TB Hackathon: Development and Comparison of Five Models to Predict Subnational Tuberculosis Prevalence in Pakistan
Abstract
:1. Introduction
2. Methods
2.1. Data Sources
2.2. Comparison of Models and Predictions
2.3. Identification of Districts with Most Under-Reporting
3. Results
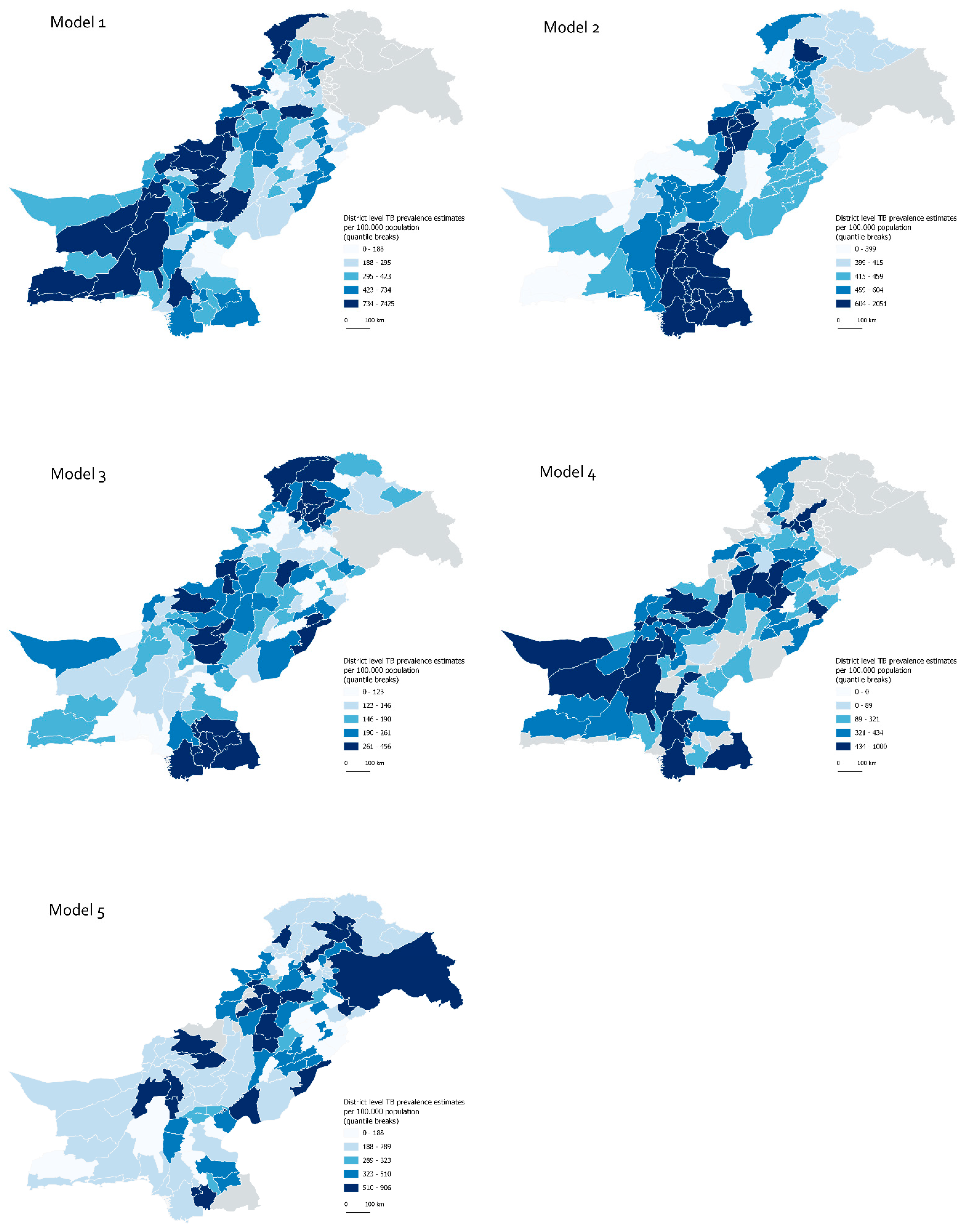
3.1. Comparison of Models
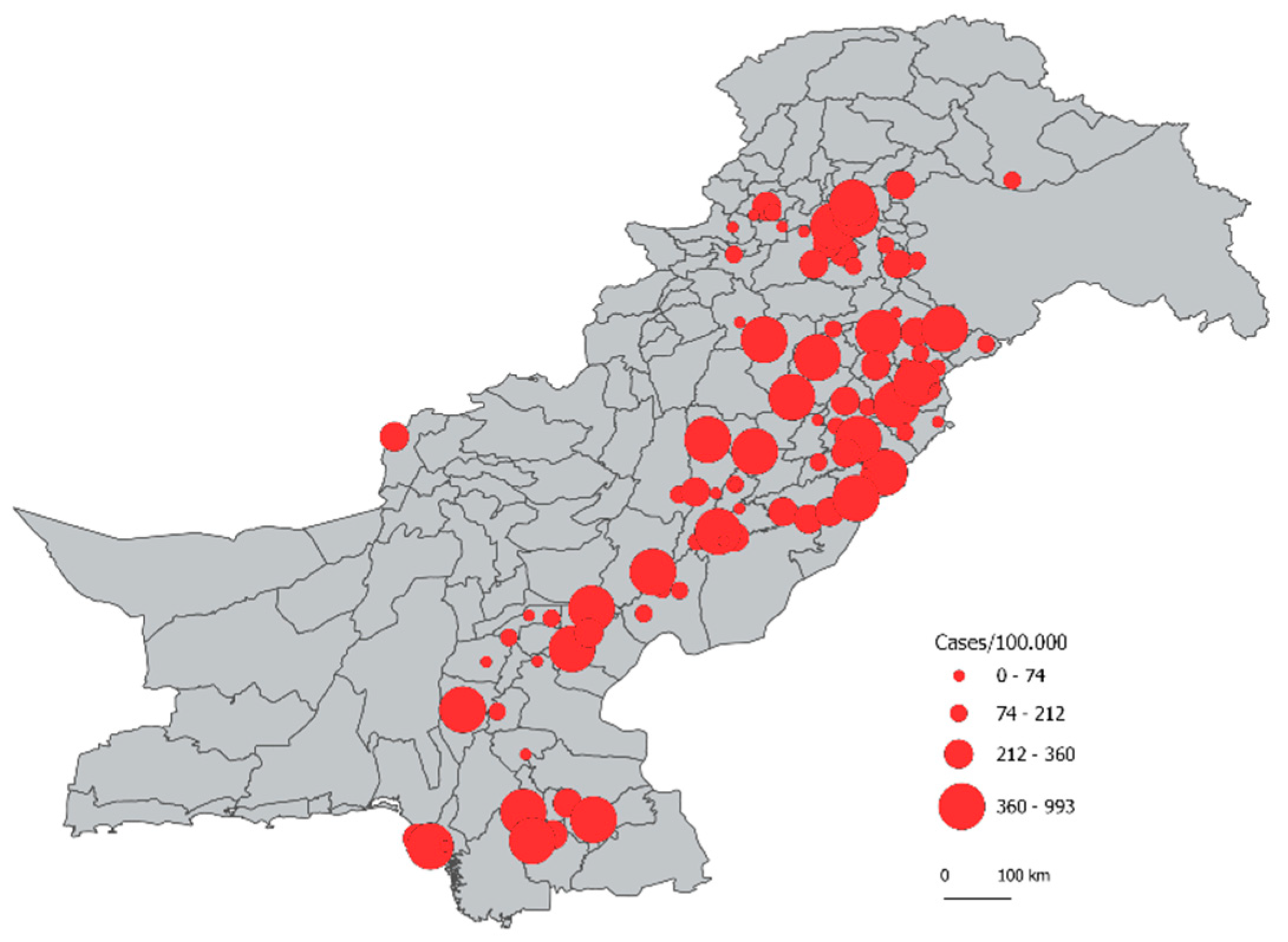
3.2. Comparison of Predictions
3.3. Identification of Districts with Most Under-Reporting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glaziou, P.; Floyd, K. Latest Developments in WHO Estimates of TB Disease Burden; World Health Organisation: Geneva, Switzerland, 2018; Available online: https://www.who.int/tb/advisory_bodies/impact_measurement_taskforce/meetings/tf7_background_4a_burden_estimates.pdf (accessed on 20 November 2020).
