Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand
Abstract
1. Introduction
2. Virus Biology and Classification
2.1. Rotavirus Biology
2.2. Dual Classification System
2.3. Full-Length Genome Classification System
2.4. Genotype Constellations
3. Molecular Epidemiology and Genetic Diversity
3.1. Rotavirus Prevalence and Age Distribution
3.2. Seasonal Patterns of Human Rotavirus Infection
3.3. Distribution of Rotavirus A Genotypes in Pediatric Patients
3.4. Prevalence and Distribution of Rotavirus A Genotypes in Animals
4. Interspecies Transmission of Rotaviruses
5. Conclusions and Further Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fields, B.N. Fields’ Virology; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1. [Google Scholar]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D.; World Health Organization–Coordinated Global Rotavirus Surveillance Network; Agocs, M.; Serhan, F.; de Oliveira, L.; Mwenda, J.M.; Mihigo, R.; et al. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin. Infect. Dis. 2016, 62 (Suppl. S2), S96–S105. [Google Scholar] [CrossRef] [PubMed]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G. Rotavirus infection. Nat. Rev. Dis. Prim. 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.; Platts-Mills, J.; Nakamura, T.; Operario, D.; Antoni, S.; Mwenda, M.; Weldegebriel, G.; Rey-Benito, G.; De Oliveira, L.H.; Ortiz, C.; et al. Aetiology and incidence of diarrhoea requiring hospitalisation in children under 5 years of age in 28 low-income and middle-income countries: Findings from the Global Pediatric Diarrhea Surveillance network. BMJ Glob. Health 2022, 7, e009548. [Google Scholar] [CrossRef] [PubMed]
- Manouana, G.P.; Nguema-Moure, P.A.; Mbong Ngwese, M.; Bock, C.-T.; Kremsner, P.G.; Borrmann, S.; Eibach, D.; Mordmüller, B.; Velavan, T.P.; Niendorf, S.; et al. Genetic diversity of enteric viruses in children under five years old in Gabon. Viruses 2021, 13, 545. [Google Scholar] [CrossRef]
- Bányai, K.; Estes, M.K.; Martella, V.; Parashar, U.D. Viral gastroenteritis. Lancet 2018, 392, 175–186. [Google Scholar] [CrossRef]
- Omatola, C.A.; Olaniran, A.O. Rotaviruses: From pathogenesis to disease control—A critical review. Viruses 2022, 14, 875. [Google Scholar] [CrossRef]
- Bonura, F.; Mangiaracina, L.; Filizzolo, C.; Bonura, C.; Martella, V.; Ciarlet, M.; Giammanco, G.M.; De Grazia, S. Impact of vaccination on rotavirus genotype diversity: A nearly two-decade-long epidemiological study before and after rotavirus vaccine introduction in Sicily, Italy. Pathogens 2022, 11, 424. [Google Scholar] [CrossRef]
