Abstract
This study aimed to characterize whole-genome sequencing (WGS) information of Mycobacterium tuberculosis (Mtb) in the Mandalay region of Myanmar. It was a cross-sectional study conducted with 151 Mtb isolates obtained from the fourth nationwide anti-tuberculosis (TB) drug-resistance survey. Frequency of lineages 1, 2, 3, and 4 were 55, 65, 9, and 22, respectively. The most common sublineage was L1.1.3.1 (n = 31). Respective multi-drug resistant tuberculosis (MDR-TB) frequencies were 1, 1, 0, and 0. Four clusters of 3 (L2), 2 (L4), 2 (L1), and 2 (L2) isolates defined by a 20-single-nucleotide variant (SNV) cutoff were detected. Simpson’s index for sublineages was 0.0709. Such high diversity suggests that the area probably had imported Mtb from many geographical sources. Relatively few genetic clusters and MDR-TB suggest there is a chance the future control will succeed if it is carried out properly.
1. Introduction
Although there are international efforts to fight tuberculosis (TB), it remains the main cause of death from a single pathogen [1]. Understanding the nature of the pathogen, Mycobacterium tuberculosis (Mtb), is crucial in the prevention and control program.
Whole-genome sequencing (WGS) is an effective way to disclose the nature of Mtb. Data from WGS can be used for the classification of Mtb into lineages and sublineages, with much better precision and accuracy than previous technologies such as restriction fragment length polymorphism (RFLP) [2]. With the evidence of certain genes associated with certain drug resistance, WGS can replace the conventional drug susceptibility test (DST) in the drug resistance survey. Moreover, the genetic distance between two or more isolates can be used to detect genetic cluster outbreaks of Mtb.
Worldwide, Mtb was classified into four major lineages (L), which are significantly linked with geography and host genetics [3]. Common areas of L1 to L4 are East Africa and Southeast Asia; Asia to Europe and Africa; South Asia, North Africa, and East Africa; and Europe and America [4,5,6,7].
Any areas with all sublineages in common would be considered as having high biodiversity, which may be due to the importing and maintenance of Mtb from various geographic sources. On the other hand, evidence of the genetic clusters would suggest the recent outbreak or the domination of transmission of a certain strain.
In Myanmar, the estimated incidence of TB was 360 per 100,000 population. The estimated proportions of TB cases with multi-drug-resistant (MDR)/rifampicin-resistant tuberculosis (RR-TB) in 2021 were 4.1% (3.8–4.3) among new TB cases and 19% (18–20) among previously treated cases [1]. The Myanmar National TB Programme (NTP) conducts the anti-TB drug resistance survey every five years. The three previous surveys revealed the proportion of MDR-TB among new and previously treated TB patients to be 4.0% and 15.5% in 2002–2003; 4.2% and 10.0% in 2007–2008; and 5.0% and 27.1% in 2012–2013 [8]. The fourth nationwide anti-TB drug resistance survey was conducted in 2019–2020 [8]. Here, we report the Mandalay regional part of this fourth survey, which was the first survey using WGS. Although the study of Phyu et al. was the first to describe the distribution of lineages and drug resistances using WGS in Upper Myanmar [9], here we provide new information about genomic clusters, biodiversity, and sublineages, in detail, that were circulating in the Mandalay region.
To broaden the understanding of Mtb genomics in Myanmar, readers are reminded that there was a similar study at Kayin State in the Myanmar Thailand Border area in 2019 [10]. The information contributed by our current report, when combined with that from the Kayin study, will provide valuable input for the management of the National TB Control Programme in Myanmar.
The objectives of this study were (1) to construct the phylogenetic tree based on the genomic sequences of Mtb isolates obtained from the survey in the Mandalay region of Myanmar; (2) to classify the genotypes of isolates and examine the possible genetic clustering among the collected isolates in Mandalay region; (3) to compare the biodiversity and sublineages of TB isolates in Mandalay region and Kayin state in Myanmar; and (4) to describe the drug-resistance mutations with sublineages in Mandalay.
