Global Prevalence of Antifungal-Resistant Candida parapsilosis: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol and Reporting Guideline
2.2. Search Strategy
2.3. Data Management and Study Selection
2.4. Data Extraction
2.5. Quality Assessment
2.6. Data Analysis
2.7. Subgroup and Sensitivity Analysis
3. Results
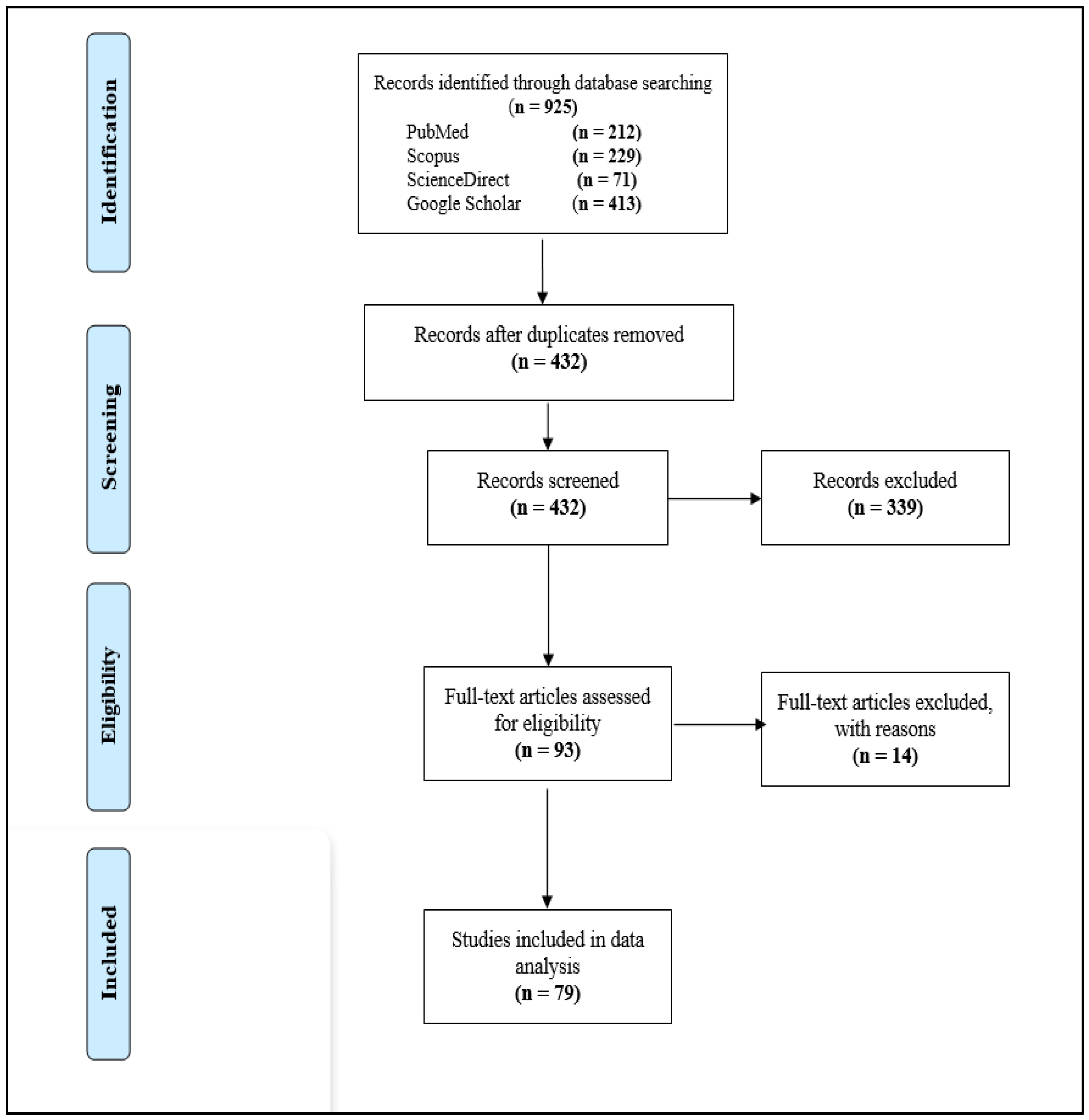
3.1. Study Selection
3.2. Characteristics of Included Studies
3.3. Prevalence of Fluconazole-Resistant C. parapsilosis Isolates
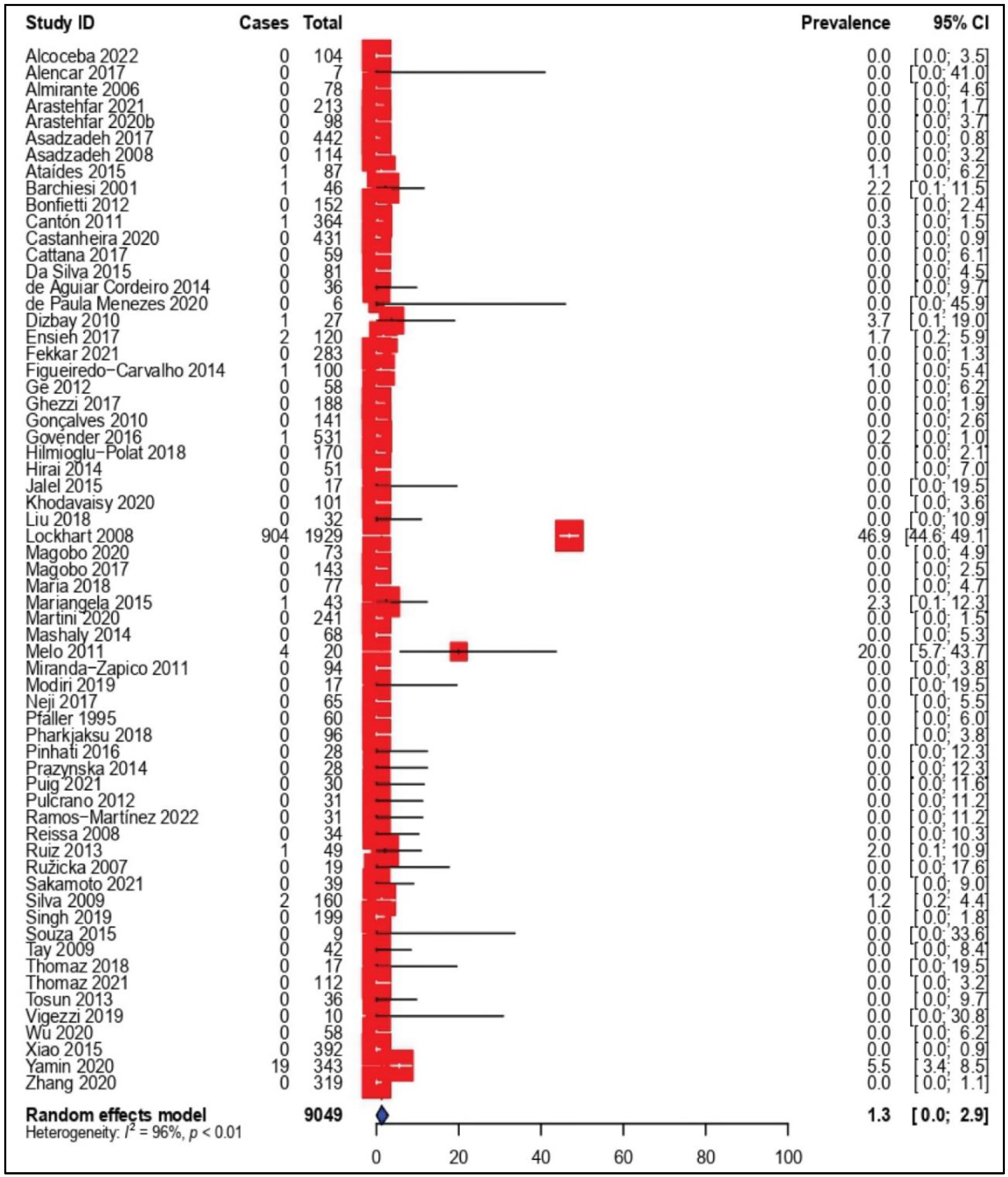
3.4. Prevalence of Amphotericin B-Resistant C. parapsilosis Isolates
3.5. Prevalence of Voriconazole-Resistant C. parapsilosis Isolates
3.6. Quality Assessment and Publication Bias
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kotey, F.C.; Dayie, N.T.; Tetteh-Uarcoo, P.B.; Donkor, E.S. Candida Bloodstream Infections: Changes in Epidemiology and Increase in Drug Resistance. Infect. Dis. Res. Treat. 2021, 14, 11786337211026927. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Santos, G.C.; Vasconcelos, C.C.; Lopes, A.J.O.; de Sousa Cartágenes, M.D.S.; Filho, A.K.D.B.; do Nascimento, F.R.F.; Ramos, R.; Pires, E.R.R.B.; de Andrade, M.S.; Rocha, F.M.G.; et al. Candida Infections and Therapeutic Strategies: Mechanisms of Action for Traditional and Alternative Agents. Front. Microbiol. 2018, 9, 1351. [Google Scholar] [CrossRef] [PubMed]
- Kett, D.H.; Azoulay, E.; Echeverria, P.M.; Vincent, J.-L. Candida bloodstream infections in intensive care units: Analysis of the extended prevalence of infection in intensive care unit study. Crit. Care Med. 2011, 39, 665–670. [Google Scholar] [CrossRef]
- Wan Ismail, W.N.A.; Jasmi, N.; Khan, T.M.; Hong, Y.H.; Neoh, C.F. The Economic Burden of Candidemia and Invasive Candidiasis: A Systematic Review. Value Health Reg. Issues 2020, 21, 53–58. [Google Scholar] [CrossRef]
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73, i4–i13. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, A.; Davidson, A.D.; Gow, N.A.; Maiden, M.C.; Odds, F.C. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 2005, 43, 284–292. [Google Scholar] [CrossRef]
- Pristov, K.E.; Ghannoum, M.A. Resistance of Candida to azoles and echinocandins worldwide. Clin. Microbiol. Infect. 2019, 25, 792–798. [Google Scholar] [CrossRef]
- Armstrong-James, D.; Brown, G.D.; Netea, M.G.; Zelante, T.; Gresnigt, M.S.; van de Veerdonk, F.L.; Levitz, S.M. Immunotherapeutic approaches to treatment of fungal diseases. Lancet Infect. Dis. 2017, 17, e393–e402. [Google Scholar] [CrossRef]
- Marak, M.B.; Dhanashree, B. Antifungal susceptibility and biofilm production of Candida spp. isolated from clinical samples. Int. J. Microbiol. 2018, 2018, 7495218. [Google Scholar] [CrossRef]
- Pahwa, N.; Kumar, R.; Nirkhiwale, S.; Bandi, A. Species distribution and drug susceptibility of Candida in clinical isolates from a tertiary care centre at Indore. Indian J. Med. Microbiol. 2014, 32, 44–48. [Google Scholar] [CrossRef]
- Ksiezopolska, E.; Gabaldón, T.J.G. Evolutionary emergence of drug resistance in Candida opportunistic pathogens. Genes 2018, 9, 461. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Munn, Z.; Moola, S.; Lisy, K.; Riitano, D.; Tufanaru, C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int. J. Evid.-Based Healthc. 2015, 13, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Hajissa, K.; Marzan, M.; Idriss, M.; Islam, A. Prevalence of Drug-Resistant Tuberculosis in Sudan: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 932. [Google Scholar] [CrossRef]
- Hajissa, K.; Islam, M.A.; Hassan, S.A.; Zaidah, A.R.; Ismail, N.; Mohamed, Z. Seroprevalence of SARS-CoV-2 Antibodies in Africa: A Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2022, 19, 7257. [Google Scholar] [CrossRef]
- Hajissa, K.; Islam, A.; Sanyang, A.M.; Mohamed, Z. Prevalence of intestinal protozoan parasites among school children in Africa: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2022, 16, e0009971. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, A.; Mahmoudi, S.; Rezaie, S.; Hashemi, S.J.; Dannaoui, E.; Badali, H.; Ghaffari, M.; Aala, F.; Izadi, A.; Maleki, A.; et al. In vitro synergy of echinocandins with triazoles against fluconazole-resistant Candida parapsilosis complex isolates. J. Glob. Antimicrob. Resist. 2020, 21, 331–334. [Google Scholar] [CrossRef]
- Alcoceba, E.; Gómez, A.; Lara-Esbrí, P.; Oliver, A.; Beltrán, A.F.; Ayestarán, I.; Muñoz, P.; Escribano, P.; Guinea, J. Fluconazole-resistant Candida parapsilosis clonally related genotypes: First report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin. Microbiol. Infect. 2022, 28, 1113–1119. [Google Scholar] [CrossRef]
- De Alencar, D.D.S.O.; de Sousa Tsujisaki, R.A.; Spositto, F.L.E.; de Oliveira Nunes, M.; de Almeida, A.A.; dos Anjos Martins, M.; de Souza CarvalhoMelhem, M.; Chang, M.R. Candidaemia due to Candida parapsilosis species complex at a hospital in Brazil: Clinical characteristics and antifungal susceptibility profile. Rev. Iberoam. Micol. 2017, 34, 106–108. [Google Scholar] [CrossRef]
- Almirante, B.; Rodríguez, D.; Cuenca-Estrella, M.; Almela, M.; Sanchez, F.; Ayats, J.; Alonso-Tarres, C.; Rodriguez-Tudela, J.L.; Pahissa, A.; Barcelona Candidemia Project Study Group. Epidemiology Risk Factors, and Prognosis of Candida parapsilosis Bloodstream Infections: Case-Control Population-Based Surveillance Study of Patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 2006, 44, 1681–1685. [Google Scholar] [CrossRef]
