Human-Altered Landscapes and Climate to Predict Human Infectious Disease Hotspots
Abstract
:1. Introduction
2. Methods
2.1. Data
2.2. Bioclimatic and Population Predictors
2.3. Model Fitting and Model Prediction
3. Results
3.1. Model Comparison
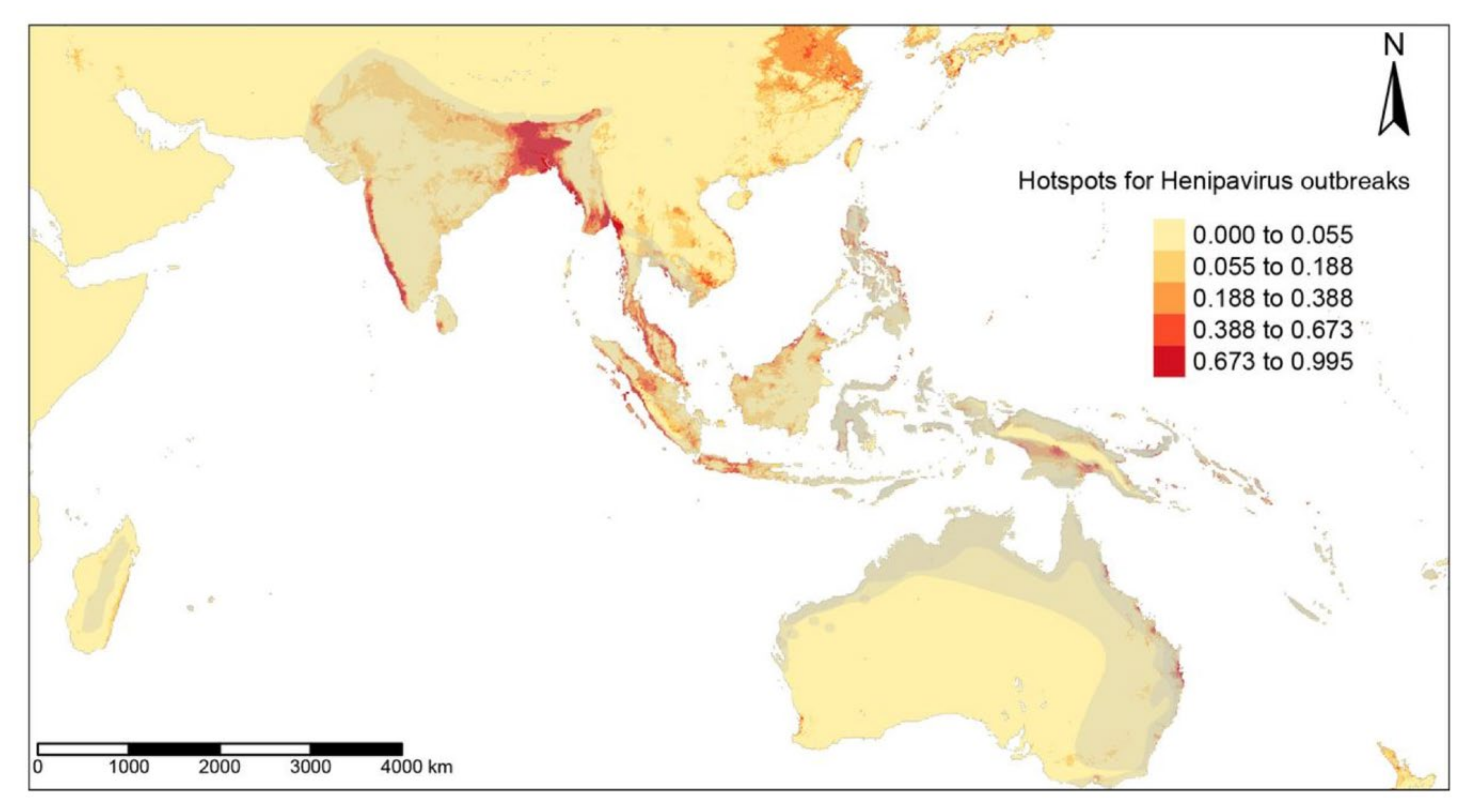
3.2. Detection of EID Hotspots
3.3. Significant Environmental Predictors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ogden, N.; AbdelMalik, P.; Pulliam, J. Emerging Infectious Diseases: Prediction and Detection. Can. Commun. Dis. Rep. 2017, 43, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global Trends in Emerging Infectious Diseases. Nature 2008, 451, 990. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E.J. Risk Factors for Human Disease Emergence. Philos. Trans. R. Soc. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Meentemeyer, R.K.; Haas, S.E.; Václavík, T. Landscape Epidemiology of Emerging Infectious Diseases in Natural and Human-Altered Ecosystems. Annu. Rev. Phytopathol. 2012, 50, 379–402. [Google Scholar] [CrossRef] [Green Version]
- Myers, S.S.; Gaffikin, L.; Golden, C.D.; Ostfeld, R.S.; Redford, K.H.; Ricketts, T.H.; Turner, W.R.; Osofsky, S.A. Human Health Impacts of Ecosystem Alteration. Proc. Natl. Acad. Sci. USA 2013, 110, 18753–18760. [Google Scholar] [CrossRef] [Green Version]
- Gibb, R.; Redding, D.W.; Chin, K.Q.; Donnelly, C.A.; Blackburn, T.M.; Newbold, T.; Jones, K.E. Zoonotic Host Diversity Increases in Human-Dominated Ecosystems. Nature 2020, 584, 398–402. [Google Scholar] [CrossRef]
- Daszak, P.; Olival, K.J.; Li, H. A Strategy to Prevent Future Epidemics Similar to the 2019-NCoV Outbreak. Biosaf. Health 2020, 2, 6–8. [Google Scholar] [CrossRef]
- Patz, J.A.; Daszak, P.; Tabor, G.M.; Aguirre, A.A.; Pearl, M.; Epstein, J.; Wolfe, N.D.; Kilpatrick, A.M.; Foufopoulos, J.; Molyneux, D.; et al. Unhealthy Landscapes: Policy Recommendations on Land Use Change and Infectious Disease Emergence. Environ. Health Perspect. 2004, 112, 1092–1098. [Google Scholar] [CrossRef] [Green Version]
- Patz, J.A.; Olson, S.H.; Uejio, C.K.; Gibbs, H.K. Disease Emergence from Global Climate and Land Use Change. Med. Clin. North Am. 2008, 92, 1473–1491. [Google Scholar] [CrossRef]