2. Materials and Methods
2.1. Study Sites
Mandalay is the major city in the middle part of Myanmar. The population of Mandalay region is around 6.2 million [11]. Notified MDR/RR-TB cases were 180 in 2020, the third rank in the whole country [12]. The Mandalay region consists of seven districts, which are subdivided into 28 townships. From the NTP survey design, our study sites covered five townships: Chanmyathazi, Kyaukpadaung, Meiktila, Sintgaing, and Singu townships. They are shown in Supplementary Figure S1.
Kayin state is situated in Lower Myanmar and adjacent to the Mandalay region. The population of Kayin state is 1.574 million [11]. It consists of five districts, which are subdivided into 7 townships. The study sites were three townships in Kayin state, and they were Hpa-An, Kawkareik, and Myawaddy [10].
2.2. Study Design
A cross-sectional study was carried out between February and August 2020 in the above five townships involving TB patients based on positive sputum culture regardless of their microscopic (for acid fast bacilli) and/or molecular study (Gene Xpert MTB/RIF Assay) results. The selected patients must not have been incarcerated at the time of selection and must never have received anti-TB treatment for more than seven days in their current regimen.
2.3. Participants
Of 28 townships of the Mandalay region, 5 were used because they could provide sputum samples from at least 10 smear positive consenting patients [8].
Morning sputum was collected on 3 consecutive days. The first specimen was sent for Gene Xpert test. The other two were sent for TB culture at the Upper Myanmar TB Laboratory if the first produced a positive Xpert test [8].
Based on the assumption that the proportion of MDR-TB was 0.05, the design effect was 1.2, and the 95% confidence limit was 0.04 from the estimate, the required sample size was 137 [13].
2.4. TB Culture Testing
At the Upper Myanmar TB Laboratory with biosafety level 3, sputum samples were decontaminated and inoculated in Mycobacterium Growth Indicator Tube (MGIT) culture immediately on arrival, and within ≤3 days of sputum sample collection. For each sample, one Löwenstein–Jensen media (LJ) tube was inoculated in parallel to MGIT as a back-up, kept at 37 °C, and the sputum sample discarded thereafter, to ensure that re-culturing in MGIT was possible if the initial MGIT culture failed. For each specimen, and for each positive MGIT culture that was confirmed as Mtb using the Capilia TB Immunochromatographic test (ICT) kit, at least one aliquot of viable culture was cryopreserved in glycerol at −80 °C, and the remaining culture tube was sub-cultured onto 2 LJ tubes. One positive sub-cultured LJ tube was used for DNA extraction for WGS, and the other tube was stored at −80 °C as a back-up, in case further DNA extractions were required [8].
Among the 221 smear positive or Xpert MTB/RIF positive samples, 206 samples were successfully cultured.
2.5. Extraction of Genomic DNA
At the Upper Myanmar TB Laboratory, genomic DNA was extracted from sputum culture isolates. MoBio Microbial DNA Isolation Kits, (Qigen DNEasy Ultraclean Microbial DNA extraction kit, Cat No: 12224-50) was used [14,15,16,17]. Both first- and second-line line probe assay (LPA) were tested by the MTB DRplus kit and the MTB DRsl kit. The extracted DNA was kept at −80 °C and sent to University of Otago, New Zealand via the National TB Reference Laboratory every 3 months for WGS or as necessary according to the international regulation to assure the stability of extracted DNA.
2.6. Sequencing at University of Otago
At University of Otago, extracted DNA was sequenced using Illumina MiSeq (https://www.illumina.com, accessed on 5 February 2021) as previously described [15,16]. The sequencing data (FASTQ) were stored on the University of Otago’s high-capacity central file storage (HCS) server.
