, X16, X17, X18 | Protein spread | |
| rv1917c | PPE34 | SNP | 8.20% | 21088X12, X13, X17 | Protein spread | |
| rv3125c | PPE49 | SNP transversion | 22.20% | 21088X3, X6, X16, X18 | Truncation | |
| rv3872 | PE35 | SNP transversion | 100% | 21088X1-18 | Truncation | |
| rv0180c | Transmembrane protein | SNP Transition | 5.60% | 21088X9 | Truncation | Membrane proteins |
| rv0446c | Transmembrane protein | SNP Transition | 72% | 21088X1, X2, X4, X5, X7, X8, X10, X11, X12, X13, X14, X15, X17 | Truncation | |
| rv1624c | Integral membrane protein | SNP Transition | 11.10% | 21088X4, X5 | Truncation | |
| rv2120c | Integral membrane protein | SNP Transition | 11.10% | 21088X10, X11 | Truncation | |
| rv2395 | Membrane protein | SNP Transition | 11.10% | 21088X12, X13 | Truncation | |
| rv3870 | EccCa1 CDS | Substitutions | 35.70% | 21088X3, X5, X6, X7, X8, X9, X12, X14, X16, X17 | Protein spread | Proteins related to the immune response |
| rv0305c | PPE6 CDS | Substitutions | 2.90% | 21088X11 | Truncation | |
| 45% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X12, X13, X14, X15, X16, X17, X18 | Protein spread | ||||
| rv0354c | PPE7 CDS | SNP Transition | 47.10% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X12, X13, X14, X15, X16, X17, X18 | Extension | |
| rv0577 | TB27.3 | SNP transversion | 5.60% | 21088X11 | Truncation | |
| rv0113 | GmhA CDS | SNP transversion | 5.90% | 21088X2 | Truncation | |
| rv2221c | GlnE CDS | SNP Transition | 5.60% | 21088X9 | Loss of start codon | |
| rv2673 | AftC CDS | SNP Transition | 33.30% | 21088X3, X4, X7, X8, X9, X12, X13, X14, X18 | Truncation | |
| rv0197 | Oxidoreductase | SNP transversion | 88.90% | 21088X1, X2, X3, X6, X7, X8, X9, X10, X11, X12, X13, X14, X15, X16, X17, X18 | Truncation | Enzymes and metabolic proteins |
| rv0389 | PurT CDS | SNP Transition | 5.60% | 21088X9 | Truncation | |
| rv0873 | FadE10 CDS | SNP Transversion | 5.60% | 21088X17 | Truncation | |
| rv0930 | PstA1 CDS | SNP Transition | 100% | 21088X1-18 | Truncation | |
| rv3287c | RsbW CDS | SNP Transversion | 5.60% | 21088X17 | Truncation | |
| rv3303c | LpdA CDS | SNP Transversion | 100% | 21088X1-18 | Truncation | |
| rv3618 | Monooxygenase | SNP Transition | 11.10% | 21088X4, X5 | Truncation | |
| rv3378c | Diterpene synthase | SNP Transversion | 11.10% | 21088X1, X14 | Truncation | |
| rv3701c | EgtD CDS | SNP Transversion | 5.60% | 21088X9 | Truncation | |
| rv3097c | LipY CDS | SNP Transition | 5.60% | 21088X6 | Truncation | |
| rv1167c | Transcriptional regulator | SNP Transversion | 5.60% | 21088X2 | Protein spread | Regulation and transcription proteins |
| rv3050c | Asn C family transcriptional regulator | SNP Transition | 5.60% | 21088X17 | Truncation | |
| rv1189 | SigI CDS | SNP Transversion | 5.60% | 21088X7 | Truncation | |
| rv1329c | DinG CDS | SNP Transversion | 5.60% | 21088X10 | Truncation | |
| rv1493 | MutB CDS | SNP Transition | 47.10% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X15, X16, X17, X18 | Truncation | |