- Smith, D.L.; Lucey, B.; Waller, L.A.; Childs, J.E.; Real, L.A. Predicting the Spatial Dynamics of Rabies Epidemics on Heterogeneous Landscapes. Proc. Natl. Acad. Sci. USA 2002, 99, 3668–3672. [Google Scholar] [CrossRef] [Green Version]
- Fenollar, F.; Mediannikov, O. Emerging Infectious Diseases in Africa in the 21st Century. New Microbes New Infect. 2018, 26, S10. [Google Scholar] [CrossRef] [PubMed]
- Biek, R.; Real, L.A. The Landscape Genetics of Infectious Disease Emergence and Spread. Mol. Ecol. 2010, 19, 3515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagadesh, S.; Combe, M.; Couppié, P.; Le Turnier, P.; Epelboin, L.; Nacher, M.; Gozlan, R.E. Emerging Human Infectious Diseases of Aquatic Origin: A Comparative Biogeographic Approach Using Bayesian Spatial Modelling. Int. J. Health Geogr. 2019, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Redding, D.W.; Lucas, T.C.D.; Blackburn, T.M.; Jones, K.E. Evaluating Bayesian Spatial Methods for Modelling Species Distributions with Clumped and Restricted Occurrence Data. PLoS ONE 2017, 12, e0187602. [Google Scholar] [CrossRef] [Green Version]
- Gelfand, A.E.; Holder, M.; Latimer, A.; Lewis, P.O.; Rebelo, A.G.; Silander, J.A., Jr.; Wu, S. Explaining Species Distribution Patterns through Hierarchical Modeling. Bayesian Anal. 2006, 1, 41–92. [Google Scholar] [CrossRef]
- Latimer, A.M.; Wu, S.; Gelfand, A.E.; Silander, J.A. Building Statistical Models To Analyze Species Distributions. Ecol. Appl. 2006, 16, 33–50. [Google Scholar] [CrossRef] [Green Version]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Broxton, P.D.; Zeng, X.; Sulla-Menashe, D.; Troch, P.A. A Global Land Cover Climatology Using MODIS Data. J. Appl. Meteorol. Climatol. 2014, 53, 1593–1605. [Google Scholar] [CrossRef]
- Kennedy, C.M.; Oakleaf, J.R.; Theobald, D.M.; Baruch-Mordo, S.; Kiesecker, J. Global Human Modification of Terrestrial Systems; NASA Socioeconomic Data and Applications Center (SEDAC): Palisades, NY, USA, 2020. [Google Scholar]
- Center for International Earth Science Information Network. Gridded Population of the World, Version 4 (GPWv4): Population Density, Revision 11; NASA Socioeconomic Data and Applications Center (SEDAC): Palisades, NY, USA, 2018. [Google Scholar]
