Leptospiral Leucine-Rich Repeat Protein-Based Lateral Flow for Assessment of Canine Leptospiral Immunoglobulin G
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. Bacteria Used in This Study
2.3. Recombinant Proteins
2.4. Line Blot Immunodetection
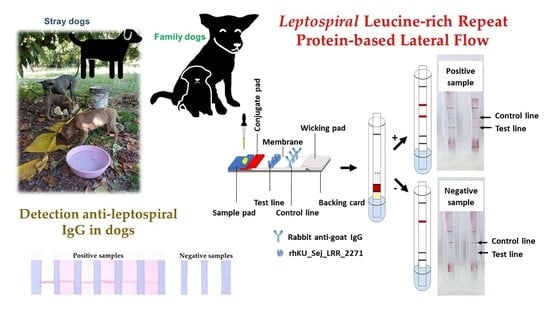
2.5. Design and Development of Lateral Flow Assay to Detect Dog IgG against Leptospira
2.6. Evaluation and Validation Methods
2.6.1. Identification of Leptospires in Dog Blood Samples by Culture
2.6.2. Molecular Analysis of rrs Real-Time PCR in Plasma Samples
2.6.3. Detection of Anti-Leptospiral IgG in Dog Using LipL32 ELISA
2.6.4. Microscopic Agglutination Test (MAT)
2.7. Statistical Analysis
3. Results
3.1. Line Blot with Rabbit Hyperimmune Sera and Dog Plasma Samples
3.2. Leptospires Detection in Blood and Plasma Samples by Culture Method and Real-Time PCR
3.3. Anti-Leptospiral IgG Detection Using LipL32 ELISA
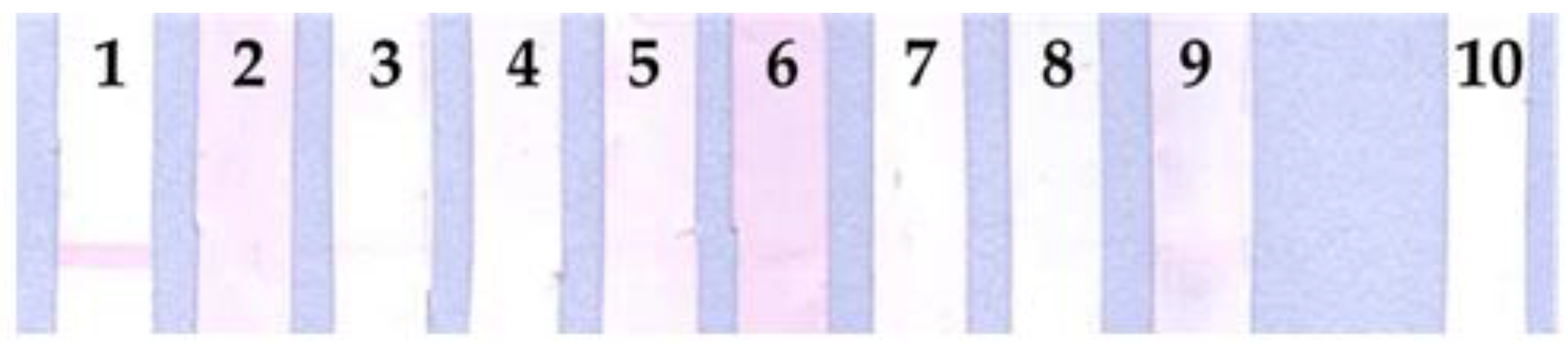
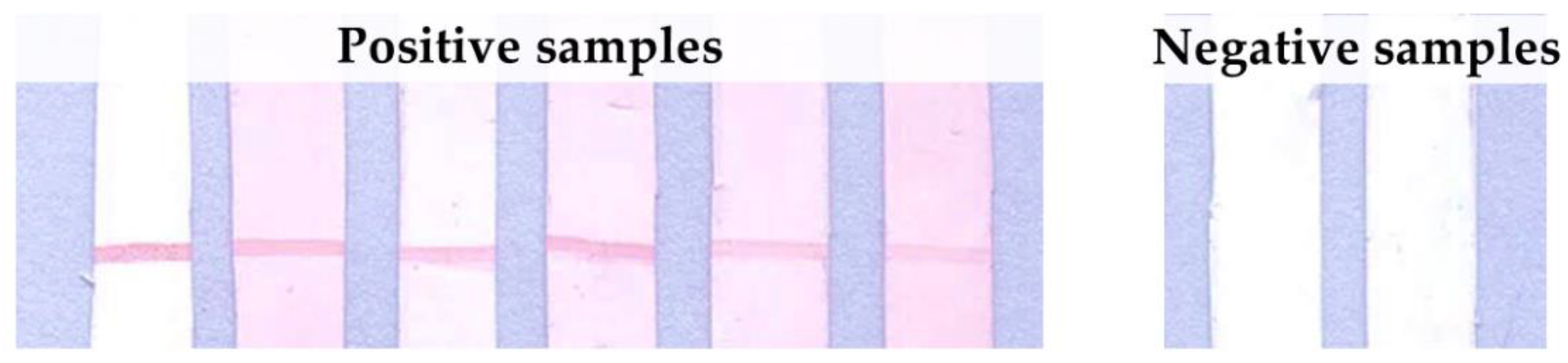
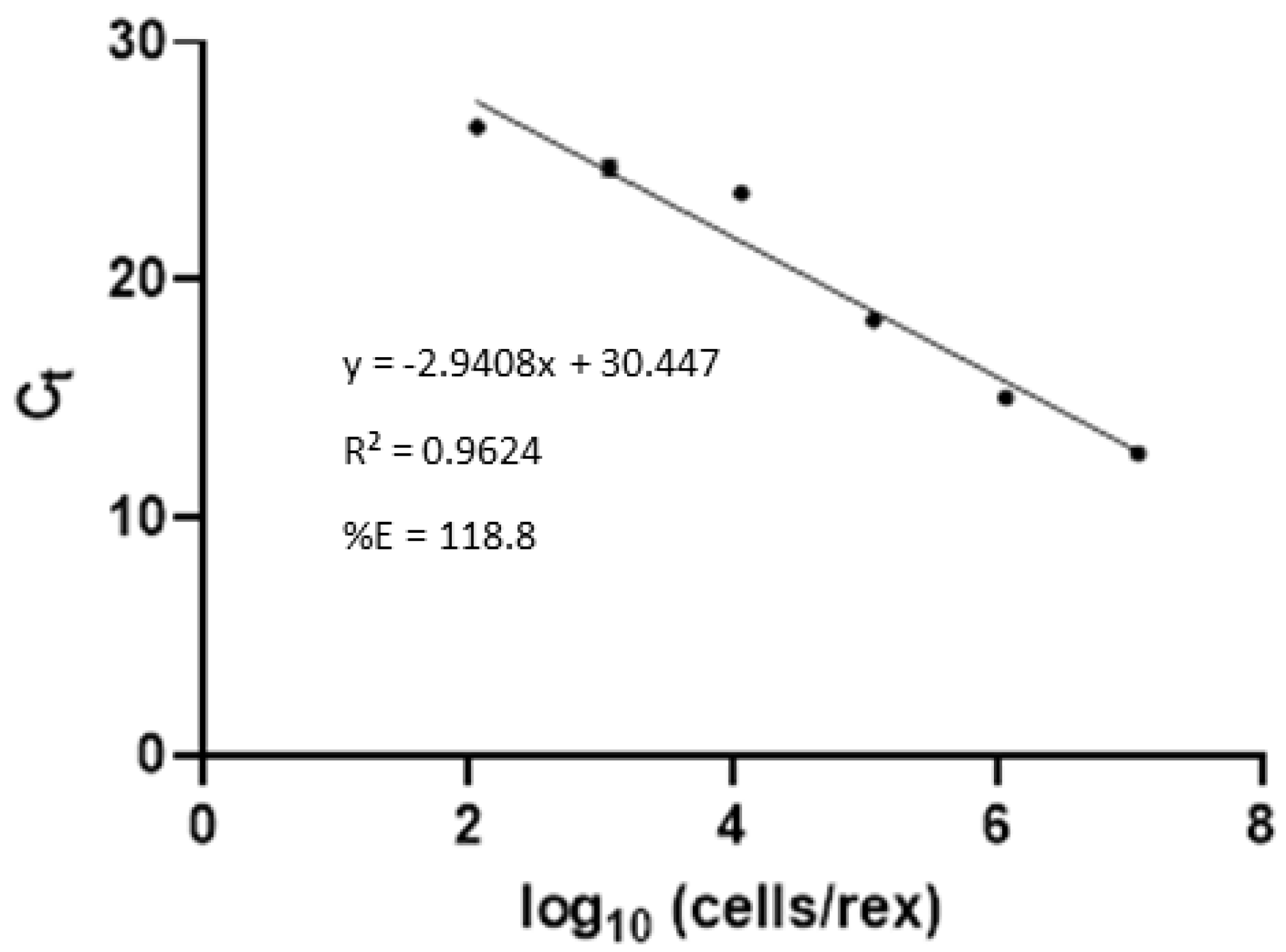
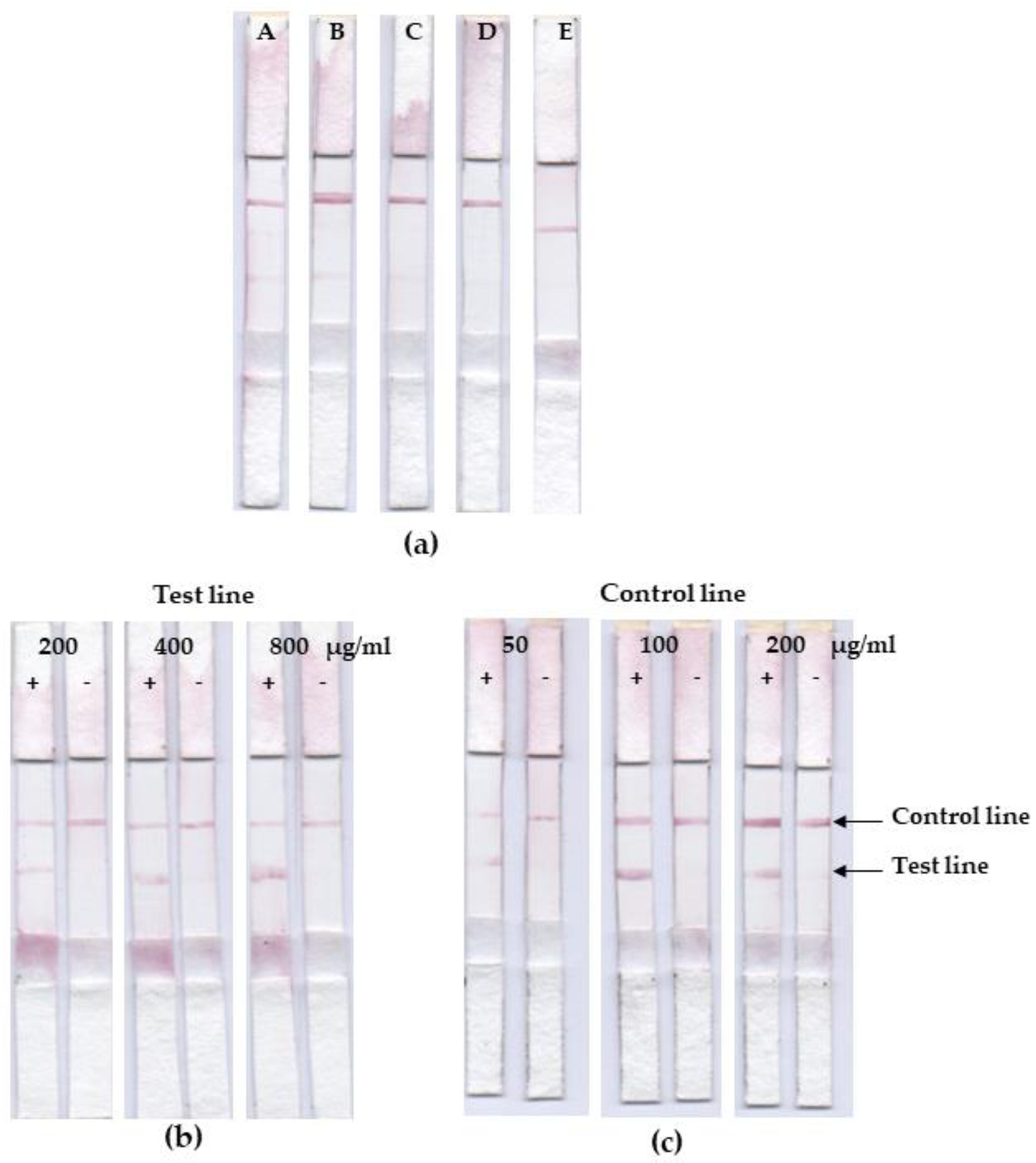
3.4. Lateral Flow Optimization
3.5. Assessment on Leptospiral IgG in Dog’s Plasma Samples
3.6. Validation with MAT
3.7. Association between Dog Types and Leptospirosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arent, Z.; Pardyak, L.; Dubniewicz, K.; Płachno, B.; Kotula-Balak, M. Leptospira taxonomy: Then and now. Med. Weter. 2022, 78, 489–496. [Google Scholar] [CrossRef]
- Brown, K.; Prescott, J. Leptospirosis in the family dog: A public health perspective. Can. Med. Assoc. J. 2008, 178, 399–401. [Google Scholar] [CrossRef] [PubMed]
- Population of Dog and Cat in 2016. Available online: http://dcontrol.dld.go.th/webnew/index.php/th/news-menu/2018-07-02-08-24-32/360-dogpop2016 (accessed on 4 August 2021).