2.7. Data Analysis Steps
2.7.1. Genomic Characterization
At the Faculty of Science, Mahidol University, Bangkok, Thailand, we used the snpplet pipeline for processing short-read sequencing data to obtain short variants (SNPs and indels). Using trimmomatic v0.39, the short reads were trimmed to remove adapter sequences and low-quality read positions (sliding-window trimming with a window size of 4 and a read quality threshold of 30) [18]. The trimmed reads were then mapped to the H37Rv reference genome (NC_000962.3) using bwa mem [19]. Picard’s MarkDuplicates was used to identify duplicate reads before a per-sample variant calling using GATK (genome analysis toolkit) HaplotypeCaller in a haploid model [20], excluding bases with a quality score below 20. To compare SNPs across samples, we performed joint genotyping of all samples using GATK GenotypeGVCFs [20], using per-sample variant calls as inputs. We used mtbtyper for genotyping Mtb isolates from WGS data (https://github.com/ythaworn/mtbtyper, accessed on 15 October 2022).
2.7.2. Phylogenetic Analysis
A maximum-likelihood (ML) phylogenetic tree of 151 isolates was inferred using IQ-TREE v2 [21] with ultrafast bootstrap supports from 1000 replications (Figure 1). For the quality of WGS, this study cut off the coverage and read depth of sequencing at 90 and 15, respectively. The phylogenetic tree was constructed from 151 isolates. The phylogenetic tree was visualized using the FigTree program version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 20 October 2022). The best-fit nucleotide substitution model was K3Pu + F+ASC + R4 chosen according to the Bayesian Information Criterion (BIC) as determined by ModelFinder [22]. The lineage 4 H37Rv reference strain (Gene Bank accession number NC_000962.3) was used as an outgroup for rooting the tree.
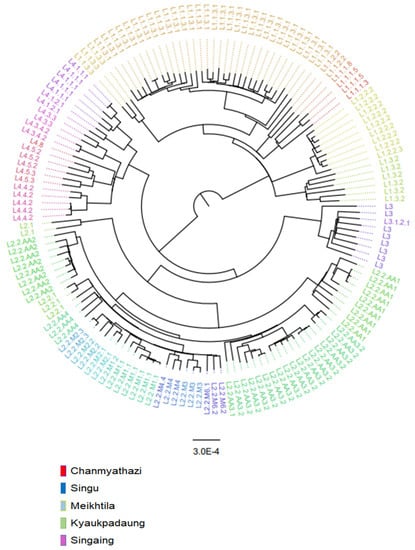
Figure 1.
A maximum-likelihood phylogenetic tree of 151 Mycobacterium tuberculosis isolates from five townships in the Mandalay region, Myanmar.
2.7.3. Identification of Genetic Clusters
Pairwise SNV distances were calculated using Molecular Evolutionary Genetics Analysis Version 11 (MEGA11) (https://doi.org/10.1093/molbev/msab120, accessed on 20 October 2022) [23]. We identified genomic clusters as clades in the phylogeny containing isolates that can be linked via pairwise single nucleotide polymorphism (SNP) distances. If the pairwise distance between two isolates was <20 SNPs, they were considered closely related or genomically linked [24]. A cluster contains an aggregate of pairs of isolates in which each one differs by fewer than 20 SNPs from at least one of the other elements of the cluster.
2.7.4. Biodiversity
At the Department of Epidemiology, Faculty of Medicine, Prince of Songkla University, Hat Yai, Thailand, Simpson’s index was calculated based on the formula:
where D is Simpson’s index, n is the total number of organisms of a particular species, N is the total number of organisms of all species, and means “sum up”. The scale ranges from 0–1, with 0 representing the highest biodiversity of a particular species and 1 representing the lowest biodiversity of a particular species [25,26].
2.7.5. Drug Resistance
We used TB-profiler software for the prediction of drug-resistance mutation [27].
2.7.6. Merging with Kayin State Data
The 109 samples of sequencing data for Kayin state were downloaded from the European Nucleotide Archive (ENA) of EMBL-EBI mirrored in the Sequence Read Archive (SRA) database [10].
2.8. Statistical Analysis
Statistical analyses were performed using R version 4.0.2 (R foundation for Statistical Computing, Vienna, Austria). The Mtb major lineages, sublineages, and their drug-resistance mutations were presented as frequency and percentage. Information on drug resistance was limited to only untreated cases in order to assess only primary drug resistance [28], which better reflected the severity of the problem. The relationship between the geographical areas of Mandalay and Mtb lineages, and between anti-TB drug resistance and major lineages, was analyzed using the Chi-squared test. The significance level was set at a p value of less than 0.05.