- Arastehfar, A.; Daneshnia, F.; Hilmioğlu-Polat, S.; Fang, W.; Yaşar, M.; Polat, F.; Metin, D.Y.; Rigole, P.; Coenye, T.; Ilkit, M.; et al. First report of candidemia clonal outbreak caused by emerging fluconazole-resistant Candida parapsilosis isolates harboring Y132F and/or Y132F + K143R in Turkey. Antimicrob. Agents Chemother. 2020, 64, e01001-20. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Hilmioglu-Polat, S.; Ilkit, M.; Yasar, M.; Polat, F.; Metin, D.Y.; Dokumcu, Ü.Z.; Pan, W.; Hagen, F.; et al. Genetically related micafungin-resistant Candida parapsilosis blood isolates harbouring novel mutation R658G in hotspot 1 of Fks1p: A new challenge? J. Antimicrob. Chemother. 2021, 76, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Najafzadeh, M.J.; Hagen, F.; Mahmoudi, S.; Salehi, M.; Zarrinfar, H.; Namvar, Z.; Zareshahrabadi, Z.; Khodavaisy, S.; et al. Evaluation of molecular epidemiology, clinical characteristics, antifungal susceptibility profiles, and molecular mechanisms of antifungal resistance of Iranian Candida parapsilosis species complex blood isolates. Front. Cell. Infect. Microbiol. 2020, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Epidemiology and molecular basis of resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb. Drug Resist. 2017, 23, 966–972. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Al-Sweih, N.A.; Ahmad, S.; Khan, Z.U. Antifungal susceptibility of clinical Candida parapsilosis isolates in Kuwait. Mycoses 2008, 51, 318–323. [Google Scholar] [CrossRef]
- Ataides, F.S.; Costa, C.R.; Souza, L.K.H.; Fernandes, O.D.L.; Jesuino, R.S.A.; Silva, M.D.R.R. Molecular identifi cation and antifungal susceptibility profi les of Candida parapsilosis complex species isolated from culture collection of clinical samples. Rev. Soc. Bras. Med. Trop. 2015, 48, 454–459. [Google Scholar] [CrossRef]
- Barchiesi, F.; Di Francesco, L.F.; Arzeni, D.; Caselli, F.; Simonetti, O.; Cellini, A.; Giacometti, A.; Offidani, A.; Scalise, G. Electrophoretic karyotyping and antifungal susceptibility patterns of Candida parapsilosis clinical isolates causing deep and superficial fungal infections. Mycopathologia 2001, 149, 117–121. [Google Scholar] [CrossRef]
- Bonfietti, L.X.; Martins, M.D.A.; Szeszs, M.W.; Pukiskas, S.B.S.; Purisco, S.U.; Pimentel, F.C.; Pereira, G.H.; Silva, D.C.; Oliveira, L.; Melhem, M.D.S.C. Prevalence, distribution and antifungal susceptibility profiles of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis bloodstream isolates. J. Med. Microbiol. 2012, 61, 1003–1008. [Google Scholar] [CrossRef]
- Cantón, E.; Pemán, J.; Quindós, G.; Eraso, E.; Miranda-Zapico, I.; Álvarez, M.; Merino, P.; Campos-Herrero, I.; Marco, F.; de la Pedrosa, E.G.G.; et al. Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia. Antimicrob. Agents Chemother. 2011, 55, 5590–5596. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 2020, 55, 105799. [Google Scholar] [CrossRef]
- Cattana, M.E.; Dudiuk, C.; Fernández, M.; Rojas, F.; Alegre, L.; Córdoba, S.; Garcia-Effron, G.; Giusiano, G. Identification of Candida parapsilosis sensu lato in pediatric patients and antifungal susceptibility testing. J. Antimicrob. Chemother. 2017, 61, e02754-16. [Google Scholar] [CrossRef] [PubMed]
- Corzo-Leon, D.E.; Peacock, M.; Rodriguez-Zulueta, P.; Salazar-Tamayo, G.J.; MacCallum, D.M. General hospital outbreak of invasive candidiasis due to azole-resistant Candida parapsilosis associated with an Erg11 Y132F mutation. Med. Mycol. 2021, 59, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, B.V.; Silva, L.B.; de Oliveira, D.B.C.; da Silva, P.R.; Ferreira-Paim, K.; Andrade-Silva, L.E.; Silva-Vergara, M.L.; Andrade, A.A. Species distribution, virulence factors, and antifungal susceptibility among Candida parapsilosis complex isolates recovered from clinical specimens. Mycopathologia 2015, 180, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Davari, A.; Haghani, I.; Hassanmoghadam, F.; Nabili, M.; Shokohi, T.; Hedayati, M.T.; Shabanzadeh, S.; Moazeni, M. Echinocandin resistance in Candida parapsilosis sensu stricto: Role of alterations in CHS3, FKS1 and Rho gene expression. J. Glob. Antimicrob. Res. 2020, 22, 685–688. [Google Scholar] [CrossRef]
- de Aguiar Cordeiro, R.; de Brito Macedo, R.; Teixeira, C.E.C.; de Farias Marques, F.J.; Bandeira, T.d.J.P.G.; Moreira, J.L.B.; Brilhante, R.S.N.; Rocha, M.F.G.; Sidrim, J.J.C. The calcineurin inhibitor cyclosporin A exhibits synergism with antifungals against Candida parapsilosis species complex. J. Med. Microb. 2014, 63, 936–944. [Google Scholar] [CrossRef]
- de Paula Menezes, R.; de Oliveira Melo, S.G.; Bessa, M.A.S.; Silva, F.F.; Alves, P.G.V.; Araújo, L.B.; Penatti, M.P.A.; Abdallah, V.O.S.; von Dollinger de Brito Röder, D.; dos Santos Pedroso, R. Candidemia by Candida parapsilosis in a neonatal intensive care unit: Human and environmental reservoirs, virulence factors, and antifungal susceptibility. Braz. J. Microb. 2020, 51, 851–860. [Google Scholar] [CrossRef]
- Demirci-Duarte, S.; Arikan-Akdagli, S.; Gülmez, D. Species distribution, azole resistance and related molecular mechanisms in invasive Candida parapsilosis complex isolates: Increase in fluconazole resistance in 21 years. Mycoses 2021, 64, 823–830. [Google Scholar] [CrossRef]
- Dizbay, M.; Fidan, I.; Kalkanci, A.; Sari, N.; Yalcin, B.; Kustimur, S.; Arman, D. High incidence of Candida parapsilosis candidaemia in non-neutropenic critically ill patients: Epidemiology and antifungal susceptibility. Scand. J. Infect. Dis. 2010, 42, 114–120. [Google Scholar] [CrossRef]
- Ensieh, L.; Ali, G.; Parivash, K.; Farideh, Z.; Hossein, M.; Rasoul, M.; Fatemeh, N.; Sassan, R. Regulation of ERG3, ERG6, and ERG11 genes in antifungal-resistant isolates of candida parapsilosis. Iran. Biomed. J. 2017, 21, 275–281. [Google Scholar]
- Fekkar, A.; Blaize, M.; Bouglé, A.; Normand, A.-C.; Raoelina, A.; Kornblum, D.; Kamus, L.; Piarroux, R.; Imbert, S. Hospital outbreak of fluconazole-resistant Candida parapsilosis: Arguments for clonal transmission and long-term persistence. Antimicrob. Ag. Chemother. 2021, 65, e02036-20. [Google Scholar] [CrossRef]