- Alba, S.; Rood, E.; Bakker, M.I.; Straetemans, M.; Glaziou, P.; Sismanidis, C. Development and validation of a predictive ecological model for TB prevalence. Int. J. Epidemiol. 2018, 47, 1645–1657. [Google Scholar] [CrossRef] [Green Version]
- Shaweno, D.; Karmakar, M.; Alene, K.A.; Ragonnet, R.; Clements, A.C.; Trauer, J.; MDenholm, J.; McBryde, E. Methods used in the spatial analysis of tuberculosis epidemiology: A systematic review. BMC Med. 2018, 16, 193. [Google Scholar] [CrossRef]
- Mulder, C.; Nkiligi, E.; Kondo, Z.; Scholten, J.N. What to look for when using SUBsET for subnational TB incidence estimates. Int. J. Tuberc. Lung. Dis. Off. J. Int. Union. Tuberc. Lung. Dis. 2020, 24, 983–984. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.M.; Henry, N.J.; Dwyer-Lindgren, L.A.; Lobo, A.D.P.; De Souza, F.M.; Biehl, M.H.; Ray, S.E.; Reiner, R.C.; Stubbs, R.W.; Wiens, K.E.; et al. Progress toward eliminating TB and HIV deaths in Brazil, 2001–2015: A spatial assessment. BMC Med. 2018, 16, 144. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, M.H.; Pelissari, D.M.; Drummond Marques da Silva, G.; Bartholomay, P.; Rocha, M.S.; Sanchez, M.; Arakaki-Sanchez, D.; Glaziou, P.; Cohen, T.; Castro, M.C.; et al. Bayesian evidence synthesis to estimate subnational TB incidence: An application in Brazil. Epidemics 2021, 35, 100443. [Google Scholar] [CrossRef]
- Prem, K.; Pheng, S.H.; Teo, A.K.J.; Evdokimov, K.; Nang, E.E.K.; Hsu, L.Y.; Saphonn, V.; Tieng, S.; Mao, T.E.; Cook, A. Spatial and temporal projections of the prevalence of active tuberculosis in Cambodia. BMJ Glob. Health 2019, 4, e001083. [Google Scholar] [CrossRef] [Green Version]
- Rood, E.; Khan, A.H.; Modak, P.K.; Mergenthaler, C.; Van Gurp, M.; Blok, L.; Bakker, M.A. Spatial Analysis Framework to Monitor and Accelerate Progress towards SDG 3 to End TB in Bangladesh. ISPRS Int. J. Geo-Inf. 2019, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- van Gurp, M.; Rood, E.; Fatima, R.; Joshi, P.; Verma, S.C.; Khan, A.H.; Blok, L.; Mergenthaler, C.; Bakker, M.I. Finding gaps in TB notifications: Spatial analysis of geographical patterns of TB notifications, associations with TB program efforts and social determinants of TB risk in Bangladesh, Nepal and Pakistan. BMC Infect. Dis. 2020, 20, 490. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation. Global Tuberculosis Report 2020; WHO: Geneva, Switzerland, 2020; Available online: https://apps.who.int/iris/bitstream/handle/10665/336069/9789240013131-eng.pdf?ua=1 (accessed on 20 November 2020).
- Fatima, R.; Harris, R.J.; Enarson, D.A.; Hinderaker, S.G.; Qadeer, E.; Ali, K.; Bassili, A.; Bassilli, A. Estimating tuberculosis burden and case detection in Pakistan. Int. J. Tuberc. Lung. Dis. Off. J. Int. Union. Tuberc. Lung. Dis. 2014, 18, 55–60. [Google Scholar] [CrossRef] [Green Version]
- DePasse, J.W.; Carroll, R.; Ippolito, A.; Yost, A.; Santorino, D.; Chu, Z.; Olson, K.R. Less noise, more hacking: How to deploy principles from MIT’s hacking medicine to accelerate health care. Int. J. Technol. Assess Health Care 2014, 30, 260–264. [Google Scholar] [CrossRef] [Green Version]
- Olson, K.R.; Walsh, M.; Garg, P.; Steel, A.; Mehta, S.; Data, S.; Petersen, R.; Guarino, A.J.; Bailey, E.; Bangsberg, D.R. Health hackathons: Theatre or substance? A survey assessment of outcomes from healthcare-focused hackathons in three countries. BMJ Innov. 2017, 3, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xiong, Y.; Sit, H.F.; Tang, W.; Hall, B.J.; Muessig, K.E.; Wei, C.; Bao, H.; Wei, S.; Zhang, D.; et al. A Men Who Have Sex With Men-Friendly Doctor Finder Hackathon in Guangzhou, China: Development of a Mobile Health Intervention to Enhance Health Care Utilization. JMIR MHealth UHealth 2020, 8, e16030. [Google Scholar] [CrossRef]
- Angelidis, P.; Berman, L.; Casas-Perez, M.D.L.L.; Celi, L.A.; Dafoulas, G.E.; Dagan, A.; Escobar, B.; Lopez, D.; Noguez, J.; Osorio-Valencia, J.S.; et al. The hackathon model to spur innovation around global mHealth. J. Med. Eng. Technol. 2016, 40, 392–399. [Google Scholar] [CrossRef] [Green Version]