3. Results
3.1. Participant Characteristics
The median age of the participants was 42 (IQR: 31.5–54.0) years. Males accounted for 70%. There were no significant association between Mtb major lineages and the age and gender of the participants; or townships (Table 1).

Table 1.
Frequency (%) of major lineages of Mycobacterium tuberculosis by demographic variables.
3.2. Phylogenetic Analysis
A maximum-likelihood phylogenetic tree of 151 Mycobacterium tuberculosis isolates from five townships is shown in Figure 1, with the names of sublineages labeled in color at the outermost part. The majority of isolates belonged to lineage 1 (L1) and L2. Intriguingly, among L1, the majority of isolates belonged to L1.1.3 (62%, 34/55), which belongs to the spoligogroup EAI6_BGD. L1.1.3.1 is the predominant subgroup of L1.1.3 (91%, 31/34), similar to the Kayin state. L1.1.2.2 and L1.3, predominant sublineages in India, were also identified, albeit in lower proportions than in the Kayin state. Among the L2.2 (Beijing, China) isolates, the majority (65%, 41/63) belonged to the ancestral group, with L2.2.AA3.2 being the highest. There were no predominant modern Beijing sublineage in the Mandalay region. L4.4. and L4.5, which are predominant L4 strains in China, constitute 45% (10/22) of the L4 strains.
3.3. Identification of Genetic Clusters
There were four clusters identified based on the pairwise distance. The number of isolates in the respective lineages were 3 (L2), 2 (L4), 2 (L1), and 2 (L2).
3.4. Distribution of Sublineages and Biodiversity in Mandalay Region and Kayin State
The distribution of sublineages and the biodiversity in Mandalay region and Kayin state are shown in Table 2. The overall Simpson’s indices were 0.0709 in Mandalay and 0.072885 in Kayin. Sublineage L1.1.3.1 was the most common (n = 31), followed by L2.2.AA3.2 (n = 17), which was available only in Mandalay. In Kayin, the most prominent sublineages were L1.1.3.1 (n = 17) and L1.3 (n = 17). The latter was only in Kayin.

Table 2.
Distribution of sublineages and biodiversity in Mandalay region and Kayin state.
3.5. Distribution of Drug-Resistance Mutation and Sublineages in Mandalay Region
The distribution of drug-resistance mutation and sublineages is shown in Table 3. Among the isoniazid-resistant strains, 70% were S315T mutations in the katG gene. Regarding the distribution of streptomycin drug resistant mutation, there were 73% resistant isolates with an rpsL K43R mutation, which was the most common occurrence in the Mandalay region.

Table 3.
Frequency distribution of drug-resistance mutation and sub-lineages in Mandalay region.
Among 126 new TB patients, MDR-TB accounted for 1.6% (2/126) and poly-drug resistance 5.6% (7/126). MDR-TB (isoniazid and rifampicin resistance) genes were found in L1 and L2. Poly-drug resistance was also found in L1 (n = 2), L2 (n = 4), and L4 (n = 1). There was no drug resistance identified in seven isolates of L3.
4. Discussion
We identified L1, L2, L3, and L4 in our samples. There were no associations between lineages and demographic variables. The most common sublineage was L1.1.3.1 (n = 31), followed by L2.2.AA3.2 (n = 17), which was exclusively found in Mandalay. In Kayin, the most prevalent sublineages were L1.1.3.1 (n = 17) and L1.3 (n = 17), with the latter being unique to Kayin. There were four small clusters, and the level of biodiversity was high. Streptomycin and isoniazid were the two most commonly observed drug-resistance mutations.
Four types of major lineages and 35 sub-lineages were identified among 151 isolates via WGS in our study. Lineage 1 is divided into five major sub-lineages, i.e., L1.1.1, L1.1.2, L1.1.3, L1.2, and L1.3. L1.1 is made up of three additional sublineages, L1.1.1–1.1.3 [3]. Specifically, L1.2.1.1 is found in Southern Taiwan and Indonesia, accounting for >90% and roughly 40% of EAI isolates, respectively [29]. In the Philippines, 80% of isolates are L1.2.2 [30]. China has mostly Modern Beijing, L4.4 and L4.5 [31].
