Spatiotemporal Patterns of the Omicron Wave of COVID-19 in the United States
Abstract
1. Introduction
2. Materials and Methods
2.1. Time Series COVID-19 Data
2.2. Space-Time Scan Statistic
2.3. The Hoover Index
2.4. Epicenter of COVID-19
3. Results
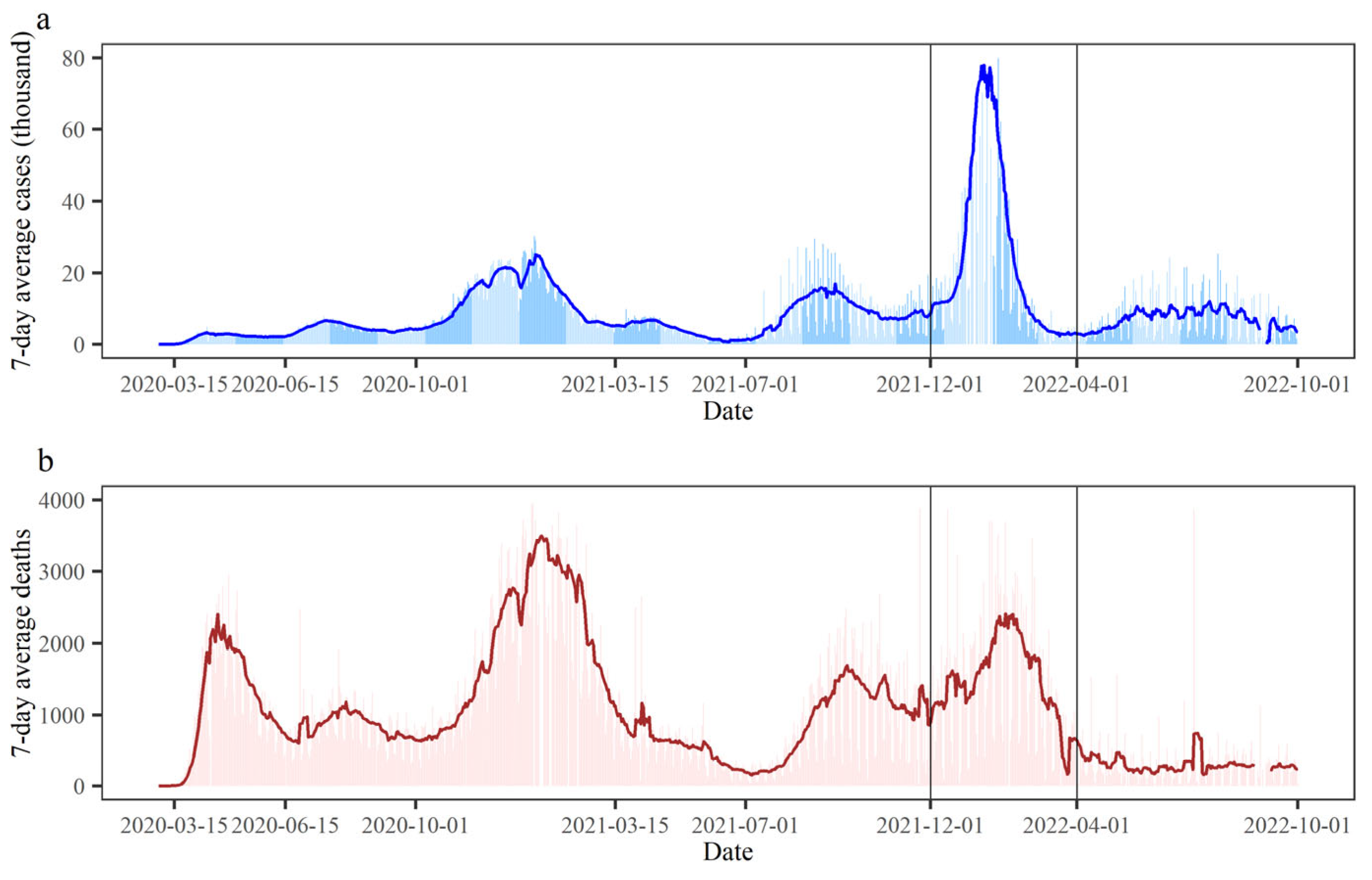
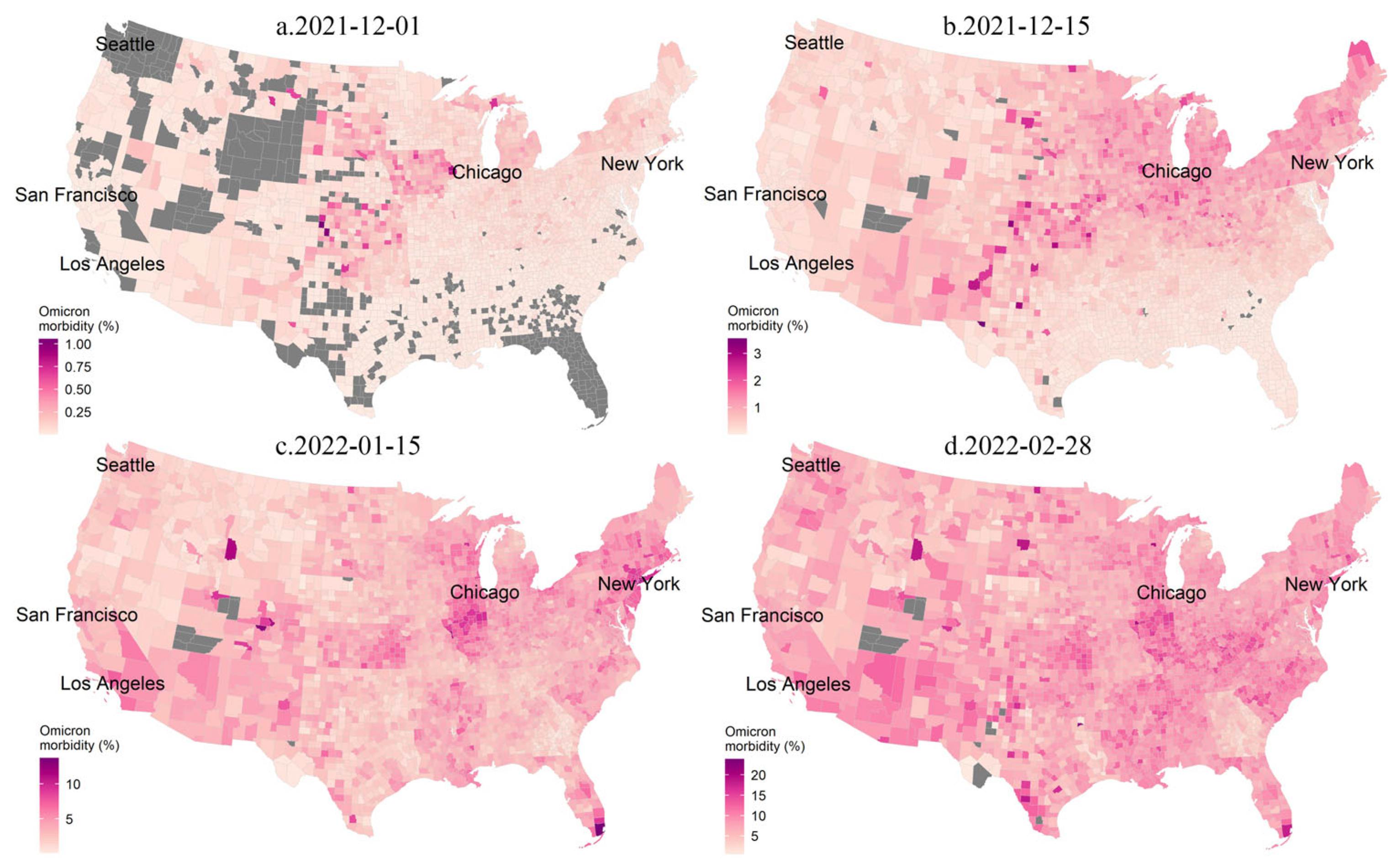
3.1. Spatiotemporal Variabilities of the COVID-19 Epidemic
3.2. Space-Time Scan Analysis
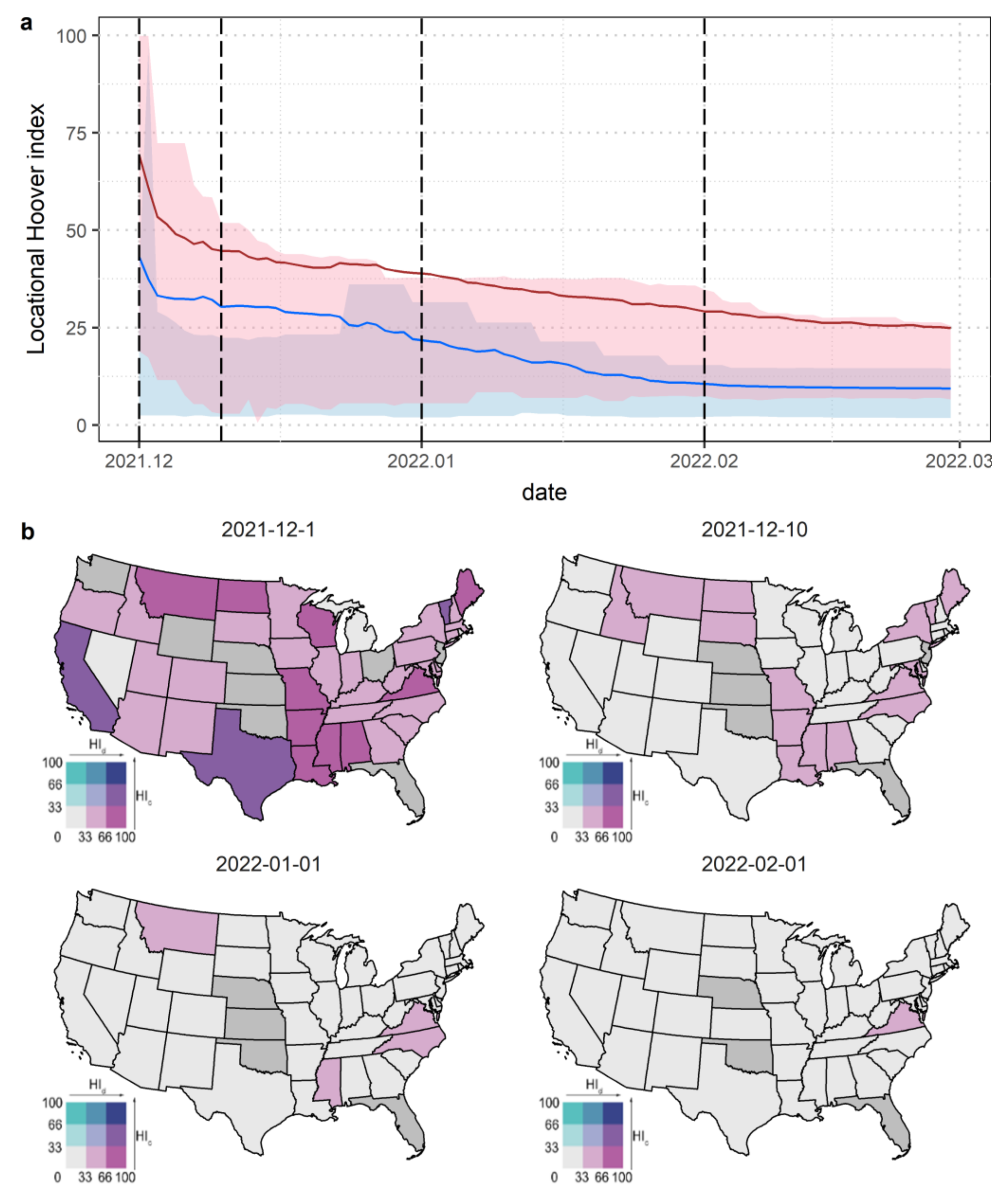
3.3. Hoover Index Analysis
3.4. Spatial Transformation of the Epicenter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, P.; Yang, S.; Dai, S.; How Jin Aik, D.; Yang, S.; Jia, P. Global spreading of Omicron variant of COVID-19. Geospat. Health 2022, 17. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.S.A.; Karim, Q.A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 2021, 398, 2126–2128. [Google Scholar] [CrossRef] [PubMed]
- Fokas, A.S.; Kastis, G.A. SARS-CoV-2: The second wave in Europe. J. Med. Internet Res. 2021, 23, e22431. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Gong, Z.; Guo, J.; Cheng, Q.; Yao, Z. Spatiotemporal analysis of the morbidity of global Omicron from November 2021 to February 2022. J. Med. Virol. 2022, 94, 5354–5362. [Google Scholar] [CrossRef]
- Del Rio, C.; Omer, S.B.; Malani, P.N. Winter of Omicron—The evolving COVID-19 pandemic. JAMA 2022, 327, 319–320. [Google Scholar] [CrossRef]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Liu, M.; Liu, M.; Li, Z.; Zhu, Y.; Liu, Y.; Wang, X.; Tao, L.; Guo, X. The spatial clustering analysis of COVID-19 and its associated factors in mainland China at the prefecture level. Sci. Total Environ. 2021, 777, 145992. [Google Scholar] [CrossRef]