- Fernández-Ruiz, M.; Aguado, J.; Almirante, B.; Lora-Pablos, D.; Padilla, B.; Puig-Asensio, M.; Montejo, M.; García-Rodríguez, J.; Pemán, J.; Ruiz Pérez de Pipaón, M. CANDIPOP Project, GEIH-GEMICOMED (SEIMC), REIPI. 2014. Initial use of echinocandins does not negatively influence outcome in Candida parapsilosis bloodstream infection: A propensity score analysis. Clin. Infect. Dis. 2014, 58, 1413–1421. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo-Carvalho, M.H.G.; Barbedo, L.S.; Oliveira, M.M.; Brito-Santos, F.; Almeida-Paes, R.; Zancopé-Oliveira, R.M. Comparison of commercial methods and the CLSI broth microdilution to determine the antifungal susceptibility of Candida parapsilosis complex bloodstream isolates from three health institutions in Rio de Janeiro, Brazil. Mycopathologia 2014, 178, 27–35. [Google Scholar] [CrossRef]
- Garcia-Effron, G.; Canton, E.; Pemán, J.; Dilger, A.; Romá, E.; Perlin, D.S. Epidemiology and echinocandin susceptibility of Candida parapsilosis sensu lato species isolated from bloodstream infections at a Spanish university hospital. J. Antimicrob. Chemother. 2012, 67, 2739–2748. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.P.; Boekhout, T.; Zhan, P.; Lu, G.X.; Shen, Y.N.; Li, M.; Shao, H.F.; Liu, W.D. Characterization of the Candida parapsilosis complex in East China: Species distribution differs among cities. Med. Mycol. 2012, 50, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, M.C.; Brunetti, G.; Visconti, V.; Giordano, A.; Raponi, G. Candidaemia in a tertiary care academic hospital in Italy. The impact of C. parapsilosis complex on the species distribution and antifungal susceptibility. J. Med. Microb. 2017, 66, 990–998. [Google Scholar] [CrossRef]
- Gonçalves, S.; Amorim, C.; Nucci, M.; Padovan, A.; Briones, M.R.; Melo, A.S.; Colombo, A.L. Prevalence rates and antifungal susceptibility profiles of the Candida parapsilosis species complex: Results from a nationwide surveillance of candidaemia in Brazil. Clin. Microb. Infect. 2010, 16, 885–887. [Google Scholar] [CrossRef]
- Govender, N.; Patel, J.; Magobo, R.; Naicker, S.; Wadula, J.; Whitelaw, A.; Coovadia, Y.; Kularatne, R.; Govind, C.; Lockhart, S. TRAC-South Africa group. Emergence of azole-resistant Candida parapsilosis causing bloodstream infection: Results from laboratory-based sentinel surveillance in South Africa. J. Antimicrob. Chemother. 2016, 71, 1994–2004. [Google Scholar] [CrossRef]
- Grossman, N.T.; Pham, C.D.; Cleveland, A.A.; Lockhart, S.R. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a US surveillance system. Antimicrob. Age. Chemother. 2015, 59, 1030–1037. [Google Scholar] [CrossRef]
- Hilmioğlu-Polat, S.; Sharifynia, S.; Öz, Y.; Aslan, M.; Gündoğdu, N.; Serin, A.; Rafati, H.; Mohammadi, F.; Yeşim-Metin, D.; Döğen, A. Genetic diversity and antifungal susceptibility of Candida parapsilosis sensu stricto isolated from bloodstream infections in Turkish patients. Mycopathologia 2018, 183, 701–708. [Google Scholar] [CrossRef]
- Hirai, Y.; Asahata, S.; Ainoda, Y.; Goto, A.; Fujita, T.; Totsuka, K. Nosocomial Candida parapsilosis candidaemia: Risk factors, antifungal susceptibility and outcome. J. Hosp. Infect. 2014, 87, 54–58. [Google Scholar] [CrossRef]
- Jalel, B.; Fatma, S. Molecular Identification and Antifungal Susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Sousse Region, Tunisia. Med. Mycol. 2015, 1, 8. [Google Scholar]
- Khan, Z.; Ahmad, S.; Joseph, L.; Chandy, R.; Theyyathel, A. Comparative In Vitro Susceptibility of Clinical Isolates of Candida parapsilosis Complex and Other Candida Species to Caspofungin and Anidulafungin by Etest. J. Chemother. 2011, 23, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Khodavaisy, S.; Badali, H.; Meis, J.F.; Modiri, M.; Mahmoudi, S.; Abtahi, H.; Salehi, M.; Dehghan Manshadi, S.A.; Aala, F.; Agha Kuchak Afshari, S.; et al. Comparative in vitro activities of seven antifungal drugs against clinical isolates of Candida parapsilosis complex. J. Mycol. Med. 2020, 30, 100968. [Google Scholar] [CrossRef]
- Liu, Y.; Kang, M.; Ye, H.; Zong, Z.; Lv, X. Analysis on clinical characteristics and drug resistance of Candida parapsilosis bloodstream infections in West China Hospital, China, from 2012 to 2015. J. Mycol. Med. 2018, 28, 222–226. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Messer, S.A.; Pfaller, M.A.; Diekema, D.J. Geographic distribution and antifungal susceptibility of the newly described species Candida orthopsilosis and Candida metapsilosis in comparison to the closely related species Candida parapsilosis. J. Clin. Microbiol. 2008, 46, 2659–2664. [Google Scholar] [CrossRef]
- Magobo, R.E.; Lockhart, S.R.; Govender, N.P. Fluconazole-resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, South Africa. Mycoses 2020, 63, 471–477. [Google Scholar] [CrossRef]
- Magobo, R.E.; Naicker, S.D.; Wadula, J.; Nchabeleng, M.; Coovadia, Y.; Hoosen, A.; Lockhart, S.R.; Govender, N.P.; van Rensburg, C.J.; TRAC-South Africa Group; et al. Detection of neonatal unit clusters of Candida parapsilosis fungaemia by microsatellite genotyping: Results from laboratory-based sentinel surveillance, South Africa, 2009–2010. Mycoses 2017, 60, 320–327. [Google Scholar] [CrossRef]
- Maria, S.; Barnwal, G.; Kumar, A.; Mohan, K.; Vinod, V.; Varghese, A.; Biswas, R. Species distribution and antifungal susceptibility among clinical isolates of Candida parapsilosis complex from India. Rev. Iberoam. Micol. 2018, 35, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Mariangela, Z.; Lucieri, O.; Rafael, M.; Anna, C.; Andrea, R. Candida parapsilosis (sensu lato) isolated from hospital located in the southeast of Brazil: Species distribution, antifungal susceptibility and virulence attributes. Int. J. Med. Microbiol. 2015, 305, 848–859. [Google Scholar]