- Charoenwat, B.; Suwannaying, K.; Paibool, W.; Laoaroon, N.; Sutra, S.; Thepsuthammarat, K. Burden and pattern of acute diarrhea in Thai children under 5 years of age: A 5-year descriptive analysis based on Thailand National Health Coverage (NHC) data. BMC Public Health 2022, 22, 1161. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Attoui, H.; Bányai, K.; Brussaard, C.P.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fāng, Q.; Johne, R. ICTV virus taxonomy profile: Sedoreoviridae 2022. J. Gen. Virol. 2022, 103, 001782. [Google Scholar] [CrossRef]
- Suzuki, H. Rotavirus replication: Gaps of knowledge on virus entry and morphogenesis. Tohoku J. Exp. Med. 2019, 248, 285–296. [Google Scholar] [CrossRef]
- Doro, R.; Farkas, S.L.; Martella, V.; Banyai, K. Zoonotic transmission of rotavirus: Surveillance and control. Expert Rev. Anti-Infect. Ther. 2015, 13, 1337–1350. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Dias, H.G.; Gonçalves, J.L.S.; Manchego, A.; Rosadio, R.; Pezo, D.; Santos, N. Genetic diversity and zoonotic potential of rotavirus A strains in the southern Andean highlands, Peru. Transbound. Emerg. Dis. 2019, 66, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Bányai, K.; Pitzer, V. Molecular epidemiology and evolution of rotaviruses. In Viral Gastroenteritis; Elsevier: London, UK, 2016; pp. 279–299. [Google Scholar]
- Maneekarn, N.; Ushijima, H. Epidemiology of rotavirus infection in Thailand. Pediatr. Int. 2000, 42, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Maneekarn, N.; Khamrin, P. Rotavirus associated gastroenteritis in Thailand. VirusDisease 2014, 25, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Adams, W.R.; Kraft, L.M. Epizootic diarrhea of infant mice: Identification of the etiologic agent. Science 1963, 141, 359–360. [Google Scholar] [CrossRef]
- Bishop, R.; Davidson, G.P.; Holmes, I.H.; Ruck, B.J. Virus particles in epithelisl cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973, 302, 1281–1283. [Google Scholar] [CrossRef]
- Bishop, R.; Davidson, G.P.; Holmes, I.H.; Ruck, B.J. Detection of an new virus by electron microscopy of faecal extrascts from children with acute gastroenteritis. Lancet 1974, 303, 149–151. [Google Scholar] [CrossRef]
- Flewett, T.; Bryden, A.; Davies, H.; Woode, G.; Bridger, J.; Derrick, J. Relation between viruses from acute gastroenteritis of children and newborn calves. Lancet 1974, 304, 61–63. [Google Scholar] [CrossRef]
- Sadiq, A.; Bostan, N.; Yinda, K.C.; Naseem, S.; Sattar, S. Rotavirus: Genetics, pathogenesis and vaccine advances. Rev. Med. Virol. 2018, 28, e2003. [Google Scholar] [CrossRef]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef]
- RCWG. List of Accepted Genotypes. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg (accessed on 3 April 2023).