- Li, W.; Zhang, P.; Zhao, K.; Zhao, S. The geographical distribution and influencing factors of COVID-19 in China. Trop. Med. Infect. Dis. 2022, 7, 45. [Google Scholar] [CrossRef]
- Desjardins, M.R.; Hohl, A.; Delmelle, E.M. Rapid surveillance of COVID-19 in the United States using a prospective space-time scan statistic: Detecting and evaluating emerging clusters. Appl. Geogr. 2020, 118, 102202. [Google Scholar] [CrossRef]
- Hohl, A.; Delmelle, E.M.; Desjardins, M.R.; Lan, Y. Daily surveillance of COVID-19 using the prospective space-time scan statistic in the United States. Spat. Spatio-Temporal Epidemiol. 2020, 34, 100354. [Google Scholar] [CrossRef]
- Xu, F.; Beard, K. A comparison of prospective space-time scan statistics and spatiotemporal event sequence based clustering for COVID-19 surveillance. PLoS ONE 2021, 16, e0252990. [Google Scholar] [CrossRef]
- Rader, B.; Scarpino, S.V.; Nande, A.; Hill, A.L.; Adlam, B.; Reiner, R.C.; Pigott, D.M.; Gutierrez, B.; Zarebski, A.E.; Shrestha, M.; et al. Crowding and the shape of COVID-19 epidemics. Nat. Med. 2020, 26, 1829–1834. [Google Scholar] [CrossRef]
- Marceló-Díaz, C.; Lesmes, M.C.; Santamaría, E.; Salamanca, J.A.; Fuya, P.; Cadena, H.; Muñoz-Laiton, P.; Morales, C.A. Spatial Analysis of Dengue Clusters at Department, Municipality and Local Scales in the Southwest of Colombia, 2014–2019. Trop. Med. Infect. Dis. 2023, 8, 262. [Google Scholar] [CrossRef]
- Xu, G.; Jiang, Y.; Wang, S.; Qin, K.; Ding, J.; Liu, Y.; Lu, B. Spatial disparities of self-reported COVID-19 cases and influencing factors in Wuhan, China. Sustain. Cities Soc. 2022, 76, 103485. [Google Scholar] [CrossRef]