- Martini, C.; Torelli, R.; De Groot, T.; De Carolis, E.; Morandotti, G.A.; De Angelis, G.; Posteraro, B.; Meis, J.F.; Sanguinetti, M. Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a large Italian hospital. Front. Cell. Infect. Microbiol. 2020, 10, 232. [Google Scholar] [CrossRef]
- Mashaly, G. Candida Parapsilosis Complex Species and Antifungal Susceptibility Profile in Patients of Intensive Care Units of Mansoura University Hospitals. Mans. Med. J. 2014, 43, 219–237. [Google Scholar] [CrossRef]
- Melo, A.S.; Bizerra, F.C.; Freymüller, E.; Arthington-Skaggs, B.A.; Colombo, A.L. Biofilm production and evaluation of antifungal susceptibility amongst clinical Candida spp. isolates, including strains of the Candida parapsilosis complex. Med. Mycol. 2011, 49, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Mesini, A.; Mikulska, M.; Giacobbe, D.R.; Del Puente, F.; Gandolfo, N.; Codda, G.; Orsi, A.; Tassinari, F.; Beltramini, S.; Marchese, A. Changing epidemiology of candidaemia: Increase in fluconazole-resistant Candida parapsilosis. Mycoses 2020, 63, 361–368. [Google Scholar] [CrossRef]
- Miranda-Zapico, I.; Eraso, E.; Carrillo-Munoz, A.J.; Hernández-Molina, J.M. Prevalence and antifungal susceptibility patterns of new cryptic species inside the species-complexes Candida parapsilosis and Candida glabrata among blood isolates from a Spanish tertiary hospital. J. Antimicrob. Chemother. 2011, 66, 2315–2322. [Google Scholar] [CrossRef][Green Version]
- Modiri, M.; Hashemi, S.J.; Ghazvini, R.D.; Khodavaisy, S.; Ahmadi, A.; Ghaffari, M.; Rezaie, S. Antifungal susceptibility pattern and biofilm-related genes expression in planktonic and biofilm cells of Candida parapsilosis species complex. Curr. Med. Mycol. 2019, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Neji, S.; Hadrich, I.; Trabelsi, H.; Abbes, S.; Cheikhrouhou, F.; Sellami, H.; Makni, F.; Ayadi, A. Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. J. Biomed. Sci. 2017, 24, 67. [Google Scholar] [CrossRef]
- Pfaller, M. Global Antifungal Surveillance Group: Geographic and temporal trends in isolation and antifungal susceptibility of Candida parapsilosis: A global assessment from the ARTEMIS DISK Antifungal Surveillance Program, 2001 to 2005. J. Clin Microb. 2008, 46, 842–849. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Messer, S.A.; Hollis, R.J. Variations in DNA subtype, antifungal susceptibility, and slime production among clinical isolates of Candida parapsilosis. Diag. Microb. Infect. Dis. 1995, 21, 9–14. [Google Scholar] [CrossRef]
- Pharkjaksu, S.; Chongtrakool, P.; Suwannakarn, K.; Ngamskulrungroj, P. Species distribution, virulence factors, and antifungal susceptibility among Candida parapsilosis complex isolates from clinical specimens at Siriraj Hospital, Thailand, from 2011 to 2015. Med. Mycol. 2018, 56, 426–433. [Google Scholar] [CrossRef]
- Pinhati, H.M.S.; Casulari, L.A.; Souza, A.C.R.; Siqueira, R.A.; Damasceno, C.M.G.; Colombo, A.L. Outbreak of candidemia caused by fluconazole resistant Candida parapsilosis strains in an intensive care unit. BMC Infect. Dis. 2016, 16, 1–6. [Google Scholar] [CrossRef]
- Prażyńska, M.; Gospodarek, E. In vitro effect of amphotericin B on Candida albicans, Candida glabrata and Candida parapsilosis biofilm formation. Mycopathologia 2014, 177, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Puig, C.R.d.A.; Merino, M.d.S.G.; Fonseca, A.D.M.P.; Balbín, J.A. Characterization, antifungal susceptibility and virulence of Candida parapsilosis complex isolates in a tertiary hospital in Cantabria, Northern Spain. Enferm. Infect. Microb. Clín. 2021. [Google Scholar] [CrossRef]
- Pulcrano, G.; Panellis, D.; De Domenico, G.; Rossano, F.; Catania, M.R. Ambroxol influences voriconazole resistance of Candida parapsilosis biofilm. FEMS Yeast Res. 2012, 12, 430–438. [Google Scholar] [CrossRef]
- Raghuram, A.; Restrepo, A.; Safadjou, S.; Cooley, J.; Orloff, M.; Hardy, D.; Butler, S.; Koval, C.E. Invasive fungal infections following liver transplantation: Incidence, risk factors, survival, and impact of fluconazole-resistant Candida parapsilosis (2003–2007). Liv. Transplant. 2012, 18, 1100–1109. [Google Scholar] [CrossRef]
- Ramos-Martínez, A.; Pintos-Pascual, I.; Guinea, J.; Gutiérrez-Villanueva, A.; Gutiérrez-Abreu, E.; Díaz-García, J.; Asensio, Á.; Iranzo, R.; Sánchez-Romero, I.; Muñoz-Algarra, M. Impact of the COVID-19 Pandemic on the Clinical Profile of Candidemia and the Incidence of Fungemia Due to Fluconazole-Resistant Candida parapsilosis. J. Fungi 2022, 8, 451. [Google Scholar] [CrossRef]
- Reiss, E.; Lasker, B.A.; Iqbal, N.J.; James, M.J.; Arthington-Skaggs, B.A. Molecular epidemiology of Candida parapsilosis sepsis from outbreak investigations in neonatal intensive care units. Infect. Genet Evol. 2008, 8, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Roberto, A.E.M.; Xavier, D.E.; Vidal, E.E.; Vidal, C.F.D.L.; Neves, R.P.; Lima-Neto, R.G.D. Rapid detection of echinocandins resistance by MALDI-TOF MS in Candida parapsilosis complex. Microorganisms 2020, 8, 109. [Google Scholar] [CrossRef]
- Ruiz, L.d.S.; Khouri, S.; Hahn, R.C.; Da Silva, E.G.; de Oliveira, V.K.P.; Gandra, R.F.; Paula, C.R. Candidemia by species of the Candida parapsilosis complex in children’s hospital: Prevalence, biofilm production and antifungal susceptibility. Mycopathologia 2013, 175, 231–239. [Google Scholar] [CrossRef]