- Jittapalapong, S.; Sittisan, P.; Sakpuaram, T.; Kabeya, H.; Maruyama, S.; Inpankaew, T. Coinfection of Leptospira spp and Toxoplasma gondii among stray dogs in Bangkok, Thailand. Southeast Asian J. Trop. Med. Public Health 2009, 40, 247–252. [Google Scholar] [PubMed]
- Altheimer, K.; Jongwattanapisan, P.; Luengyosluechakul, S.; Pusoonthornthum, R.; Prapasarakul, N.; Kurilung, A.; Broens, E.M.; Wagenaar, J.A.; Goris, M.G.A.; Ahmed, A.A.; et al. Leptospira infection and shedding in dogs in Thailand. BMC Vet. Res. 2020, 16, 89. [Google Scholar] [CrossRef]
- Ngasaman, R.; Saechan, V.; Prachantasena, S.; Yingkajorn, M.; Sretrirutchai, S. Investigation of Leptospira Infection in Stray Animals in Songkhla, Thailand: Leptospirosis Risk Reduction in Human. Vector Borne Zoonotic Dis. 2020, 20, 432–435. [Google Scholar] [CrossRef] [PubMed]
- Chotivanich, S.; Pinprasong, B.; Suwancharoen, D. Serologic survey of leptospirosis in pigs, dogs, beef cattle, buffaloes and dairy cattle in Chaiyaphum Province. DLD Tech Paps. 2000, 5, 1–13. [Google Scholar]
- Pumipuntu, N.; Suwannarong, K. Seroprevalence of Leptospira Spp. in Cattle and Dogs in Mahasarakham Province, Thailand. J. Health Res. 2016, 30, 223–226. [Google Scholar]
- Niwetpathomwat, A.; Assarasakorn, S. Preliminary investigation of canine leptospirosis in a rural area of Thailand. Med. Weter. 2007, 63, 59–61. [Google Scholar]
- Sakpuaram, T.; Wajjwalku, W.; Sirinarumithra, T.; Kasemsuwan, S.; Sithisara, P.; Chanthick, N. Monitoring of Leptospirosis in healthy vaccinated dogs in Hua-Hin district, Prachuapkhirikhan province between January to December, 2004. In Proceedings of the 44th Kasetsart University Annual Conference: Animal, Veterinary Medicine, Bangkok, Thailand, 30 January–2 February 2006; Kasetsart University: Bangkok, Thailand, 2006; pp. 439–445. [Google Scholar]
- Paungpin, W.; Chaiwattanarungruengpaisan, S.; Mongkolphan, C.; Wiriyarat, W.; Thongdee, M. Genotyping of the causative Leptospira in symptomatic dogs in Thailand. Korean J. Vet. Res. 2020, 60, 1–7. [Google Scholar] [CrossRef]
- Romero-Vivas, C.M.; Cuello-Pérez, M.; Agudelo-Flórez, P.; Thiry, D.; Levett, P.N.; Falconar, A.K. Cross-sectional study of Leptospira seroprevalence in humans, rats, mice, and dogs in a main tropical sea-port city. Am. J. Trop. Med. Hyg. 2013, 88, 178–183. [Google Scholar] [CrossRef]
- Venkataraman, K.S.; Nedunchelliyan, S. Epidemiology of an outbreak of leptospirosis in man and dog. Comp. Immunol. Microbiol. Infect. Dis. 1992, 15, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chierakul, W.; Smythe, L.D.; Petkanchanapong, W.; Limpaiboon, R.; Apiwatanaporn, A.; Slack, A.T.; Suputtamongkol, Y.; White, N.J.; et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLOS Negl. Trop. Dis. 2007, 1, e56. [Google Scholar] [CrossRef] [PubMed]
- Tangkanakul, W.; Smits, H.L.; Jatanasen, S.; Ashford, D.A. Leptospirosis: An emerging health problem in Thailand. Southeast Asian J. Trop. Med. Public Health 2005, 36, 281–288. [Google Scholar] [PubMed]