- Isberg, S.; Balaguera-Reina, S.A.; Ross, J.P. The IUCN Red List of Threatened Species; IUCN Red List: London, UK, 2017. [Google Scholar] [CrossRef]
- Plumptre, A.J.; Nixon, S.; Kujirakwinja, D.K.; Vieilledent, G.; Critchlow, R.; Williamson, E.A.; Nishuli, R.; Kirkby, A.E.; Hall, J.S. Catastrophic Decline of World’s Largest Primate: 80% Loss of Grauer’s Gorilla (Gorilla Beringei Graueri) Population Justifies Critically Endangered Status. PLoS ONE 2016, 11, e0162697. [Google Scholar] [CrossRef] [Green Version]
- Wilson, A.M.; Jetz, W. Remotely Sensed High-Resolution Global Cloud Dynamics for Predicting Ecosystem and Biodiversity Distributions. PLoS Biol. 2016, 14, e1002415. [Google Scholar] [CrossRef]
- Besag, J. Spatial Interaction and the Statistical Analysis of Lattice Systems. J. R. Stat. Soc. Ser. B Methodol. 1974, 36, 192–236. [Google Scholar] [CrossRef]
- Liu, C.; White, M.; Newell, G. Measuring and Comparing the Accuracy of Species Distribution Models with Presence-Absence Data. Ecography 2011, 34, 232–243. [Google Scholar] [CrossRef]
- Wolfe, N.D.; Daszak, P.; Kilpatrick, A.M.; Burke, D.S. Bushmeat Hunting, Deforestation, and Prediction of Zoonotic Disease Emergence. In Emerging Infectious Diseases; Centers for Disease Control and Prevention (CDC): Atlanta, GA, USA, 2005; pp. 1822–1827. [Google Scholar] [CrossRef]
- García, F.C.; Bestiona, E.; Warfielda, R.; Yvon-Durochera, G. Changes in Temperature Alter the Relationship between Biodiversity and Ecosystem Functioning. Proc. Natl. Acad. Sci. USA 2018, 115, 10989–10994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sayed, A.; Kamel, M. Climatic Changes and Their Role in Emergence and Re-Emergence of Diseases. Environ. Sci. Pollut. Res. Int. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Patz, J.A.; Olson, S.H. Malaria Risk and Temperature: Influences from Global Climate Change and Local Land Use Practices. Proc. Natl. Acad. Sci. USA. 2006, 103, 5635. [Google Scholar] [CrossRef] [Green Version]
- Folland, C.K.; Karl, T.R.; Salinger, M.J. Observed Climate Variability and Change. Weather 2002, 57, 269–278. [Google Scholar] [CrossRef]