- Steger, C.L.; Boudreaux, C.E.; LaConte, L.E.; Pease, J.B.; McDonald, S.M. Group A rotavirus VP1 polymerase and VP2 core shell proteins: Intergenotypic sequence variation and in vitro functional compatibility. J. Virol. 2019, 93, e01642-18. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Van Ranst, M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr. Opin. Virol. 2012, 2, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-Like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef] [PubMed]
- Nakagomi, O.; Ohshima, A.; Aboudy, Y.; Shif, I.; Mochizuki, M.; Nakagomi, T.; Gotlieb-Stematsky, T. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J. Clin. Microbiol. 1990, 28, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef]
- Janko, M.M.; Joffe, J.; Michael, D.; Earl, L.; Rosettie, K.L.; Sparks, G.W.; Albertson, S.B.; Compton, K.; Velandia, P.P.; Stafford, L. Cost-effectiveness of rotavirus vaccination in children under five years of age in 195 countries: A meta-regression analysis. Vaccine 2022, 40, 3903–3917. [Google Scholar] [CrossRef]
- Jayavasu, C.; Hoonniwat, Y.; Srijamorn, S.; Dumavibhat, B.; Wongpanich, K. Prevalence of rotavirus antibody in Bangkok Metropolis 1982. J. Diarrhoeal Dis. Res. 1983, 1, 29–31. [Google Scholar]
- Sakpaisal, P.; Silapong, S.; Yowang, A.; Boonyasakyothin, G.; Yuttayong, B.; Suksawad, U.; Sornsakrin, S.; Lertsethtakarn, P.; Bodhidatta, L.; Crawford, J.M.; et al. Prevalence and genotypic distribution of rotavirus in Thailand: A multicenter study. Am. J. Trop. Med. Hyg. 2019, 100, 1258–1265. [Google Scholar] [CrossRef]
- Yodmeeklin, A.; Khamrin, P.; Kumthip, K.; Malasao, R.; Ukarapol, N.; Ushijima, H.; Maneekarn, N. Increasing predominance of G8P[8] species A rotaviruses in children admitted to hospital with acute gastroenteritis in Thailand, 2010–2013. Arch. Virol. 2018, 163, 2165–2178. [Google Scholar] [CrossRef]
- Chieochansin, T.; Vutithanachot, V.; Phumpholsup, T.; Posuwan, N.; Theamboonlers, A.; Poovorawan, Y. The prevalence and genotype diversity of human rotavirus A circulating in Thailand, 2011–2014. Infect. Genet. Evol. 2016, 37, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Guntapong, R.; Tacharoenmuang, R.; Singchai, P.; Upachai, S.; Sutthiwarakom, K.; Komoto, S.; Tsuji, T.; Tharmaphornpilas, P.; Yoshikawa, T.; Sangkitporn, S.; et al. Predominant prevalence of human rotaviruses with the G1P[8] and G8P[8] genotypes with a short RNA profile in 2013 and 2014 in Sukhothai and Phetchaboon provinces, Thailand. J. Med. Virol. 