Many studies have suggested that the L1.1.3 sublineage is more virulent and more resistant to anti-TB drugs. According to the research done in South Africa, active TB cases infected with L1.1.3 strains were at a higher risk of developing drug-resistant strains and treatment failure than those infected with other strains [32]. Another study conducted in Ethiopia found that the L1.1.3 sublineage was associated with a higher risk of treatment failure and death among TB patients, compared to other Mtb strains [33]. The prominence of the L2.2.AA3.2 sublineage in Mandalay is not fully understood.
Our current study revealed relatively few outbreaks of TB genetic clusters. These findings imply the simultaneous transmission of different lineages imported from different sources. Simpson’s index (D) was 0.0709 in our study and 0.07 reported by Maung et al. in Kayin state [10], much lower than that of 0.68 in the Philippines [30] and 0.21 in China [31]. It has been proposed that the primary causes of genetic diversity within the Mycobacterium tuberculosis complex (Mtbc) may be attributed to the relative fitness and adaptability of imported genotypes to ecological and other unknown environmental factors [34].
The two most common drug-resistance mutations found were streptomycin (n = 11, 7%) and isoniazid (n = 10, 7%), which have been extensively used in the past. The first Mtb drug resistance to be identified was to streptomycin [35]. Of the streptomycin-resistant isolates, 73% had a rpsL K43R mutation, which confers high-level resistance to streptomycin by reducing the binding affinity of the drug to the ribosome [36]. This was the most common occurrence in the Mandalay region. A previous study from Upper Myanmar also reported resistant streptomycin 32 isolates with a rpsL K43R mutation [9]. In Asia, rpsl mutations were prevalent [37,38,39,40]. In Myanmar, streptomycin is utilized in developing specialized treatment for hepatotoxic TB patients who are not suitable for the standard anti-TB treatment regimen [9]. S315T mutations in the katG gene accounted for 70% of the isoniazid-resistant strains in our study. There were 22 resistant isoniazid isolates with the katG S315T mutation in a previous study from Upper Myanmar [9]. The katG gene encodes for the enzyme catalase-peroxidase, which is involved in the activation of isoniazid, a major component of anti-TB treatment [41]. A significant mechanism for isoniazid resistance in Mtb is via mutations in the katG gene [41].
Our study only obtained isolates from culture-positive TB patients. The findings cannot be generalized to smear negative and extra-pulmonary TB patients. We also did not have metadata on ethnicity, socio-economic status, traveling history, and treatment compliance. These limitations should be taken into account for TB control planning.
5. Conclusions
The current data suggest that the area probably had imported Mtb from many geographical sources. With relatively few genetic clusters and MDR-TB, there is a good chance that the future control will succeed if it is carried out properly.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/tropicalmed8040239/s1, Figure S1: Selected five townships in Mandalay region.
Author Contributions
Conceptualization, A.N.P., S.T.A., P.P. (Prasit Palittapongarnpim) and V.C.; methodology, A.N.P., S.T.A., P.P. (Prasit Palittapongarnpim) and V.C.; software, A.N.P., K.K.K.H. and B.J.; validation, A.N.P., S.T.A., P.P., K.K.K.H. and V.C.; formal analysis, A.N.P., P.P. (Prasit Palittapongarnpim), K.K.K.H., B.J., H.M.W.M. and V.C.; writing—original draft preparation, A.N.P. and V.C.; writing—review and editing, A.N.P., S.T.A., P.P. (Prasit Palittapongarnpim), K.K.K.H., S.M., W.R., B.J., H.L.A., H.M.W.M., A.C., P.P. (Petchawan Pungrassami) and V.C.; and visualization, A.N.P., K.K.K.H., W.R. and B.J. All authors have read and agreed to the published version of the manuscript.