| rv1494 | MazE4 CDS | SNP Transversion | 47.10% | 21088X2, X3, X4, X5, X6, X7, X8, X9, X10, X11, X12, X13, X15, X16, X17, X18 | Truncation | |
| rv1574 | Phage protein | SNP Transversion | 50% | 21088X3, X6, X9, X16, X18 | Protein spread | |
| rv1575 | Phage protein | SNP Transversion | 50% | 21088X3, X6, X9, X16, X18 | Truncation |
| Gene Involved | Protein Coding | Type of Modification | Variant Frequency | Protein Effect | Biological Function |
|---|---|---|---|---|---|
| rv1917c | PPE34 CDS | Nucleotide insertion | 100% | Protein spread | Virulence and evasion of the host immune response |
| rv0355c | PPE8 CDS | ||||
| rv1039c | PPE15 CDS | ||||
| rv19c | PPE59 CDS | ||||
| rv0297 | PE PGRS5 CDS | ||||
| rv2487c | PE PGRS42 CDS | ||||
| rv0376 | PE PGRS59 CDS | ||||
| rv3652 | PE PGRS60 CDS | ||||
| rv356 | PE PGRS51 CDS | ||||
| ppsA | PpsA CDS | Substitution | 100% | No effect | Lipid biosynthesis and stress response |
| kdtB | KdtB CDS | Substitution | |||
| sigJ | SigJ CDS | Substitution | |||
| sigM | SigM CDS | Tandem deletion | Change in reading frame | ||
| aldA | AldA CDS | SNPs | 100% | Amino acide residue change No effects of truncation or change in reading frames have been observed | Lipid and carbohydrate metabolism |
| rocA | RocA CDS | SNPs | |||
| cyp144 | Cyp144 CDS | SNPs | |||
| pykA | PykA CDS | SNPs | |||
| gnd1 | Gnd1 CDS | SNPs | |||
| gnd1 | Gnd1 CDS | Insertion | Change in reading frame | ||
| glpQ1 | GlpQ1 CDS | Tandem deletion | |||
| plsB1 | PlsB1 CDS | SNPs | 100% | Amino acide residue change No effects of truncation or change in reading frames have been observed | Lipid and carbohydrate metabolism |
| pks1 | Pks1 CDS | ||||
| pks5 | Pks5 CDS | ||||
| pks7 | Pks7 CDS | ||||
| pks9 | Pks9 CDS | ||||
| fadB3 | FadB3 CDS | ||||
| fadE28 | FadE28 CDS | ||||
| fadD35 | FadD35 CDS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohada-Lizarazo, D.P.; Bravo-Sanabria, K.D.; Cárdenas-Malpica, P.; Rodríguez, R. Comparative Genomic Analysis of Mycobacterium tuberculosis Isolates Circulating in North Santander, Colombia. Trop. Med. Infect. Dis. 2024, 9, 197. https://doi.org/10.3390/tropicalmed9090197
Bohada-Lizarazo DP, Bravo-Sanabria KD, Cárdenas-Malpica P, Rodríguez R. Comparative Genomic Analysis of Mycobacterium tuberculosis Isolates Circulating in North Santander, Colombia. Tropical Medicine and Infectious Disease. 2024; 9(9):197. https://doi.org/10.3390/tropicalmed9090197
Chicago/Turabian StyleBohada-Lizarazo, Diana Patricia, Karen Dayana Bravo-Sanabria, Paola Cárdenas-Malpica, and Raúl Rodríguez. 2024. "Comparative Genomic Analysis of Mycobacterium tuberculosis Isolates Circulating in North Santander, Colombia" Tropical Medicine and Infectious Disease 9, no. 9: 197. https://doi.org/10.3390/tropicalmed9090197
APA StyleBohada-Lizarazo, D. P., Bravo-Sanabria, K. D., Cárdenas-Malpica, P., & Rodríguez, R. (2024). Comparative Genomic Analysis of Mycobacterium tuberculosis Isolates Circulating in North Santander, Colombia. Tropical Medicine and Infectious Disease, 9(9), 197. https://doi.org/10.3390/tropicalmed9090197