- Yatbantoong, N.; Chaiyarat, R. Factors Associated with Leptospirosis in Domestic Cattle in Salakphra Wildlife Sanctuary, Thailand. Int. J. Environ. Res. Public Health 2019, 16, 1042. [Google Scholar] [CrossRef] [PubMed]
- Sundharagiati, B.; Harinasuta, C.; Photha, U. Human leptospirosis in Thailand. Trans. R. Soc. Trop. Med. Hyg. 1966, 60, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, C.; Camacho, G.; Osuna, I.; Enríquez Verdugo, I.; Castro del Campo, N.; López-Moreno, H. Prevalence and risk factors associated with serovars of Leptospira in dogs from Culiacan, Sinaloa. Vet. Mex. OA 2017, 4, 1–12. [Google Scholar]
- Ramírez, C. Prevalence and Risk Factors Associated with Serovars of Leptospira in Dogs, Related Human Seropositive. J. Dairy Vet. Anim. Res. 2017, 6, 275–279. [Google Scholar]
- Aslantaş, O.; Ozdemir, V.; Kiliç, S.; Babür, C. Seroepidemiology of leptospirosis, toxoplasmosis, and leishmaniosis among dogs in Ankara, Turkey. Vet. Parasitol. 2005, 129, 187–191. [Google Scholar] [CrossRef]
- Weekes, C.C.; Everard, C.O.; Levett, P.N. Seroepidemiology of canine leptospirosis on the island of Barbados. Vet. Microbiol. 1997, 57, 215–222. [Google Scholar] [CrossRef]
- De Paula Dreer, M.K.; Gonçalves, D.D.; da Silva Caetano, I.C.; Gerônimo, E.; Menegas, P.H.; Bergo, D.; Ruiz Lopes-Mori, F.M.; Benitez, A.; de Freitas, J.C.; Evers, F.; et al. Toxoplasmosis, leptospirosis and brucellosis in stray dogs housed at the shelter in Umuarama municipality, Paraná, Brazil. J. Venom. Anim. Toxins Incl. Trop. Dis. 2013, 19, 23. [Google Scholar] [CrossRef]
- Pinto, G.V.; Senthilkumar, K.; Rai, P.; Kabekkodu, S.P.; Karunasagar, I.; Kumar, B.K. Current methods for the diagnosis of leptospirosis: Issues and challenges. J. Microbiol. Methods 2022, 195, 106438. [Google Scholar] [CrossRef] [PubMed]
- Ghazaei, C. Leptospiral major outer membrane protein: LipL32. Rev. Med. Microbiol. 2015, 26, 65–69. [Google Scholar] [CrossRef]
- Ahmed, A.A.; Goris, M.G.A.; Meijer, M.C. Development of lipL32 real-time PCR combined with an internal and extraction control for pathogenic Leptospira detection. PLoS ONE 2020, 15, e0241584. [Google Scholar] [CrossRef] [PubMed]
- Nagraik, R.; Sethi, S.; Sharma, A.; Kumar, D.; Kumar, D.; Kumar, A.P. Ultrasensitive nanohybrid electrochemical sensor to detect LipL32 gene of Leptospira interrogans. Chem. Pap. 2021, 75, 5453–5462. [Google Scholar] [CrossRef]
- Sapna, K.; Tarique, M.; Asiamma, A.; Ravi Kumar, T.N.; Shashidhar, V.; Arun, A.B.; Prasad, K.S. Early detection of leptospirosis using Anti-LipL32 carbon nanotube immunofluorescence probe. J. Biosci. Bioeng. 2020, 130, 424–430. [Google Scholar] [CrossRef]
- Joseph, S.; Mini, M.; Sriram, V.; Sathiadevan, A.; Ambily, R.; Aravindakshan, T.; Kumar, S. Evaluation of real-time PCR, MAT, and recombinant LipL32-based ELISA for the diagnosis of canine leptospirosis in a disease-endemic South Indian state, Kerala. Turkish J. Vet. Anim. Sci. 2018, 42, 191–197. [Google Scholar]
- Ye, C.; Yan, W.; Xiang, H.; He, H.; Yang, M.; Ijaz, M.; Useh, N.; Hsieh, C.-L.; McDonough, P.L.; McDonough, S.P.; et al. Recombinant antigens rLipL21, rLoa22, rLipL32 and rLigACon4-8 for serological diagnosis of leptospirosis by enzyme-linked immunosorbent assays in dogs. PLoS ONE 2014, 9, e111367. [Google Scholar] [CrossRef]