- Ambat, A.S.; Zubair, S.M.; Prasad, N.; Pundir, P.; Rajwar, E.; Patil, D.S.; Mangad, P. Nipah Virus: A Review on Epidemiological Characteristics and Outbreaks to Inform Public Health Decision Making. J. Infect. Public Health 2019, 12, 634–639. [Google Scholar] [CrossRef]
- Royce, K.; Fu, F. Mathematically Modeling Spillovers of an Emerging Infectious Zoonosis with an Intermediate Host. PLoS ONE 2020, 15, e0237780. [Google Scholar] [CrossRef]
- Epstein, J.H.; Field, H.E.; Luby, S.; Pulliam, J.R.; Daszak, P. Nipah Virus: Impact, Origins, and Causes of Emergence. Curr. Infect. Dis. Rep. 2006, 8, 59–65. [Google Scholar] [CrossRef]
- Olivero, J.; Fa, J.E.; Real, R.; Márquez, A.L.; Farfán, M.A.; Vargas, J.M.; Gaveau, D.; Salim, M.A.; Park, D.; Suter, J.; et al. Recent Loss of Closed Forests Is Associated with Ebola Virus Disease Outbreaks. Sci. Rep. 2017, 7, 14291. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Chavez, C.; Curtiss, R.; Daszak, P.; Levin, S.A.; Patterson-Lomba, O.; Perrings, C.; Poste, G.; Towers, S. Beyond Ebola: Lessons to Mitigate Future Pandemics. Lancet Glob. Health 2015, 3, e354–e355. [Google Scholar] [CrossRef] [Green Version]
- Redding, D.W.; Atkinson, P.M.; Cunningham, A.A.; Lo Iacono, G.; Moses, L.M.; Wood, J.L.N.; Jones, K.E. Impacts of Environmental and Socio-Economic Factors on Emergence and Epidemic Potential of Ebola in Africa. Nat. Commun. 2019, 10, 4531. [Google Scholar] [CrossRef] [PubMed]
- Rahalkar, M.C.; Bahulikar, R.A. Lethal Pneumonia Cases in Mojiang Miners (2012) and the Mineshaft Could Provide Important Clues to the Origin of SARS-CoV-2. Front. Public Health 2020, 8, 638. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Hu, Z. A Review of Studies on Animal Reservoirs of the SARS Coronavirus. Virus Res. 2008, 133, 74. [Google Scholar] [CrossRef]
- Emhj. An Outbreak of Middle East Respiratory Syndrome (MERS) due to Coronavirus in Al-Ahssa Region, Saudi Arabia. 2015. Available online: http://www.emro.who.int/emhj-volume-22-2016/volume-22-issue-7/an-outbreak-of-middle-east-respiratory-syndrome-mers-due-to-coronavirus-in-al-ahssa-region-saudi-arabia-2015.html (accessed on 29 June 2022).