- Tian, H.; Liu, Y.; Li, Y.; Wu, C.-H.; Chen, B.; Kraemer, M.U.; Li, B.; Cai, J.; Xu, B.; Yang, Q. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in China. Science 2020, 368, 638–642. [Google Scholar] [CrossRef]
- Kraemer, M.U.; Yang, C.-H.; Gutierrez, B.; Wu, C.-H.; Klein, B.; Pigott, D.M.; Open COVID-19 Data Working Group; Du Plessis, L.; Faria, N.R.; Li, R.; et al. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science 2020, 368, 493–497. [Google Scholar] [CrossRef]
- Xu, G.; Wang, W.; Lu, D.; Lu, B.; Qin, K.; Jiao, L. Geographically varying relationships between population flows from Wuhan and COVID-19 cases in Chinese cities. Geo-Spat. Inf. Sci. 2022, 25, 121–131. [Google Scholar] [CrossRef]
- Zhu, D.; Ye, X.; Manson, S. Revealing the spatial shifting pattern of COVID-19 pandemic in the United States. Sci. Rep. 2021, 11, 8396. [Google Scholar] [CrossRef]
- Ives, A.R.; Bozzuto, C. Estimating and explaining the spread of COVID-19 at the county level in the USA. Commun. Biol. 2021, 4, 60. [Google Scholar] [CrossRef]
- Maroko, A.R.; Nash, D.; Pavilonis, B.T. COVID-19 and Inequity: A Comparative Spatial Analysis of New York City and Chicago Hot Spots. J. Urban Health 2020, 97, 461–470. [Google Scholar] [CrossRef]
- McMahon, T.; Chan, A.; Havlin, S.; Gallos, L.K. Spatial correlations in geographical spreading of COVID-19 in the United States. Sci. Rep. 2022, 12, 699. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Darzi, A.; Kabiri, A.; Zhao, G.; Luo, W.; Xiong, C.; Zhang, L. Quantifying human mobility behaviour changes during the COVID-19 outbreak in the United States. Sci. Rep. 2020, 10, 20742. [Google Scholar] [CrossRef]