- Eshghi, A.; Gaultney, R.A.; England, P.; Brûlé, S.; Miras, I.; Sato, H.; Coburn, J.; Bellalou, J.; Moriarty, T.J.; Haouz, A.; et al. An extracellular Leptospira interrogans leucine-rich repeat protein binds human E- and VE-cadherins. Cell. Microbiol. 2019, 21, e12949. [Google Scholar] [CrossRef]
- Miras, I.; Saul, F.; Nowakowski, M.; Weber, P.; Haouz, A.; Shepard, W.; Picardeau, M. Structural characterization of a novel subfamily of leucine-rich repeat proteins from the human pathogen Leptospira interrogans. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1351–1359. [Google Scholar] [CrossRef]
- Hsu, S.H.; Yang, C.W. Insight into the Structure, Functions, and Dynamics of the Leptospira Outer Membrane Proteins with the Pathogenicity. Membranes 2022, 12, 300. [Google Scholar] [CrossRef]
- Hniman, A.; Prapong, S. Development of leptospira molecular markers by using bioinformation from predicted leucine-rich repeat (LRR) protein genes. J. Thail. Vet. Med. Assoc. 2007, 58, 65–78. [Google Scholar]
- Nitipan, S. Molecular Cloning and Sequencing Analysis of the Genes Encoding Leucine-Rich Repeat Proteins of Pathogenic Leptospira Borgpetersenii. Ph.D. Thesis, Kasetsart University, Bangkok, Thailand, 2013. [Google Scholar]
- Nitipan, S.; Sritrakul, T.; Kunjantarachot, A.; Prapong, S. Identification of epitopes in Leptospira borgpetersenii leucine-rich repeat proteins. Infect. Genet. Evol. 2013, 14, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Sritrakul, T.; Nitipan, S.; Wajjwalku, W.; La-Ard, A.; Suphatpahirapol, C.; Petkarnjanapong, W.; Ongphiphadhanakul, B.; Prapong, S. Leptospira borgpetersenii hybrid leucine-rich repeat protein: Cloning and expression, immunogenic identification and molecular docking evaluation. J. Microbiol. Methods 2017, 142, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Tansiri, Y.; Sritrakul, T.; Saparpakorn, P.; Boondamnern, T.; Chimprasit, A.; Sripattanakul, S.; Hannongbua, S.; Prapong, S. New potent epitopes from Leptospira borgpetersenii for the stimulation of humoral and cell-mediated immune responses: Experimental and theoretical studies. Inform. Med. Unlocked 2021, 25, 100649. [Google Scholar] [CrossRef]
- Mérien, F.; Amouriaux, P.; Perolat, P.; Baranton, G.; Saint Girons, I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J. Clin. Microbiol. 1992, 30, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.A.; Sandai, D.A.; Musa, S.; Hoe, C.H.; Riadzi, M.; Lau, K.L.; Tang, T.H. Rapid diagnosis of leptospirosis by multiplex PCR. Malays. J. Med. Sci. 2012, 19, 9–16. [Google Scholar]
- Sripattanakul, S.; Prapong, T.; Kamlangdee, A.; Katzenmeier, G.; Haltrich, D.; Hongprayoon, R.; Prapong, S. Leptospira borgpetersenii Leucine-Rich Repeat Proteins and Derived Peptides in an Indirect ELISA Development for theDiagnosis of Canine Leptospiral Infections. Trop. Med. Int. Health 2022, 7, 311. [Google Scholar]
- Stangroom, J. Easy Fisher Exact Test Calculator. Available online: https://www.socscistatistics.com/tests/fisher/default2.aspx (accessed on 20 October 2022).