Funding
Research reported in this publication was supported by the Fogarty International Center and the National Institute of Allergy and Infectious Diseases, of the National Institutes of Health under Awarded Number D43TW009522. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
The National Tuberculosis Programme, Department of Public Health, Ministry of Health, Myanmar, as well as the Institutional Ethics Committee of the Faculty of Medicine, Prince of Songkla University, Hat Yai, Thailand (REC number 65-324-18-9), approved this study.
Informed Consent Statement
Not applicable.
Data Availability Statement
All of the analyzed data are included in this published article.
Acknowledgments
This study was a part of the thesis of Aye Nyein Phyu in partial fulfillment of the requirement of a degree of Doctor of Philosophy (PhD) in epidemiology at Prince of Songkla University, Hat Yai, Songkhla, Thailand. The authors thank the National Tuberculosis Reference Laboratory; the Upper Myanmar Tuberculosis Laboratory; University of Otago; and Mahidol University for their technical support. We also thank Alan Frederick Geater for his expert grammar check.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization. Global Tuberculosis Report. 2022. Available online: https://www.who.int/teams/globaltuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 30 November 2022).
- Sacchi, F.P.C.; Tatara, M.B.; de Lima, C.C.; da Silva, L.F.; Cunha, E.A.; Simonsen, V.; Ferrazoli, L.; Gomes, H.M.; Vasconcellos, S.E.G.; Suffys, P.N.; et al. Genetic clustering of tuberculosis in an indigenous community of Brazil. Am. J. Trop. Med. Hyg. 2018, 98, 372–375. [Google Scholar] [CrossRef]
- Netikul, T.; Thawornwattana, Y.; Mahasirimongkol, S.; Yanai, H.; Maung, H.M.W.; Chongsuvivatwong, V.; Palittapongarnpim, P. Whole-genome single nucleotide variant phylogenetic analysis of Mycobacterium tuberculosis Lineage 1 in endemic regions of Asia and Africa. Sci. Rep. 2022, 12, 1565. [Google Scholar] [CrossRef] [PubMed]
- Palittapongarnpim, P.; Ajawatanawong, P.; Viratyosin, W.; Smittipat, N.; Disratthakit, A.; Mahasirimongkol, S.; Yanai, H.; Yamada, N.; Nedsuwan, S.; Imasanguan, W.; et al. Evidence for host-bacterial co-evolution via genome sequence analysis of 480 Thai Mycobacterium tuberculosis lineage 1 isolates. Sci. Rep. 2018, 8, 11597. [Google Scholar] [CrossRef] [PubMed]
- Niemann, S.; Diel, R.; Khechinashvili, G.; Gegia, M.; Mdivani, N.; Tang, Y.-W. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J. Clin. Microbiol. 2010, 48, 3544–3550. [Google Scholar] [CrossRef]
- Shuaib, Y.A.; Utpatel, C.; Kohl, T.A.; Barilar, I.; Diricks, M.; Ashraf, N.; Wieler, L.H.; Kerubo, G.; Mesfin, E.A.; Diallo, A.B.; et al. Origin and global expansion of Mycobacterium tuberculosis Complex lineage 3. Genes 2022, 13, 990. [Google Scholar] [CrossRef]
- Napier, G.; Campino, S.; Merid, Y.; Abebe, M.; Woldeamanuel, Y.; Aseffa, A.; Hibberd, M.L.; Phelan, J.; Clark, T.G. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020, 12, 114. [Google Scholar] [CrossRef]
- National Tuberculosis Program. Fourth Nationwide Anti-Tuberculosis Drug Resistance Survey in Myanmar Protocol; National Tuberculosis Programme: Naypyitaw, Myanmar, 2019. [Google Scholar]