- Růžička, F.; Holá, V.; Votava, M.; Tejkalová, R. Importance of biofilm inCandida parapsilosis and evaluation of its susceptibility to antifungal agents by colorimetric method. Folia Microb. 2007, 52, 209–214. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Kawabe, K.; Suzuki, T.; Sano, K.; Ide, K.; Nishigaki, T.; Enoki, Y.; Taguchi, K.; Koike, H.; Kato, H. Species distribution of candidemia and their susceptibility in a single japanese university hospital: Prior micafungin use affects the appearance of candida parapsilosis and elevation of micafungin mics in non-parapsilosis candida species. J. Fungi 2021, 7, 596. [Google Scholar] [CrossRef]
- Sarvikivi, E. Lyytik inen O, Soll DR, Pujol C, Pfaller MA, ä Richardson M, et al. Emergence of fluconazole resistance in a Candida parapsilosis strain that caused infections in a neonatal intensive care unit. J. Clin. Microb. 2005, 43, 2729–2735. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.P.; Miranda, I.M.; Lisboa, C.; Pina-Vaz, C.; Rodrigues, A.G. Prevalence, distribution, and antifungal susceptibility profiles of Candida parapsilosis, C. orthopsilosis, and C. metapsilosis in a tertiary care hospital. J. Clin. Microb. 2009, 47, 2392–2397. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, P.K.; de Groot, T.; Kumar, A.; Mathur, P.; Tarai, B.; Sachdeva, N.; Upadhyaya, G.; Sarma, S.; Meis, J.F. Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J. Antimicrob. Chemother. 2019, 74, 1260–1268. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.C.R.; Fuchs, B.B.; Pinhati, H.M.; Siqueira, R.A.; Hagen, F.; Meis, J.F.; Mylonakis, E.; Colombo, A.L. Candida parapsilosis resistance to fluconazole: Molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob. Agents Chemother. 2015, 59, 6581–6587. [Google Scholar] [CrossRef] [PubMed]
- Tay, S.T.; Na, S.L.; Chong, J. Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J. Med. Microb. 2009, 58, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Thomaz, D.; de Almeida Jr, J.; Lima, G.; Nunes, M.; Camargo, C.; Grenfell, R. An azole-resistant Candida parapsilosis outbreak: Clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front. Microbiol. 2018, 9, 2997. [Google Scholar] [CrossRef] [PubMed]
- Thomaz, D.Y.; de Almeida, J.N.; Sejas, O.N.; Del Negro, G.; Carvalho, G.O.; Gimenes, V.M.; de Souza, M.E.B.; Arastehfar, A.; Camargo, C.H.; Motta, A.L. Environmental clonal spread of azole-resistant Candida parapsilosis with Erg11-Y132F mutation causing a large candidemia outbreak in a Brazilian Cancer Referral Center. J. Fungi 2021, 7, 259. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; Del Negro, G.; Ribeiro, L.B.; da Silva, M.; Carvalho, G.O.; Camargo, C.H.; de Almeida, J.N.; Motta, A.L.; Siciliano, R.F.; Sejas, O.N. A Brazilian Inter-Hospital Candidemia Outbreak Caused by Fluconazole-Resistant Candida parapsilosis in the COVID-19 Era. J. Fungi 2022, 8, 100. [Google Scholar] [CrossRef]
- Tosun, I.; Akyuz, Z.; Guler, N.C.; Gulmez, D.; Bayramoglu, G.; Kaklikkaya, N.; Arikan-Akdagli, S.; Aydin, F. Distribution, virulence attributes and antifungal susceptibility patterns of Candida parapsilosis complex strains isolated from clinical samples. Med. Mycol. 2013, 51, 483–492. [Google Scholar] [CrossRef]
- Treviño-Rangel, R.d.J.; Garza-González, E.; González, J.G.; Bocanegra-García, V.; Llaca, J.M.; González, G.M. Molecular characterization and antifungal susceptibility of the Candida parapsilosis species complex of clinical isolates from Monterrey, Mexico. Med. Mycol. 2012, 50, 781–784. [Google Scholar] [CrossRef]
- Vigezzi, C.; Icely, P.A.; Dudiuk, C.; Rodríguez, E.; Miró, M.S.; Castillo, G.D.V.; Azcurra, A.I.; Abiega, C.; Caeiro, J.P.; Riera, F.O.; et al. Frequency, virulence factors and antifungal susceptibility of Candida parapsilosis species complex isolated from patients with candidemia in the central region of Argentina. J. Mycol. Med. 2019, 29, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wei, D.; Gong, X.; Shen, Y.; Zhu, Y.; Wang, J.; Gao, Z. Initial use of voriconazole positively affects outcome of Candida parapsilosis bloodstream infection: A retrospective analysis. Transl. Pediatr. 2020, 9, 480. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Fan, X.; Chen, S.; Wang, H.; Sun, Z.; Liao, K.; Chen, S.; Yan, Y.; Kang, M. Hu 190 ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Kong F, Xu YC. 2015. Antifungal 191 susceptibilities of Candida glabrata species complex, Candida krusei, Candida 192 parapsilosis species complex and Candida tropicalis causing invasive candidiasis in 193 China: 3 year national surveillance. J. Antimicrob. Chemother. 2015, 70, 802–810. [Google Scholar] [PubMed]
- Yamin, D.; Husin, A.; Harun, A. Distribution of candidemia in malaysian tertiary care hospital revealed predominance of candida parapsilosis. Trop. Biomed. 2020, 37, 903–910. [Google Scholar]
- Zhang, L.; Yu, S.-Y.; Chen, S.C.-A.; Xiao, M.; Kong, F.; Wang, H.; Ning, Y.-T.; Lu, M.-Y.; Sun, T.-S.; Hou, X. Molecular characterization of Candida parapsilosis by microsatellite typing and emergence of clonal antifungal drug resistant strains in a multicenter surveillance in China. Front. Microb. 2020, 11, 1320. [Google Scholar] [CrossRef]
- Gauna, T.T.; Oshiro, E.; Luzio, Y.C.; Paniago, A.M.M.; Pontes, E.R.J.C.; Chang, M.R. Bloodstream infection in patients with end-stage renal disease in a teaching hospital in central-western Brazil. Rev. Soc. Bras. Med. Trop. 2013, 46, 426–432. [Google Scholar] [CrossRef]
- Girmenia, C.; Martino, P.; De Bernardis, F.; Gentile, G.; Boccanera, M.; Monaco, M.; Antonucci, G.; Cassone, A. Rising incidence of Candida parapsilosis fungemia in patients with hematologic malignancies: Clinical aspects, predisposing factors, and differential pathogenicity of the causative strains. Clin. Infect. Dis. 1996, 23, 506–514. [Google Scholar] [CrossRef][Green Version]