2017, 89, 615–620. [Google Scholar] [CrossRef]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Upachai, S.; Singchai, P.; Ide, T.; Fukuda, S.; Ruchusatsawast, K.; Sriwantana, B.; Tatsumi, M.; et al. High prevalence of equine-like G3P [8] rotavirus in children and adults with acute gastroenteritis in Thailand. J. Med. Virol. 2020, 92, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Chan-It, W.; Chanta, C. Emergence of G9P [8] rotaviruses in children with acute gastroenteritis in Thailand, 2015–2016. J. Med. Virol. 2018, 90, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Chan-It, W.; Chanta, C.; Ushijima, H. Predominance of DS-1-like G8P [8] rotavirus reassortant strains in children hospitalized with acute gastroenteritis in Thailand, 2018–2020. J. Med. Virol. 2023, 95, e28870. [Google Scholar] [CrossRef]
- Jampanil, N.; Kumthip, K.; Yodmeeklin, A.; Kanai, Y.; Okitsu, S.; Kobayashi, T.; Ukarapol, N.; Ushijima, H.; Maneekarn, N.; Khamrin, P. Epidemiology and genetic diversity of group A rotavirus in pediatric patients with acute gastroenteritis in Thailand, 2018–2019. Infect. Genet. Evol. 2021, 95, 104898. [Google Scholar] [CrossRef]
- Pasittungkul, S.; Lestari, F.B.; Puenpa, J.; Chuchaona, W.; Posuwan, N.; Chansaenroj, J.; Mauleekoonphairoj, J.; Sudhinaraset, N.; Wanlapakorn, N.; Poovorawan, Y. High prevalence of circulating DS-1-like human rotavirus A and genotype diversity in children with acute gastroenteritis in Thailand from 2016 to 2019. PeerJ 2021, 9, e10954. [Google Scholar] [CrossRef] [PubMed]
- Burnett, E.; Parashar, U.D.; Winn, A.; Curns, A.T.; Tate, J.E. Major Changes in Spatiotemporal Trends of US Rotavirus Laboratory Detections After Rotavirus Vaccine Introduction—2009–2021. Pediatr. Infect. Dis. J. 2022, 41, 759–763. [Google Scholar] [CrossRef]
- Burnett, E.; Parashar, U.D.; Winn, A.; Tate, J.E. Trends in Rotavirus Laboratory Detections and Internet Search Volume Before and After Rotavirus Vaccine Introduction and in the Context of the Coronavirus Disease 2019 Pandemic—United States, 2000–2021. J. Infect. Dis. 2022, 226, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, C.; Zhang, X.; Yan, D.; Jiang, D.; Liu, X.; Yang, M.; Ding, C.; Lan, L.; Hecht, R.; et al. Global burden and trends of rotavirus infection-associated deaths from 1990 to 2019: An observational trend study. Virol. J. 2022, 19, 166. [Google Scholar] [CrossRef]
- Lestari, F.B.; Vongpunsawad, S.; Wanlapakorn, N.; Poovorawan, Y. Rotavirus infection in children in Southeast Asia 2008–2018: Disease burden, genotype distribution, seasonality, and vaccination. J. Biomed. Sci. 2020, 27, 66. [Google Scholar] [CrossRef] [PubMed]