- Lee, T.M.; Sigouin, A.; Pinedo-Vasquez, M.; Nasi, R. The Harvest of Tropical Wildlife for Bushmeat and Traditional Medicine. Annu. Rev. Environ. Resour. 2020, 45, 145–170. [Google Scholar] [CrossRef]


| Model | Deviance | Percentage of Deviance Explained |
|---|---|---|
| Filoviridae | ||
| NULL | 1891.41 | 0 |
| Binomial | 839.24 | 65 |
| ZIB | 764.38 | 69 |
| Binomial.iCAR | 268.52 | 100 |
| ZIB.iCAR | 267.14 | 100 |
| Coronaviridae | ||
| NULL | 1889.18 | 0 |
| Binomial | 629.11 | 69 |
| ZIB | 548.47 | 74 |
| Binomial.iCAR | 63.09 | 100 |
| ZIB.iCAR | 67.01 | 100 |
| Henipavirus | ||
| NULL | 1862.74 | 0 |
| Binomial | 635.28 | 70 |
| ZIB | 627.49 | 71 |
| Binomial.iCAR | 113.44 | 100 |
| ZIB.iCAR | 116.76 | 100 |
| Variable | 2.50% | 25% | 50% | 75% | 97.50% |
|---|---|---|---|---|---|
| ZIB iCAR Filoviridae outbreak model | |||||
| Min. temperature | 1.41 | 2.14 | 2.49 | 2.87 | 3.62 |
| Max. temperature | −2.13 | −1.55 | −1.26 | −0.98 | −0.48 |
| Mean precipitation | 1.01 | 1.31 | 1.49 | 1.67 | 2.05 |
| Land cover | −1.04 | −0.77 | −0.62 | −0.45 | −0.16 |
| Elevation | 0.59 | 1.06 | 1.3 | 1.57 | 2.14 |
| Land-use changes | 0.75 | 1 | 1.15 | 1.33 | 1.64 |
| Population density | 0.48 | 1 | 1.33 | 1.7 | 2.5 |
| ZIB iCAR Coronaviridae outbreak model | |||||
| Min. temperature | −1.09 | 2.75 | 4.35 | 5.92 | 9.36 |
| Max. temperature | −5.07 | −2.42 | −0.97 | 0.34 | 3.56 |
| Mean precipitation | −3.21 | −2.33 | −1.89 | −1.37 | −0.38 |
| Land cover | −1.14 | −0.56 | −0.27 | 0.02 | 0.64 |
| Elevation | 0.95 | 1.77 | 2.2 | 2.63 | 3.51 |
| Land-use changes | 1.47 | 2.15 | 2.5 | 2.9 | 3.69 |
| Population density | 2.15 | 3.19 | 3.75 | 4.31 | 5.43 |
| ZIB iCAR Henipavirus outbreak model | |||||
| Min. temperature | −6.26 | −4.49 | −3.67 | −2.88 | −0.66 |
| Max. temperature | −0.02 | 1.67 | 2.35 | 3.1 | 4.53 |
| Mean precipitation | 1.62 | 2.13 | 2.43 | 2.71 | 3.12 |
| Land cover | −0.23 | 0.11 | 0.29 | 0.47 | 0.83 |
| Elevation | −16.27 | −10.32 | −8.84 | −7.6 | −4.95 |
| Land-use changes | 1.41 | 1.89 | 2.13 | 2.39 | 2.88 |
| Population density | −0.31 | −0.1 | 0 | 0.11 | 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jagadesh, S.; Combe, M.; Gozlan, R.E. Human-Altered Landscapes and Climate to Predict Human Infectious Disease Hotspots. Trop. Med. Infect. Dis. 2022, 7, 124. https://doi.org/10.3390/tropicalmed7070124
Jagadesh S, Combe M, Gozlan RE. Human-Altered Landscapes and Climate to Predict Human Infectious Disease Hotspots. Tropical Medicine and Infectious Disease. 2022; 7(7):124. https://doi.org/10.3390/tropicalmed7070124
Chicago/Turabian StyleJagadesh, Soushieta, Marine Combe, and Rodolphe Elie Gozlan. 2022. "Human-Altered Landscapes and Climate to Predict Human Infectious Disease Hotspots" Tropical Medicine and Infectious Disease 7, no. 7: 124. https://doi.org/10.3390/tropicalmed7070124
APA StyleJagadesh, S., Combe, M., & Gozlan, R. E. (2022). Human-Altered Landscapes and Climate to Predict Human Infectious Disease Hotspots. Tropical Medicine and Infectious Disease, 7(7), 124. https://doi.org/10.3390/tropicalmed7070124