- Huang, Y.-C.; Lin, T.-Y.; Leu, H.-S.; Peng, H.-L.; Wu, J.-H.; Chang, H.-Y. Outbreak ofCandida parapsilosis fungemia in neonatal intensive care units: Clinical implications and genotyping analysis. Infection 1999, 27, 97–102. [Google Scholar] [CrossRef]
- Cisneros Herreros, J.; Cordero Matía, E. Therapeutic armamentarium against systemic fungal infections. Clin. Microb. Infect. 2006, 12, 53–64. [Google Scholar] [CrossRef]
- Kohno, S.; Izumikawa, K.; Yoshida, M.; Takesue, Y.; Oka, S.; Kamei, K.; Miyazaki, Y.; Yoshinari, T.; Kartsonis, N.A.; Niki, Y. A double-blind comparative study of the safety and efficacy of caspofungin versus micafungin in the treatment of candidiasis and aspergillosis. Eur. J. Clin. Microb. Infect. Dis. 2013, 32, 387–397. [Google Scholar] [CrossRef]
- Kawai, A.; Yamagishi, Y.; Mikamo, H. In vitro efficacy of liposomal amphotericin B, micafungin and fluconazole against non-albicans Candida species biofilms. J. Infect. Chemother. 2015, 21, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, L. Analysis of the utilization of antifungal agents in our hospital during 2007–2009. Chin. Gen. Pract. 2012, 15, 324–327. [Google Scholar]
- Qin, J.; Yang, H.; Shan, Z.; Jiang, L.; Zhang, Q. Clinical efficacy and safety of antifungal drugs for the treatment of Candida parapsilosis infections: A systematic review and network meta-analysis. J. Med. Microb. 2021, 70. [Google Scholar] [CrossRef] [PubMed]
- Zeng, r.; Li, M.; Chen, Q.; Wang, L.; Lv, G.; Shen, Y.; Cai, Q.; Li, C.; Tang, R.; Liu, W. Dynamic study on the susceptibilities of caspofungin and micafungin to Candida species in vitro. Chin. J. Mycol. 2011, 6, 276. [Google Scholar]
- Nawaz, A.; Pärnänen, P.; Kari, K.; Meurman, J.H. Proteolytic activity and cytokine up-regulation by non-albicans Candida albicans. Arch. Microb. 2015, 197, 533–537. [Google Scholar] [CrossRef]
- Verma, R.; Pradhan, D.; Hasan, Z.; Singh, H.; Jain, A.K.; Khan, L.A. A systematic review on distribution and antifungal resistance pattern of Candida species in the Indian population. Med. Mycol. 2021, 59, 1145–1165. [Google Scholar] [CrossRef]



| No | Study ID [References] | Study Design | Country | No. of Patients | Clinical Isolates Identification Method | AFST Method | Total Isolates Tested | Tested Antifungal | |
|---|---|---|---|---|---|---|---|---|---|
| Male (n) | Female (n) | ||||||||
| 1 | Ahmadi 2020 [17] | NR | NR | NR | NR | Molecular methods | BMD | 15 | FLC |
| 2 | Alcoceba 2022 [18] | NR | Spain | 53 | 17 | Molecular methods | BMD | 104 | FLC, AMB, POS, VOR, ANF and MCF. |
| 3 | Alencar 2017 [19] | Cross sectional | Brazil | NR | NR | Molecular methods | Vitek-2, BMD | 7 | FLC, AMB, ITC and VOR. |
| 4 | Almirante 2006 [20] | Prospective Case Control | Spain | 43 | 35 | Conventional methods | BMD | 78 | FLC, AMB, ITC, VOR and CAS. |
| 5 | Arastehfar 2020a [21] | Cross sectional | Turkey | 123 | 91 | Both conventional and molecular methods | BMD | 225 | FLC and VOR. |
| 6 | Arastehfar 2021 [22] | Cross sectional | Turkey | NR | NR | Molecular methods | BMD | 213 | AMB, ANF and MCF. |
| 7 | Arastehfar 2020b [23] | Cross sectional | Iran | 45 | 45 | Molecular methods | BMD | 98 | FLC, AMB, ITC, ANF and MCF. |
| 8 | Asadzadeh 2017 [24] | Cross sectional | Kuwait | NR | NR | Both conventional and molecular methods | E-test, Vitek-2, BMD | 442 | FLC, AMB, VOR, CAS and MCF. |
| 9 | Asadzadeh 2008 [25] | Cross sectional | Kuwait | NR | NR | Both conventional and molecular methods | E-test | 114 | FLC, AMB, POS and CAS. |
| 10 | Ataídes 2015 [26] | Cross sectional | Brazil | NR | NR | Both conventional and molecular methods | E-test | 87 | FLC, AMB, ITC, POS, VOR and CAS. |
| 11 | Barchiesi 2001 [27] | Cross sectional | Italy | NR | NR | Conventional methods | BMD | 46 | FLC, AMB and ITC. |
| 12 | Bonfietti 2012 [28] | Cross sectional | Brazil | NR | NR | Both conventional and molecular methods | BMD | 152 | FLC, AMB and ITC. |
| 13 | Cantón 2011 [29] | Prospective Cohort | Spain | 231 | 169 | Both conventional and molecular methods | Sensititre YeastOne BMD | 364 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 14 | Castanheira 2020 [30] | Cross sectional | 25 countries | NR | NR | Both conventional and molecular methods | BMD | 431 | FLC, AMB, POS, VOR, CAS, ANF and MCF. |
| 15 | Cattana 2017 [31] | Cross sectional | Argentina | NR | NR | Both conventional and molecular methods | BMD | 59 | FLC, AMB, ITC, VOR, CAS and ANF. |
| 16 | Corzo-Leon 2021 [32] | Cross sectional | Mexico | 45 | 29 | Both conventional and molecular methods | Vitek-2, BMD | 29 | FLC and VOR. |
| 17 | Da Silva 2015 [33] | Cross sectional | Brazil | 27 | 54 | Both conventional and molecular methods | BMD | 81 | FLC, AMB, ITC and VOR. |
| 18 | Davari 2020 [34] | Cross sectional | Iran | NR | NR | Both conventional and molecular methods | BMD | 105 | CAS, ANF and MCF. |
| 19 | de Aguiar Cordeiro 2014 [35] | NR | Italy | NR | NR | Both conventional and molecular methods | BMD | 36 | FLC, AMB, VOR and CAS. |
| 20 | de Paula Menezes 2020 [36] | Cross sectional | Brazil | NR | NR | Both conventional and molecular methods | BMD | 6 | FLC, AMB and MCF. |
| 21 | Demirci-Duarte 2021 [37] | Cross sectional | Turkey | NR | NR | Both conventional and molecular methods | BMD | 181 | FLC, POS and VOR. |
| 22 | Dizbay 2010 [38] | Cross sectional | Turkey | 13 | 14 | Conventional methods | BMD | 27 | FLC, AMB, VOR and CAS. |
| 23 | Ensieh 2017 [39] | Cross sectional | Iran | NR | NR | NR | BMD | 120 | FLC, AMB and ITC. |
| 24 | Fekkar 2021 [40] | Cross sectional | France | NR | NR | Molecular methods | E-test, BMD | 283 | FLC, AMB, ITC, POS, VOR, CAS and MCF. |
| 25 | Fernández-Ruiz 2014 [41] | Cross sectional | Spain | 127 | 63 | Both conventional and molecular methods | BMD | 189 | FLC, VOR, ANF and MCF. |