- Stangroom, J. Chi Square Calculator for 2 × 2. Available online: https://www.socscistatistics.com/tests/chisquare/default.aspx (accessed on 20 October 2022).
- Bonhomme, D.; Werts, C. Host and Species-Specificities of Pattern Recognition Receptors Upon Infection with Leptospira interrogans. Front. Cell. Infect. Microbiol. 2022, 12, 932137. [Google Scholar] [CrossRef]
- Doxey, A.C.; McConkey, B.J. Prediction of molecular mimicry candidates in human pathogenic bacteria. Virulence 2013, 4, 453–466. [Google Scholar] [CrossRef]
- Lizer, J.; Velineni, S.; Weber, A.; Krecic, M.; Meeus, P. Evaluation of 3 Serological Tests for Early Detection of Leptospira-specific Antibodies in Experimentally Infected Dogs. J. Vet. Intern. Med. 2018, 32, 201–207. [Google Scholar] [CrossRef]
- Dey, S.; Mohan, C.M.; Kumar, T.M.; Ramadass, P.; Nainar, A.M.; Nachimuthu, K. Recombinant LipL32 antigen-based single serum dilution ELISA for detection of canine leptospirosis. Vet. Microbiol. 2004, 103, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Sinha, D.; Chaudhury, P.; Shankar, H.; Srivastava, S.K. Comparative studies on seroepidemiology of canine leptospirosis by micro agglutination test (MAT) and recombinant Lip L32 ELISA. Indian J. Anim. Sci. 2009, 79, 1089–1094. [Google Scholar]
- Kurilung, A.; Chanchaithong, P.; Lugsomya, K.; Niyomtham, W.; Wuthiekanun, V.; Prapasarakul, N. Molecular detection and isolation of pathogenic Leptospira from asymptomatic humans, domestic animals and water sources in Nan province, a rural area of Thailand. Res. Vet. Sci. 2017, 115, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Thongdee, M.; Chaiwattanarungruengpaisan, S.; Lekcharoen, P.; Yimchoho, N.; Buathong, R.; Wiriyarat, W. A Novel Genotype of Leptospira interrogans Recovered from Leptospirosis Outbreak Samples from Southern Thailand. Jpn. J. Infect. Dis. 2019, 72, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.D.; Gravekamp, C.; Carrington, D.G.; van de Kemp, H.; Hartskeerl, R.A.; Edwards, C.N.; Everard, C.O.; Terpstra, W.J.; Levett, P.N. Evaluation of the polymerase chain reaction for early diagnosis of leptospirosis. J. Med. Microbiol. 1995, 43, 110–114. [Google Scholar] [CrossRef]
- Meeyam, T.; Tablerk, P.; Petchanok, B.; Pichpol, D.; Padungtod, P. Seroprevalence and risk factors associated with leptospirosis in dogs. Southeast Asian J. Trop. Med. Public Health 2006, 37, 148–153. [Google Scholar]
- Goldstein, R.E. Canine leptospirosis. Vet. Clin. North Am. Small Anim. Pract. 2010, 40, 1091–1101. [Google Scholar] [CrossRef]
- Reagan, K.L.; Sykes, J.E. Diagnosis of Canine Leptospirosis. Vet. Clin. Small Anim. Pract. 2019, 49, 719–731. [Google Scholar] [CrossRef]
- Minke, J.M.; Bey, R.; Tronel, J.P.; Latour, S.; Colombet, G.; Yvorel, J.; Cariou, C.; Guiot, A.L.; Cozette, V.; Guigal, P.M. Onset and duration of protective immunity against clinical disease and renal carriage in dogs provided by a bi-valent inactivated leptospirosis vaccine. Vet. Microbiol. 2009, 137, 137–145. [Google Scholar] [CrossRef]
- Riediger, I.N.; Stoddard, R.A.; Ribeiro, G.S.; Nakatani, S.M.; Moreira, S.D.R.; Skraba, I.; Biondo, A.W.; Reis, M.G.; Hoffmaster, A.R.; Vinetz, J.M.; et al. Rapid, actionable diagnosis of urban epidemic leptospirosis using a pathogenic Leptospira lipL32-based real-time PCR assay. PLoS Negl. Trop. Dis. 2017, 11, e0005940. [Google Scholar] [CrossRef]
- IDEXX Laboratories. Diagnosing and Managing Canine Leptospirosis. Available online: https://www.idexx.com/files/canine-leptospirosis-test-dx-update.pdf (accessed on 14 April 2022).