- Tharmaphornpilas, P.; Jiamsiri, S.; Boonchaiya, S.; Rochanathimoke, O.; Thinyounyong, W.; Tuntiwitayapun, S.; Guntapong, R.; Riewpaiboon, A.; Rasdjarmrearnsook, A.O.; Glass, R.I. Evaluating the first introduction of rotavirus vaccine in Thailand: Moving from evidence to policy. Vaccine 2017, 35, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Lappe, B.L.; Wikswo, M.E.; Kambhampati, A.K.; Mirza, S.A.; Tate, J.E.; Kraay, A.N.M.; Lopman, B.A. Predicting norovirus and rotavirus resurgence in the United States following the COVID-19 pandemic: A mathematical modelling study. BMC Infect. Dis. 2023, 23, 254. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef]
- Bwogi, J.; Karamagi, C.; Byarugaba, D.K.; Tushabe, P.; Kiguli, S.; Namuwulya, P.; Malamba, S.S.; Jere, K.C.; Desselberger, U.; Iturriza-Gomara, M. Co-surveillance of rotaviruses in humans and domestic animals in Central Uganda reveals circulation of wide genotype diversity in the animals. Viruses 2023, 15, 738. [Google Scholar] [CrossRef]
- Yodmeeklin, A.; Khamrin, P.; Chuchaona, W.; Saikruang, W.; Kongkaew, A.; Vachirachewin, R.; Kumthip, K.; Okitsu, S.; Ushijima, H.; Maneekarn, N. Great genetic diversity of rotaviruses detected in piglets with diarrhea in Thailand. Arch. Virol. 2016, 161, 2843–2849. [Google Scholar] [CrossRef]
- Tuanthap, S.; Phupolphan, C.; Luengyosluechakul, S.; Duang-In, A.; Theamboonlers, A.; Wattanaphansak, S.; Vongpunsawad, S.; Amonsin, A.; Poovorawan, Y. Porcine rotavirus C in pigs with gastroenteritis on Thai swine farms, 2011–2016. PeerJ 2018, 6, e4724. [Google Scholar] [CrossRef]
- Tuanthap, S.; Vongpunsawad, S.; Luengyosluechakul, S.; Sakkaew, P.; Theamboonlers, A.; Amonsin, A.; Poovorawan, Y. Genome constellations of 24 porcine rotavirus group A strains circulating on commercial Thai swine farms between 2011 and 2016. PLoS ONE 2019, 14, e0211002. [Google Scholar] [CrossRef]
- Charoenkul, K.; Janetanakit, T.; Bunpapong, N.; Boonyapisitsopa, S.; Tangwangvivat, R.; Suwannakarn, K.; Theamboonlers, A.; Poovorawan, Y.; Amonsin, A. Molecular characterization identifies intra-host recombination and zoonotic potential of canine rotavirus among dogs from Thailand. Transbound. Emerg. Dis. 2021, 68, 1240–1252. [Google Scholar] [CrossRef]
- Lestari, F.B.; Chandranoi, K.; Chuchaona, W.; Vongpunsawad, S.; Poovorawan, Y. A G3P [9] rotavirus strain with an unusual genome constellation in a diarrheic cat in Thailand. Arch. Virol. 2023, 168, 24. [Google Scholar] [CrossRef]
- Boene, S.S.; João, E.D.; Strydom, A.; Munlela, B.; Chissaque, A.; Bauhofer, A.F.L.; Nabetse, E.; Latifo, D.; Cala, A.; Mapaco, L. Prevalence and genome characterization of porcine rotavirus A in southern Mozambique. Infect. Genet. Evol. 2021, 87, 104637. [Google Scholar] [CrossRef] [PubMed]