| 26 | Figueiredo-Carvalho 2014 [42] | Cross sectional | Brazil | NR | NR | Both conventional and molecular methods | E-test, Vitek-2, BMD | 100 | FLC, AMB, ITC, VOR and CAS. |
| 27 | Garcia-Effron 2012 [43] | Cross sectional | Spain | 179 | 108 | Both conventional and molecular methods | BMD | 287 | CAS, ANF and MCF. |
| 28 | Ge 2012 [44] | Cross sectional | China | NR | NR | Both conventional and molecular methods | BMD | 58 | FLC, AMB, ITC, VOR and MCF.0 |
| 29 | Ghezzi 2017 [45] | Retrospective cohort | Italy | 264 | 188 | Both conventional and molecular methods | BMD | 188 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 30 | Gonçalves 2010 [46] | Cross sectional | Brazil | 86 | 60 | Both conventional and molecular methods | BMD | 141 | FLC, AMB, ITC, VOR and CAS. |
| 31 | Govender 2016 [47] | Cross sectional | South Africa | 279/513 | 234/513 | Both conventional and molecular methods | BMD | 531 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 32 | Grossman 2015 [48] | Cross sectional | USA | NR | NR | Both conventional and molecular methods | E-test, BMD | 706 | FLC. |
| 33 | Hilmioğlu-Polat 2018 [49] | Cross sectional | Turkey | NR | NR | Molecular methods | BMD | 170 | FLC, AMB, VOR, CAS and ANF. |
| 34 | Hirai 2014 [50] | Cross sectional | Japan | 37/51 | 14/51 | Conventional methods | DP-Eiken | 51 | FLC, AMB, ITC, VOR and MCF. |
| 35 | Jalel 2015 [51] | Cross sectional | Tunisia | NR | NR | Both conventional and molecular methods | E-test | 17 | FLC, AMB, ITC and VOR. |
| 36 | Khan 2011 [52] | Cross sectional | Kuwait | NR | NR | Both conventional and molecular methods | E-test | 86 | CAS and ANF. |
| 37 | Khodavaisy 2020 [53] | Cross sectional | Iran | 34 | 67 | Molecular methods | BMD | 101 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 38 | Liu 2018 [54] | Cross sectional | China | 22 | 10 | NR | E-test | 32 | FLC, AMB, VOR and CAS. |
| 39 | Lockhart 2008 [55] | Cross sectional | 25 countries | NR | NR | Both conventional and molecular methods | E-test, BMD | 1929 | FLC, AMB, CAS, ANF and MCF. |
| 40 | Magobo 2020 [56] | Cross sectional | South Africa | NR | NR | Both conventional and molecular methods | NR | 73 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 41 | Magobo 2017 [57] | Cross sectional | South Africa | NR | NR | Both conventional and molecular methods | E-test, BMD | 143 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 42 | Maria 2018 [58] | Cross sectional | India | 42 | 35 | Both conventional and molecular methods | E-test, Vitek-2, BMD | 77 | FLC, AMB, VOR, CAS and MCF. |
| 43 | Mariangela 2015 [59] | Cross sectional | Brazil | NR | NR | Both conventional and molecular methods | Vitek-2, BMD | 43 | FLC, AMB, ITC, VOR and CAS. |
| 44 | Martini 2020 [60] | Cross sectional | Italy | NR | NR | Molecular methods | Sensititre YeastOne, BMD | 241 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 45 | Mashaly 2014 [61] | Cross sectional | Egypt | 29 | 39 | Both conventional and molecular methods | E-test | 68 | FLC, AMB and ITC. |
| 46 | Melo 2011 [62] | NR | Brazil | NR | NR | Both conventional and molecular methods | NR | 20 | FLC and AMB. |
| 47 | Mesini 2020 [63] | Cross sectional | Italy | 386 | 274 | Both conventional and molecular methods | Sensititre YeastOne, BMD | 194 | FLC, VOR, CAS, ANF and MCF. |
| 48 | Miranda-Zapico 2011 [64] | Cross sectional | Spain | NR | NR | Both conventional and molecular methods | Sensititre YeastOne, BMD | 94 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 49 | Modiri 2019 [65] | NR | Iran | NR | NR | Molecular methods | BMD | 17 | FLC, AMB, ITC, POS, VOR and CAS. |
| 50 | Neji 2017 [66] | Cross sectional | Tunisia | NR | NR | Both conventional and molecular methods | Sensititre YeastOne, BMD | 65 | FLC, AMB, ITC, VOR and CAS. |
| 51 | Pfaller 2008 [67] | Surveillance | Many countries | NR | NR | NR | E-test, BMD | 2834 | FLC VOR, CAS, ANF and MCF. |
| 52 | Pfaller 1995 [68] | NR | USA | NR | NR | Molecular methods | BMD | 60 | FLC, AMB and ITC. |
| 53 | Pharkjaksu 2018 [69] | Cross sectional | Thailand | NR | NR | Molecular methods | Sensititre YeastOne, BMD | 96 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 54 | Pinhati 2016 [70] | Cross sectional | Brazil | 25 | 15 | Both conventional and molecular methods | Vitek-2, BMD | 28 | FLC, AMB and ANF. |
| 55 | Prażyńska 2014 [71] | Cross sectional | Poland | NR | NR | Conventional methods | BMD | 28 | AMB. |
| 56 | Puig 2021 [72] | Cross sectional | Spain | NR | NR | Both conventional and molecular methods | MALDI-TOF | 30 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 57 | Pulcrano 2012 [73] | NR | Italy | NR | NR | Both conventional and molecular methods | BMD | 31 | AMB and VOR. |
| 58 | Raghuram 2012 [74] | Cross sectional | USA | NR | NR | NR | NR | 16 | FLC and CAS. |
| 59 | Ramos-Martínez 2022 [75] | Cross sectional | Spain | 61 | 27 | Both conventional and molecular methods | BMD | 31 | FLC, AMB, POS, VOR and CAS. |
| 60 | Reissa 2008 [76] | NR | USA | NR | NR | Both conventional and molecular methods | E-test, BMD | 34 | FLC, AMB, ITC, POS, VOR and CAS. |
| 61 | Roberto 2020 [77] | NR | Brazil | NR | NR | Molecular methods | MALDI-TOF-MS, BMD | 20 | CAS, ANF and MCF. |
| 62 | Ruiz 2013 [78] | Cross sectional | Brazil | NR | NR | Molecular methods | E-test | 49 | FLC, AMB, ITC, VOR and CAS. |
| 63 | Růžička 2007 [79] | NR | Czechia | NR | NR | Conventional methods | BMD | 19 | AMB, ITC and VOR. |
| 64 | Sakamoto 2021 [80] | Cross sectional | Japan | 96 | 51 | Conventional methods | DP-Eiken | 39 | FLC, AMB, ITC, VOR, CAS and MCF. |
| 65 | Sarvikivi 2005 [81] | Cross sectional | Finland | NR | NR | Both conventional and molecular methods | BMD | 26 | FLC. |
| 66 | Silva 2009 [82] | Cross sectional | Portugal | NA | NA | Both conventional and molecular methods | BMD | 160 | FLC, AMB, POS, VOR, CAS and ANF. |