- Verma, R.; Yabe, T.; Ukkusuri, S.V. Spatiotemporal contact density explains the disparity of COVID-19 spread in urban neighborhoods. Sci. Rep. 2021, 11, 10952. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.; Prentice, C.R. Using spatial and temporal modeling to visualize the effects of U.S. state issued stay at home orders on COVID-19. Sci. Rep. 2021, 11, 13939. [Google Scholar] [CrossRef] [PubMed]
- Oraby, T.; Tyshenko, M.G.; Maldonado, J.C.; Vatcheva, K.; Elsaadany, S.; Alali, W.Q.; Longenecker, J.C.; Al-Zoughool, M. Modeling the effect of lockdown timing as a COVID-19 control measure in countries with differing social contacts. Sci. Rep. 2021, 11, 3354. [Google Scholar] [CrossRef]
- Xu, G.; Xiu, T.; Li, X.; Liang, X.; Jiao, L. Lockdown induced night-time light dynamics during the COVID-19 epidemic in global megacities. Int. J. Appl. Earth Obs. 2021, 102, 102421. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Q. Analysis of distribution characteristics of COVID-19 in America based on space-time scan statistic. Front. Public Health 2022, 10, 897784. [Google Scholar] [CrossRef]
- Kulldorff, M.; Nagarwalla, N. Spatial Disease Clusters: Detection and Inference. Stat. Med. 1995, 14, 799–810. [Google Scholar] [CrossRef]
- Angel, S.; Blei, A. Covid-19 Thrives in Larger Cities, Not Denser Ones. J. Extrem. Events 2021, 7, 2150004. [Google Scholar] [CrossRef]
- Stier, A.J.; Berman, M.G.; Bettencourt, L.M.A. Early pandemic COVID-19 case growth rates increase with city size. Npj Urban Sustain. 2021, 1, 31. [Google Scholar] [CrossRef]