- German, A.; Iturriza-Gómara, M.; Dove, W.; Sandrasegaram, M.; Nakagomi, T.; Nakagomi, O.; Cunliffe, N.; Radford, A.; Morgan, K. Molecular epidemiology of rotavirus in cats in the United Kingdom. J. Clin. Microbiol. 2015, 53, 455–464. [Google Scholar] [CrossRef]
- Matthijnssens, J.; De Grazia, S.; Piessens, J.; Heylen, E.; Zeller, M.; Giammanco, G.M.; Bányai, K.; Buonavoglia, C.; Ciarlet, M.; Martella, V. Multiple reassortment and interspecies transmission events contribute to the diversity of feline, canine and feline/canine-like human group A rotavirus strains. Infect. Genet. Evol. 2011, 11, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Sinchai, P.; Upachai, S.; Fukuda, S.; Yoshikawa, T.; Tharmaphornpilas, P.; Sangkitporn, S.; et al. Identification and characterization of a human G9P[23] rotavirus strain from a child with diarrhoea in Thailand: Evidence for porcine-to-human interspecies transmission. J. Gen. Virol. 2017, 98, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Tacharoenmuang, R.; Guntapong, R.; Upachai, S.; Singchai, P.; Fukuda, S.; Ide, T.; Hatazawa, R.; Sutthiwarakom, K.; Kongjorn, S.; Onvimala, N.; et al. Full genome-based characterization of G4P[6] rotavirus strains from diarrheic patients in Thailand: Evidence for independent porcine-to-human interspecies transmission events. Virus Genes 2021, 57, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Malasao, R.; Khamrin, P.; Kumthip, K.; Ushijima, H.; Maneekarn, N. Complete genome sequence analysis of rare G4P[6] rotavirus strains from human and pig reveals the evidence for interspecies transmission. Infect. Genet. Evol. 2018, 65, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Ide, T.; Haga, K.; Katayama, K.; Kato, T.; Ouchi, Y.; Kurahashi, H.; Tsuji, T.; et al. Whole genomic analysis of an unusual human G6P[14] rotavirus strain isolated from a child with diarrhea in Thailand: Evidence for bovine-to-human interspecies transmission and reassortment events. PLoS ONE 2015, 10, e0139381. [Google Scholar] [CrossRef]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Ide, T.; Singchai, P.; Upachai, S.; Fukuda, S.; Yoshida, Y.; Murata, T.; Yoshikawa, T.; et al. Characterization of a G10P[14] rotavirus strain from a diarrheic child in Thailand: Evidence for bovine-to-human zoonotic transmission. Infect. Genet. Evol. 2018, 63, 43–57. [Google Scholar] [CrossRef]