- Phyu, A.N.; Aung, S.T.; Palittapongarnpim, P.; Htet, K.K.K.; Mahasirimongkol, S.; Aung, H.L.; Chaiprasert, A.; Chongsuvivatwong, V. Distribution of Mycobacterium tuberculosis Lineages and Drug Resistance in Upper Myanmar. Trop. Med. Infect. Dis. 2022, 7, 448. [Google Scholar] [CrossRef]
- Maung, H.M.W.; Palittapongarnpim, P.; Aung, H.L.; Surachat, K.; Nyunt, W.W.; Chongsuvivatwong, V. Geno-Spatial Distribution of Mycobacterium tuberculosis and Drug Resistance Profiles in Myanmar–Thai Border Area. Trop. Med. Infect. Dis. 2020, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Department of Population Ministry of Immigration and Population. The 2014 Myanmar Population and Housing Census, Mandalay Region; Department of Population Ministry of Immigration and Population: Naypyitaw, Myanmar, 2015. [Google Scholar]
- National Tuberculosis Programme; Ministry of Health and Sports. Annual National Tuberculosis Report; National Tuberculosis Programme: Naypyitaw, Myanmar, 2021. [Google Scholar]
- Chongsuvivatwong, V. Analysis of Epidemiological Data Using R and Epicalc; Prince of Songkla University: Hat Yai, Thailand, 2012; p. 328. [Google Scholar]
- Rivière, E.; Heupink, T.H.; Ismail, N.; Dippenaar, A.; Clarke, C.; Abebe, G.; Heusden, P.; Warren, R.; Meehan, C.J.; Van Rie, A. Capacity building for whole genome sequencing of Mycobacterium tuberculosis and bioinformatics in high TB burden countries. Brief. Bioinform. 2021, 22, bbaa246. [Google Scholar] [CrossRef]
- Aung, H.L.; Tun, T.; Moradigaravand, D.; Köser, C.U.; Nyunt, W.W.; Aung, S.T.; Lwin, T.; Thinn, K.K.; Crump, J.A.; Parkhill, J.; et al. Whole-genome sequencing of multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. J. Glob. Antimicrob. Resist. 2016, 6, 113–117. [Google Scholar] [CrossRef]
- Aung, H.L.; Nyunt, W.W.; Fong, Y.; Biggs, P.J.; Winkworth, R.C.; Lockhart, P.J.; Yeo, T.W.; Hill, P.C.; Cook, G.M.; Aung, S.T. Genomic Profiling of Mycobacterium tuberculosis strains, Myanmar. Emerg. Infect. Dis. 2021, 27, 2847–2855. [Google Scholar] [CrossRef]
- Zade, A.; Shah, S.; Hirani, N.; Kondabagil, K.; Joshi, A.; Chatterjee, A. Whole-genome sequencing of presumptive MDR-TB isolates from a tertiary healthcare setting in Mumbai. J. Glob. Antimicrob. Resist. 2022, 31, 256–262. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Poplin, R.; Ruano-Rubio, V.; DePristo, M.A.; Fennell, T.J.; Carneiro, M.O.; Van der Auwera, G.A.; Kling, D.E.; Gauthier, L.D.; Levy-Moonshine, A.; Roazen, D.; et al. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv 2017. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Walker, T.M.; Ip, C.L.; Harrell, R.H.; Evans, J.T.; Kapatai, G.; Dedicoat, M.J.; Eyre, D.W.; Wilson, D.J.; Hawkey, P.M.; Crook, D.W.; et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: A retrospective observational study. Lancet Infect. Dis. 2013, 13, 137–146. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]
- Phelan, J.E.; O’Sullivan, D.M.; Machado, D.; Ramos, J.; Oppong, Y.E.; Campino, S.; O’Grady, J.; McNerney, R.; Hibberd, M.L.; Viveiros, M.; et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Song, W.M.; Li, Y.F.; Ma, X.B.; Liu, J.Y.; Tao, N.N.; Liu, Y.; Zhang, Q.Y.; Xu, T.T.; Li, S.J.; Yu, C.B.; et al. Primary drug resistance of Mycobacterium tuberculosis in Shandong, China, 2004–2018. Respir. Res. 2019, 20, 223. [Google Scholar] [CrossRef] [PubMed]