- Smallman-Raynor, M.R.; Cliff, A.D.; Consortium, C.-G.U. Spatial growth rate of emerging SARS-CoV-2 lineages in England, September 2020–December 2021. Epidemiol. Infect. 2022, 150, e145. [Google Scholar] [CrossRef]
- Kremer, C.; Braeye, T.; Proesmans, K.; Andre, E.; Torneri, A.; Hens, N. Serial Intervals for SARS-CoV-2 Omicron and Delta Variants, Belgium, November 19–December 31, 2021. Emerg. Infect. Dis. 2022, 28, 1699–1702. [Google Scholar] [CrossRef]
- Tong, C.; Shi, W.; Zhang, A.; Shi, Z. Tracking and controlling the spatiotemporal spread of SARS-CoV-2 Omicron variant in South Africa. Travel Med. Infect. Dis. 2022, 46, 102252. [Google Scholar] [CrossRef]
- Andersen, L.M.; Harden, S.R.; Sugg, M.M.; Runkle, J.D.; Lundquist, T.E. Analyzing the spatial determinants of local Covid-19 transmission in the United States. Sci. Total Environ. 2021, 754, 142396. [Google Scholar] [CrossRef]
- Lancet, T. Genomic sequencing in pandemics. Lancet 2021, 397, 445. [Google Scholar] [CrossRef]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef]
- Kortessis, N.; Simon, M.W.; Barfield, M.; Glass, G.E.; Singer, B.H.; Holt, R.D. The interplay of movement and spatiotemporal variation in transmission degrades pandemic control. Proc. Natl. Acad. Sci. USA 2020, 117, 30104–30106. [Google Scholar] [CrossRef]
- Zhang, W.; Ning, K. Spatiotemporal heterogeneities in the causal effects of mobility intervention policies during the COVID-19 outbreak: A spatially interrupted time-series (SITS) analysis. Ann. Am. Assoc. Geogr. 2023, 113, 1112–1134. [Google Scholar] [CrossRef]