- Yodmeeklin, A.; Khamrin, P.; Chuchaona, W.; Kumthip, K.; Kongkaew, A.; Vachirachewin, R.; Okitsu, S.; Ushijima, H.; Maneekarn, N. Analysis of complete genome sequences of G9P[19] rotavirus strains from human and piglet with diarrhea provides evidence for whole-genome interspecies transmission of nonreassorted porcine rotavirus. Infect. Genet. Evol. 2017, 47, 99–108. [Google Scholar] [CrossRef]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Ide, T.; Sinchai, P.; Upachai, S.; Yoshikawa, T.; Tharmaphornpilas, P.; Sangkitporn, S.; Taniguchi, K. Full genome characterization of novel DS-1-like G8P[8] rotavirus strains that have emerged in Thailand: Reassortment of bovine and human rotavirus gene segments in emerging DS-1-like intergenogroup reassortant strains. PLoS ONE 2016, 11, e0165826. [Google Scholar] [CrossRef] [PubMed]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Haga, K.; Katayama, K.; Kato, T.; Ouchi, Y.; Kurahashi, H.; Tsuji, T.; et al. Emergence and characterization of unusual DS-1-Like G1P[8] rotavirus strains in children with diarrhea in Thailand. PLoS ONE 2015, 10, e0141739. [Google Scholar] [CrossRef] [PubMed]
- Simsek, C.; Corman, V.M.; Everling, H.U.; Lukashev, A.N.; Rasche, A.; Maganga, G.D.; Binger, T.; Jansen, D.; Beller, L.; Deboutte, W. At least seven distinct rotavirus genotype constellations in bats with evidence of reassortment and zoonotic transmissions. mBio 2021, 12, e02755-20. [Google Scholar] [CrossRef] [PubMed]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Upachai, S.; Singchai, P.; Ide, T.; Fukuda, S.; Hatazawa, R.; Sutthiwarakom, K.; Kongjorn, S.; et al. Genomic characterization of a novel G3P[10] rotavirus strain from a diarrheic child in Thailand: Evidence for bat-to-human zoonotic transmission. Infect. Genet. Evol. 2021, 87, 104667. [Google Scholar] [CrossRef] [PubMed]
- Lestari, F.B.; Vongpunsawad, S.; Poovorawan, Y. Diverse human and bat-like rotavirus G3 strains circulating in suburban Bangkok. PLoS ONE 2022, 17, e0268465. [Google Scholar] [CrossRef] [PubMed]
- Khamrin, P.; Maneekarn, N.; Peerakome, S.; Malasao, R.; Thongprachum, A.; Chan-it, W.; Mizuguchi, M.; Okitsu, S.; Ushijima, H. Molecular characterization of VP4, VP6, VP7, NSP4, and NSP5/6 genes identifies an unusual G3P[10] human rotavirus strain. J. Med. Virol. 2009, 81, 176–182. [Google Scholar] [CrossRef]
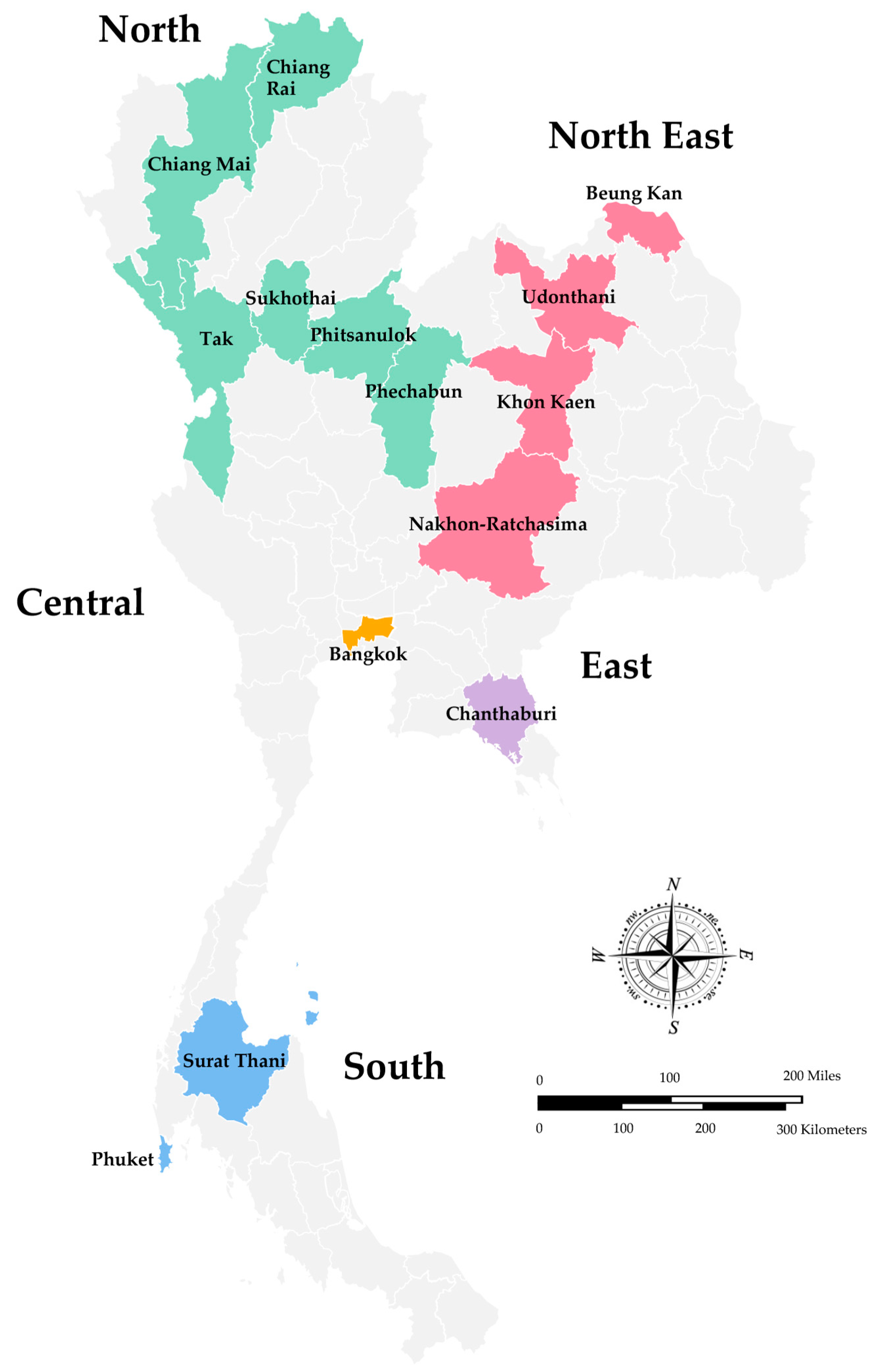
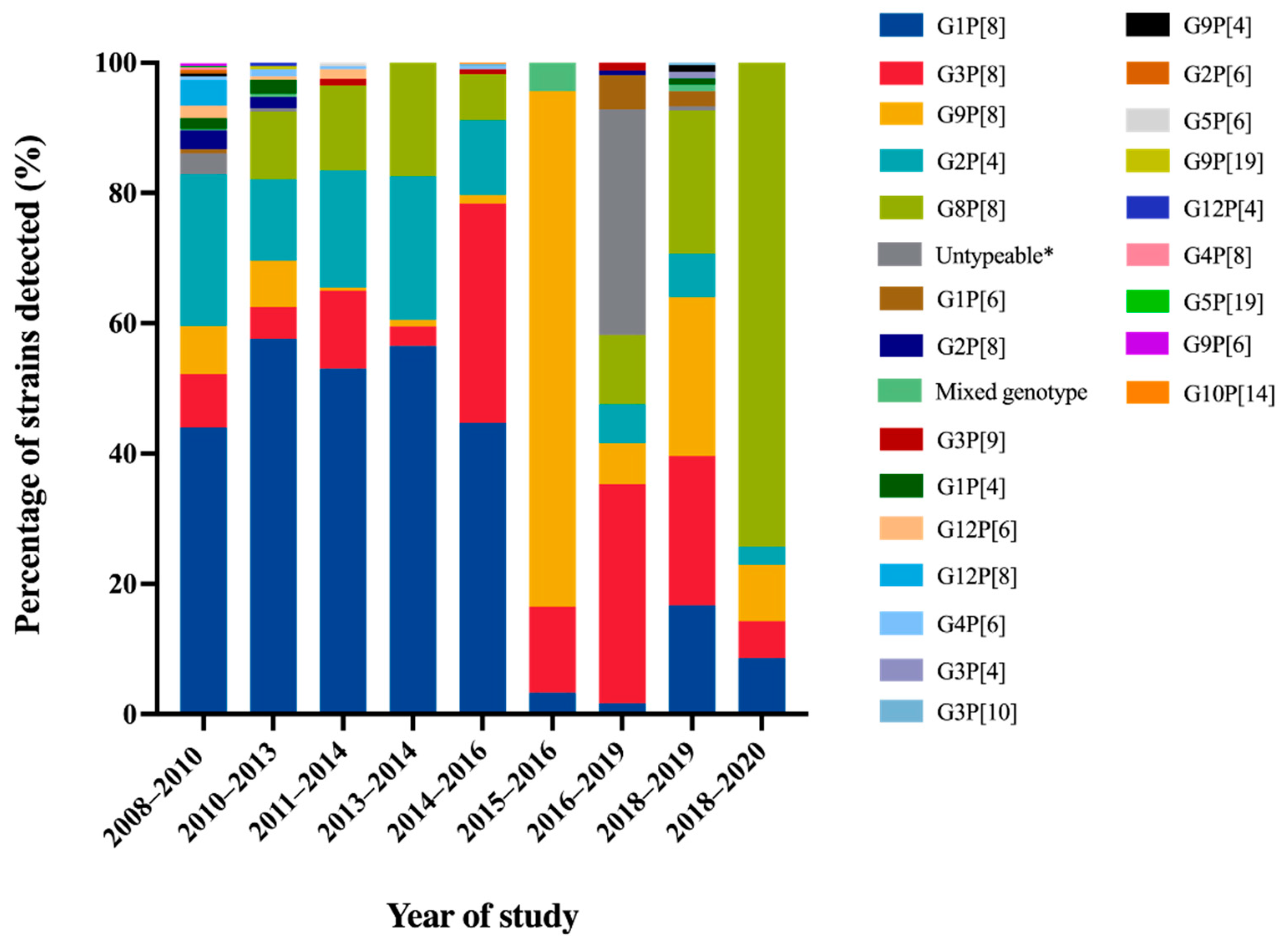
| Year of Study | Place of Study | No. of Specimens Tested | No. of Rotavirus A Positive (%) | Detection Method | Age | Age Group with High Infection Rate | Seasonal Pattern | References |
|---|---|---|---|---|---|---|---|---|
| 2008–2010 | Chiang Rai, Nakhon- Ratchasima, Surat Thani, Phitsanulok | 3470 | 458 (26.8) | qRT-PCR, ELISA | 3–60 months | <2 years | January–March (52.0–69.0%) | [32] |
| 2010–2013 | Chiang Mai | 1032 | 184 (17.8) | RT-PCR | 0–15 years | 12–24 months (54%; 100 of 184) | - | [33] |