| Species | Serogroup | Serovar | Strain |
|---|---|---|---|
| L. interrogans | Australis | Australis | Ballico |
| Bataviae | Bataviae | Swart | |
| Canicola | Canicola | Hond Utrecht lV | |
| Hebdomadis | Hebdomadis | Hebdomadis | |
| Icterohaemorrhagiae | Icterohaemorrhagiae | Ictero l | |
| Pomona | Pomona | Pomona | |
| L. biflexa | Semaranga | Patoc | Patoc l |
| L. borgpetersenii | Mini | Mini | Sari |
| Tarassovi | Tarassovi | Perepelitsin |
| ELISA Positive (Dogs) | ELISA Negative (Dogs) | Total (Dogs) | |
|---|---|---|---|
| rhKU_Sej_LRR_2271 LFA positive | 35 | 24 | 59 |
| rhKU_Sej_LRR_2271 LFA negative | 15 | 110 | 125 |
| Total (dogs) | 50 | 134 | 184 |
| Methods | Results (Number of Dogs with Positive Results) | |||
|---|---|---|---|---|
| Real-Time PCR Positive (6) | LipL32 ELISA Positive (5) | Vaccinated Dogs (20) | ||
| MAT (serovar with titer ≥1:100) | Autumnalis | 1 | - | - |
| Bratislava | - | 1 | 8 | |
| Canicola | 4 | 2 | 11 | |
| Icterohaemorrhagiae | 3 | 1 | 13 * | |
| Pomona | - | - | 6 | |
| Ballum | - | 1 | - | |
| Sejroe | 5 | 4 | - | |
| Tarassovi | 4 | 1 | - | |
| Grippotyphosa | - | - | 3 | |
| Patoc | 3 | 3 | - | |
| LipL32 ELISA | 6 | 5 | 19 | |
| Culture method | 0 | 0 | 0 | |
| rhKU_Sej_LRR_2271-based LFA | 6 | 5 | 0 | |
| Method | Dog Type | Result (Dogs) | p-Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Culture | stray dog | 0 | 91 | 1.000 * |
| family dog | 0 | 93 | ||
| Real-time PCR | stray dog | 0 | 91 | 0.029 * |
| family dog | 6 | 87 | ||
| ELISA | stray dog | 21 | 70 | 0.217 |
| family dog | 29 | 64 | ||
| Lateral flow | stray dog | 24 | 67 | 0.102 |
| family dog | 35 | 58 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sripattanakul, S.; Boonchuay, K.; Prapong, T.; Wajjwalku, W.; Katzenmeier, G.; Haltrich, D.; Hongprayoon, R.; Prapong, S. Leptospiral Leucine-Rich Repeat Protein-Based Lateral Flow for Assessment of Canine Leptospiral Immunoglobulin G. Trop. Med. Infect. Dis. 2022, 7, 427. https://doi.org/10.3390/tropicalmed7120427
Sripattanakul S, Boonchuay K, Prapong T, Wajjwalku W, Katzenmeier G, Haltrich D, Hongprayoon R, Prapong S. Leptospiral Leucine-Rich Repeat Protein-Based Lateral Flow for Assessment of Canine Leptospiral Immunoglobulin G. Tropical Medicine and Infectious Disease. 2022; 7(12):427. https://doi.org/10.3390/tropicalmed7120427
Chicago/Turabian StyleSripattanakul, Sineenat, Kanpapat Boonchuay, Teerasak Prapong, Worawidh Wajjwalku, Gerd Katzenmeier, Dietmar Haltrich, Ratchanee Hongprayoon, and Siriwan Prapong. 2022. "Leptospiral Leucine-Rich Repeat Protein-Based Lateral Flow for Assessment of Canine Leptospiral Immunoglobulin G" Tropical Medicine and Infectious Disease 7, no. 12: 427. https://doi.org/10.3390/tropicalmed7120427
APA StyleSripattanakul, S., Boonchuay, K., Prapong, T., Wajjwalku, W., Katzenmeier, G., Haltrich, D., Hongprayoon, R., & Prapong, S. (2022). Leptospiral Leucine-Rich Repeat Protein-Based Lateral Flow for Assessment of Canine Leptospiral Immunoglobulin G. Tropical Medicine and Infectious Disease, 7(12), 427. https://doi.org/10.3390/tropicalmed7120427