- Ahmed, F.; Ahmed, N.e.; Pissarides, C.; Stiglitz, J. Why inequality could spread COVID-19. Lancet Public Health 2020, 5, e240. [Google Scholar] [CrossRef]
- Glaeser, E.L.; Jin, G.Z.; Leyden, B.T.; Luca, M. Learning from deregulation: The asymmetric impact of lockdown and reopening on risky behavior during COVID-19. J. Reg. Sci. 2021, 61, 696–709. [Google Scholar] [CrossRef]
- Gatto, M.; Bertuzzo, E.; Mari, L.; Miccoli, S.; Carraro, L.; Casagrandi, R.; Rinaldo, A. Spread and dynamics of the COVID-19 epidemic in Italy: Effects of emergency containment measures. Proc. Natl. Acad. Sci. USA 2020, 117, 10484–10491. [Google Scholar] [CrossRef] [PubMed]
- Champlin, C.; Sirenko, M.; Comes, T. Measuring social resilience in cities: An exploratory spatio-temporal analysis of activity routines in urban spaces during COVID-19. Cities 2023, 135, 104220. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Prakash, A.; Mehmet Baskonus, H. The epidemic COVID-19 model via Caputo–Fabrizio fractional operator. Waves Random Complex Media 2022, 1–15. [Google Scholar] [CrossRef]
- Srinivasa, K.; Baskonus, H.M.; Guerrero Sánchez, Y. Numerical solutions of the mathematical models on the digestive system and covid-19 pandemic by hermite wavelet technique. Symmetry 2021, 13, 2428. [Google Scholar] [CrossRef]
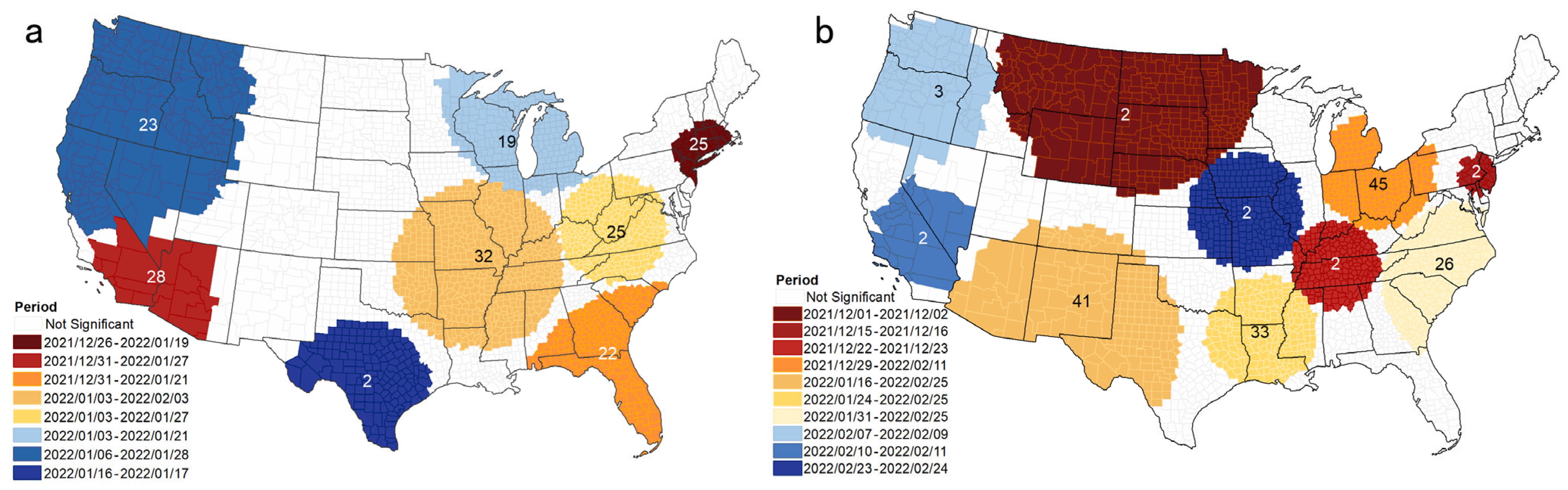

| Cluster | Duration (Days) | Number of Counties | p | Observed | Expected | RR | Number of Counties with RR > 1 |
|---|---|---|---|---|---|---|---|
| 1 | 26 December 2021–19 January 2022 | 65 | <0.001 | 2,449,866 | 806,725 | 3.22 | 45 |