| 2011–2014 | Bangkok, Khon Kaen | 688 | 204 (30.0) | RT-PCR | <15 years | 1–5 years | January–May | [34] |
| 2013–2014 | Phechaboon, Sukothai | 2754 | 666 (24.2) | PAGE | - | - | - | [35] |
| 2014–2016 | Bangkok, Udonthani, Beung Kan, Phuket, Tak, Chanthaburi | 1867 | 514 (27.5) | RT-PCR | 0–>60 years | 5–10 years (43%; 59 of 137) | November–April | [36] |
| 2015–2016 | Chiang Rai | 1867 | 91 (33.7) | Multiplex RT-PCR | 0–5 years | 12–23 months (45.1%; 41 of 91) | March (64.8%) | [37] |
| 2016–2019 | Bangkok | 2001 | 301 (15.0) | qRT-PCR, | 0–<15 years | 0–24 months | December–March | [40] |
| 2018–2019 | Chiang Mai | 1170 | 209 (17.9) | Nested RT-PCR | 0–5 years | - | January–March (39.8–63.3%) | [39] |
| 2018–2020 | Chiang Rai | 302 | 35 (11.5) | qRT-PCR | <5 years | 6–11 months (37.1%; 13 of 35) | March–May | [38] |
| Year of Study | Place | Hosts | No. of Specimens Tested | No. of Rotavirus A Positive (%) | Detection Method | Age of Animals | Age Group with High Infection Rate | References |
|---|---|---|---|---|---|---|---|---|
| 2011–2014 | Chiang Mai, Lamphun | Piglets | 491 | 113 (23.0) | RT-PCR | 0–4 weeks | - | [50] |
| 2011–2016 | Kanchanaburi, Prachuap Khiri Khan, Phechaboon, Ratchaburi, Lop Buri, Samut Songkhram, Suphan Buri, Saraburi, Phra Nakhon Si Ayutthaya, Nakhon Pathom, Chon Buri, Chachoengsao, Ubon Ratchathani, Udon Thani, Nakhon Ratchasima, Trang, and Nakhon Si Thammarat | Piglets | 769 | 73 (9.5) | RT-PCR | 0–>12 weeks | ≥4–8 weeks | [51] |
| 2016–2019 | Nakhon Si Ayutthaya, Bangkok, Suphan Buri, Nakhon Rachasima, Tak | Dogs | 710 | 5 (0.7) | RT-PCR | 0–>1 years | <1 year | [53] |
| 2021 | Bangkok | Cat | 1 * | 1 (100.0) | qRT-PCR | 2 years | - | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jampanil, N.; Kumthip, K.; Maneekarn, N.; Khamrin, P. Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand. Trop. Med. Infect. Dis. 2023, 8, 347. https://doi.org/10.3390/tropicalmed8070347
Jampanil N, Kumthip K, Maneekarn N, Khamrin P. Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand. Tropical Medicine and Infectious Disease. 2023; 8(7):347. https://doi.org/10.3390/tropicalmed8070347
Chicago/Turabian StyleJampanil, Nutthawadee, Kattareeya Kumthip, Niwat Maneekarn, and Pattara Khamrin. 2023. "Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand" Tropical Medicine and Infectious Disease 8, no. 7: 347. https://doi.org/10.3390/tropicalmed8070347
APA StyleJampanil, N., Kumthip, K., Maneekarn, N., & Khamrin, P. (2023). Genetic Diversity of Rotaviruses Circulating in Pediatric Patients and Domestic Animals in Thailand. Tropical Medicine and Infectious Disease, 8(7), 347. https://doi.org/10.3390/tropicalmed8070347