- Séraphin, M.N.; Norman, A.; Rasmussen, E.M.; Gerace, A.M.; Chiribau, C.B.; Rowlinson, M.C.; Lillebaek, T.; Lauzardo, M. Direct transmission of within-host Mycobacterium tuberculosis diversity to secondary cases can lead to variable between-host heterogeneity without de novo mutation: A genomic investigation. EBioMedicine 2019, 47, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Phelan, J.E.; Lim, D.R.; Mitarai, S.; de Sessions, P.F.; Tujan, M.A.A.; Reyes, L.T.; Medado, I.A.P.; Palparan, A.G.; Naim, A.N.M.; Jie, S.; et al. Mycobacterium tuberculosis whole genome sequencing provides insights into the Manila strain and drug-resistance mutations in the Philippines. Sci. Rep. 2019, 9, 9305. [Google Scholar] [CrossRef]
- Liu, Q.; Ma, A.; Wei, L.; Pang, Y.; Wu, B.; Luo, T.; Zhou, Y.; Zheng, H.X.; Jiang, Q.; Gan, M.; et al. China’s tuberculosis epidemic stems from historical expansion of four strains of Mycobacterium tuberculosis. Nat. Ecol. Evol. 2018, 2, 1982–1992. [Google Scholar] [CrossRef]
- Klopper, M.; Warren, R.M.; Hayes, C.; van Pittius, N.C.G.; Streicher, E.M.; Müller, B.; Sirgel, F.A.; Chabula-Nxiweni, M.; Hoosain, E.; Coetzee, G.; et al. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg. Infect. Dis. 2013, 19, 449–455. [Google Scholar] [CrossRef]
- Tessema, B.; Beer, J.; Emmrich, F.; Sack, U.; Rodloff, A.C. First insights into circulating Mycobacterium tuberculosis complex lineages and drug resistance in West Gojjam, Northwest Ethiopia. PLoS ONE 2013, 8, e77691. [Google Scholar] [CrossRef]
- Panaiotov, S.; Madzharov, D.; Hodzhev, Y. Biodiversity of Mycobacterium tuberculosis in Bulgaria Related to Human Migrations or Ecological Adaptation. Microorganisms 2022, 10, 146. [Google Scholar] [CrossRef]
- Crofton, J.; Mitchison, D.A. Streptomycin resistance in pulmonary tuberculosis. Br. Med. J. 1948, 2, 1009–1015. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Gao, H.; Zhang, Z.; Liu, Y.; Lu, J.; Dai, E. The roles of rpsL, rrs, and gidB mutations in predicting streptomycin-resistant drugs used on clinical Mycobacterium tuberculosis isolates from Hebei Province, China. Int. J. Clin. Exp. Pathol. 2019, 12, 2713–2721. [Google Scholar] [PubMed]
- Wong, S.Y.; Lee, J.S.; Kwak, H.K.; Via, L.E.; Boshoff, H.I.M.; Barry, C.E. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2011, 55, 2515–2522. [Google Scholar] [CrossRef] [PubMed]
- Nath, H.; Heekyung, J.; Sungweon, Y.; Koh, K.; Kim, C.; Kil, Y. Molecular genetics of Mycobacterium tuberculosis resistant to aminoglycosides and cyclic peptide capreomycin antibiotics in Korea. World J. Microbiol. Biotechnol. 2013, 29, 975–982. [Google Scholar]
- Huang, H.; Han, J.; Wang, J.; Song, C.; Liang, H.; Zhang, Z.L. rpsL gene analysis associated with Streptomycin resistance in Mycobacterium tuberculosis. Yi Chuan Xue Bao 2003, 30, 376–381. [Google Scholar] [PubMed]
- Ramaswamy, S.V.; Dou, S.J.; Rendon, A.; Yang, Z.; Cave, M.D.; Graviss, E.A. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J. Med. Microbiol. 2004, 53, 107–113. [Google Scholar] [CrossRef]
- Purkan, P.; Ihsanawati, I.; Natalia, D.; Syah, Y.M.; Retnoningrum, D.S.; Siswanto, I. Molecular analysis of katG encoding catalase-peroxidase from clinical isolate of isoniazid-resistant Mycobacterium tuberculosis. J. Med. Life 2018, 11, 160–167. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).