| 67 | Singh 2019 [83] | Surveillance | India | NR | NR | Both conventional and molecular methods | BMD | 199 | FLC, AMB, ITC, POS and VOR. |
| 68 | Souza 2015 [84] | Surveillance | Brazil | NR | NR | Both conventional and molecular methods | Vitek-2, BMD | 9 | FLC, AMB, VOR and ANF. |
| 69 | Tay 2009 [85] | NR | Malaysia | NR | NR | Both conventional and molecular methods | E-test | 42 | FLC, AMB, ITC, KET and VOR. |
| 70 | Thomaz 2018 [86] | NR | Brazil | NR | NR | Both conventional and molecular methods | BMD | 17 | FLC, AMB, VOR, ANF and MCF. |
| 71 | Thomaz 2021 [87] | NR | Brazil | NR | NR | Molecular methods | E-test, BMD | 112 | FLC, AMB, VOR, ANF and MCF. |
| 72 | Thomaz 2022 [88] | NR | Brazil | NR | NR | Molecular methods | Disk Diffusion | 65 | FLC. |
| 73 | Tosun 2013 [89] | NR | Turkey | NR | NR | Both conventional and molecular methods | BMD | 36 | FLC, AMB, VOR, CAS and ANF. |
| 74 | Treviño-Rangel 2012 [90] | NR | Mexico | NR | NR | Both conventional and molecular methods | BMD | 344 | FLC CAS, ANF and MCF. |
| 75 | Vigezzi 2019 [91] | NR | Argentina | NR | NR | Both conventional and molecular methods | BMD | 10 | FLC, AMB, ITC, POS, VOR, CAS and ANF. |
| 76 | Wu 2020 [92] | Cross sectional | China | 33 | 25 | NR | NR | 58 | FLC, AMB, ITC, VOR, and MCF. |
| 77 | Xiao 2015 [93] | Surveillance | China | NR | NR | Conventional methods | E-test, Sensititre YeastOne BMD | 392 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| 78 | Yamin 2020 [94] | Cross sectional | Malaysia | NR | NR | Conventional methods | NA | 343 | FLC, AMB, VOR, and CAS. |
| 79 | Zhang 2020 [95] | Surveillance | China | 232 | 87 | Molecular methods | Sensititre YeastOne BMD | 319 | FLC, AMB, ITC, POS, VOR, CAS, ANF and MCF. |
| Subgroups | Prevalence of Antifungal Resistance [95% CIs] (%) | No. of Studies Analysed | Total No. of Subjects | Heterogeneity | Publication Bias, Egger’s Test (p-Value) | ||
|---|---|---|---|---|---|---|---|
| I2 | p-Value | ||||||
| Fluconazole | |||||||
| Total | 15.2 [9.2; 21.2] | 71 | 13,582 | 98% | <0.0001 | <0.0001 | |
| Enrolment time | Before 2016 | 11.6 [4.9; 18.3] | 43 | 10,244 | 97% | <0.01 | 0.0002 |
| 2016–2022 | 36.7 [10.9; 62.6] | 8 | 1126 | 99% | <0.01 | NA | |
| Continent | Europe | 13.3 [1.3–25.3] | 15 | 2064 | 98% | <0.01 | 0.0439 |
| America | 21.2 [7.6–34.7] | 23 | 1831 | 97% | <0.01 | 0.0116 | |
| Asia | 6.0 [2.9–9.1] | 23 | 3237 | 90% | <0.01 | 0.0116 | |
| Africa | 27.7 [2.7–52.8] | 6 | 897 | 98% | <0.01 | NA | |
| AFST method | BrothMicrodilution | 16.5 [8.5–24.5] | 43 | 5107 | 98% | <0.0001 | <0.0001 |
| E-test and Broth Microdilution | 13.0 [0.5–25.6] | 12 | 7371 | 98% | <0.01 | 0.0315 | |
| E-test | 11.3 [0.0–30.2] | 8 | 474 | 97% | <0.01 | NA | |
| DP-Eiken | 0.6 [0.0–2.9] | 2 | 90 | 0% | 0.37 | NA | |
| MALDI-TOF | 0.0 [0.0–11.6] | 1 | 30 | NA | NA | NA | |
| Amphotericin B | |||||||
| Total | 1.3 [0.0–2.9] | 63 | 9049 | 96% | <0.01 | 0.1828 | |
| Enrolment time | Before 2016 | 1.6 [0.0–4.1] | 40 | 6023 | 98% | <0.0001 | 0.2710 |
| 2016–2022 | 0.0 [0.0–0.2] | 8 | 1138 | 0 | 1 | NA | |
| Continent | Europe | 0.1 [0.0–0.4] | 15 | 1733 | 0 | 1 | 0.3617 |
| America | 0.2 [0.0–0.7] | 18 | 1015 | 0 | 0.95 | 0.0419 | |
| Asia | 0.0 [0.0–0.2] | 22 | 3044 | 9% | 0.34 | 0.1135 | |
| Africa | 0.2 [0.0–0.05] | 6 | 897 | 0% | 1 | NA | |
| AFST method | Broth Microdilution | 0.1 [0.0–0.2] | 40 | 4514 | 0 | 1 | 0.0936 |
| E-test and Broth Microdilution | 5.3 [0.0–15.5] | 9 | 3512 | 100 | <0.0001 | NA | |
| E-test | 5.3 [0.0–1.1] | 7 | 409 | 0 | 0.95 | NA | |
| DP-Eiken | 0.0 [0.0–2.1] | 2 | 90 | 0 | 1 | NA | |
| MALDI-TOF | 0.0 [0.0–11.6] | 1 | 30 | NA | Na | NA | |
| Voriconazole | |||||||
| Total | 4.7 [2.2; 7.3] | 58 | 10,031 | 91% | <0.01 | <0.0001 | |
| Enrolment time | Before 2016 | 3.2 [1.2–5.2] | 37 | 8030 | 93% | <0.01 | 0.0078 |
| 2016–2022 | 17.9 [0.2–35.6] | 7 | 1132 | 98% | <0.01 | NA | |
| Continent | Europe | 5.3 [0.8–9.7] | 15 | 2042 | 90% | <0.01 | 0.0054 |
| America | 9.2 [0.0–19.2] | 14 | 778 | 94% | <0.01 | 0.0569 | |
| Asia | 1.2 [0.3–2.0] | 22 | 3117 | 67% | <0.01 | 0.0120 | |
| Africa | 12.0 [2.4–21.6] | 5 | 829 | 96% | <0.01 | NA | |
| AFST method | Broth Microdilution | 4.4 [2.1–6.8] | 37 | 4679 | 90% | <0.01 | 0.0002 |
| E-test and Broth Microdilution | 9.2 [0.0–22.1] | 9 | 4417 | 97% | <0.01 | NA | |
| E-test | 0.0 [0.0–0.8] | 6 | 341 | 0% | 1 | NA | |
| DP-Eiken | 0.0 [0.0–2.1] | 2 | 90 | 0 | 1 | NA | |
| MALDI-TOF | 0.0 [0.0–11.6] | 1 | 30 | NA | NA | Na | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamin, D.; Akanmu, M.H.; Al Mutair, A.; Alhumaid, S.; Rabaan, A.A.; Hajissa, K. Global Prevalence of Antifungal-Resistant Candida parapsilosis: A Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2022, 7, 188. https://doi.org/10.3390/tropicalmed7080188
Yamin D, Akanmu MH, Al Mutair A, Alhumaid S, Rabaan AA, Hajissa K. Global Prevalence of Antifungal-Resistant Candida parapsilosis: A Systematic Review and Meta-Analysis. Tropical Medicine and Infectious Disease. 2022; 7(8):188. https://doi.org/10.3390/tropicalmed7080188
Chicago/Turabian StyleYamin, Dina, Mutiat Hammed Akanmu, Abbas Al Mutair, Saad Alhumaid, Ali A. Rabaan, and Khalid Hajissa. 2022. "Global Prevalence of Antifungal-Resistant Candida parapsilosis: A Systematic Review and Meta-Analysis" Tropical Medicine and Infectious Disease 7, no. 8: 188. https://doi.org/10.3390/tropicalmed7080188
APA StyleYamin, D., Akanmu, M. H., Al Mutair, A., Alhumaid, S., Rabaan, A. A., & Hajissa, K. (2022). Global Prevalence of Antifungal-Resistant Candida parapsilosis: A Systematic Review and Meta-Analysis. Tropical Medicine and Infectious Disease, 7(8), 188. https://doi.org/10.3390/tropicalmed7080188