| 2 | 31 December 2021–27 January 2022 | 20 | <0.001 | 2,316,904 | 897,684 | 2.72 | 6 |
| 3 | 31 December 2021–21 January 2022 | 246 | <0.001 | 1,821,212 | 684,190 | 2.77 | 43 |
| 4 | 3 January 2022–3 February 2022 | 644 | <0.001 | 2,209,112 | 1,034,411 | 2.23 | 341 |
| 5 | 3 January 2022–27 January 2022 | 382 | <0.001 | 1,726,603 | 806,957 | 2.21 | 217 |
| 6 | 3 January 2022–21 January 2022 | 239 | <0.001 | 1,326,786 | 594,240 | 2.29 | 83 |
| 7 | 6 January 2022–28 January 2022 | 218 | <0.001 | 1,570,202 | 742,166 | 2.18 | 28 |
| 8 | 16 January 2022–17 January 2022 | 150 | <0.001 | 287,673 | 38,676 | 7.5 | 29 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Liu, L.; Meng, Q.; Zhang, Y.; Yang, H.; Xu, G. Spatiotemporal Patterns of the Omicron Wave of COVID-19 in the United States. Trop. Med. Infect. Dis. 2023, 8, 349. https://doi.org/10.3390/tropicalmed8070349
Zhang S, Liu L, Meng Q, Zhang Y, Yang H, Xu G. Spatiotemporal Patterns of the Omicron Wave of COVID-19 in the United States. Tropical Medicine and Infectious Disease. 2023; 8(7):349. https://doi.org/10.3390/tropicalmed8070349
Chicago/Turabian StyleZhang, Siyuan, Liran Liu, Qingxiang Meng, Yixuan Zhang, He Yang, and Gang Xu. 2023. "Spatiotemporal Patterns of the Omicron Wave of COVID-19 in the United States" Tropical Medicine and Infectious Disease 8, no. 7: 349. https://doi.org/10.3390/tropicalmed8070349
APA StyleZhang, S., Liu, L., Meng, Q., Zhang, Y., Yang, H., & Xu, G. (2023). Spatiotemporal Patterns of the Omicron Wave of COVID-19 in the United States. Tropical Medicine and Infectious Disease, 8(7), 349. https://doi.org/10.3390/tropicalmed8070349