- Ghouila, A.; Siwo, G.H.; Entfellner, J.-B.D.; Panji, S.; Button-Simons, K.; Davis, S.Z.; Fadlelmola, F.M.; Ferdig, M.T.; Mulder, N.; Participants, T.D.O.M.H. Hackathons as a means of accelerating scientific discoveries and knowledge transfer. Genome Res. 2018, 28, 759–765. [Google Scholar] [CrossRef]
- Ferreira, G.C.; Oberstaller, J.; Fonseca, R.; Keller, T.E.; Adapa, S.R.; Gibbons, J.; Wang, C.; Liu, X.; Li, C.; Pham, M.; et al. Iron Hack—A symposium/hackathon focused on porphyrias, Friedreich’s ataxia, and other rare iron-related diseases. F1000Research 2019, 8, 1135. [Google Scholar] [CrossRef]
- Ramadi, K.; Srinavasan, S.; Atun, R. Health diplomacy through health entrepreneurship: Using hackathons to address Palestinian-Israeli health concerns. BMJ Glob. Health 2019, 4, e001548. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ong, J.J.; Tang, W.; Ritchwood, T.D.; Walker, J.S.; Iwelunmor, J.; Tucker, J.D. Crowdsourcing Methods to Enhance HIV and Sexual Health Services: A Scoping Review and Qualitative Synthesis. J. Acquir. Immune Defic. Syndr. 2019, 82 (Suppl. S3), 271–278. [Google Scholar] [CrossRef] [Green Version]
- Qadeer, E.; Fatima, R.; Tahseen, S.; Samad, Z.; Kalisvaart, N.; Tiemersma, E. Prevalence of Pulmonary Tuberculosis among the Adult Populiation of Pakistan 2010–2011; Islamabad TB Care I: Islamabad, Pakistan, 2013. [Google Scholar]
- Qadeer, E.; Fatima, R.; Yaqoob, A.; Tahseen, S.; Haq, M.U.; Ghafoor, A.; Asif, M.; Straetemans, M.; Tiemersma, E.W. Population Based National Tuberculosis Prevalence Survey among Adults (>15 Years) in Pakistan, 2010–2011. PLoS ONE 2016, 11, e0148293. [Google Scholar] [CrossRef] [PubMed]
- Pakistan Bureau of Statistics. Provisional Province Wise Population by Sex and Rural/Urban—Census 2017 Pakistan. Islamabad. Available online: http://www.pbs.gov.pk/sites/default/files//DISTRICT_WISE_CENSUS_RESULTS_CENSUS_2017.pdf (accessed on 20 November 2020).
- Pakistan Bureau of Statistics. Provisional Province Wise Population—Census 2017 Pakistan. Islamabad. Available online: http://www.pbs.gov.pk/sites/default/files//DISTRICT_WISE_CENSUS_2017.pdf (accessed on 20 November 2020).
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Briggs, A.M. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 393, e44. [Google Scholar] [CrossRef] [Green Version]
- Bertarelli, G.; Ranalli, G.; Bartolucci, F.; d’Alò, M.; Solari, F. Small area estimation for unemployment using latent Markov models. Surv. Methodol. 2018, 44, 167–192. [Google Scholar]
- Philemon, M.D.; Ismail, Z.; Dare, J. A Review of Epidemic Forecasting Using Artificial Neural Networks. Int. J. Epidemiol. Res. 2019, 6, 132–143. [Google Scholar]
- López-Martínez, F.; Núñez-Valdez, E.R.; Crespo, R.G.; García-Díaz, V. An artificial neural network approach for predicting hypertension using NHANES data. Sci. Rep. 2020, 10, 10620. [Google Scholar] [CrossRef]
- Mollalo, A.; Mao, L.; Rashidi, P.; Glass, G.E. A GIS-Based Artificial Neural Network Model for Spatial Distribution of Tuberculosis across the Continental United States. Int. J. Environ. Res. Public Health 2019, 16, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuberculosis Prevalence Surveys: A Handbook; World Health Organization: Geneva, Switzerland, 2011; Available online: http://www.who.int/tb/advisory_bodies/impact_measurement_taskforce/resources_documents/thelimebook/en/ (accessed on 30 March 2021).
- Alba, S.; Verdonck, K.; Lenglet, A.; Rumisha, S.F.; Wienia, M.; Teunissen, I.; Straetemans, M.; Mendoza, W.; Jeannetot, D.; Weibel, D.; et al. Bridging research integrity and global health epidemiology (BRIDGE) statement: Guidelines for good epidemiological practice. BMJ Glob. Health 2020, 5, e003236. [Google Scholar] [CrossRef] [PubMed]

| Dataset | Disaggregation | Time Period |
|---|---|---|
| 1. Prevalence survey data 1 | Individual | 2010–2011 |
| 2. TB notifications | District | quarterly 2009–2018 |
| 3. Laboratory External Quality Assessment data | District | quarterly 2013–2017 |
| 4. Drug-sensitive TB treatment outcomes data | District | quarterly 2009–2015 |
| 5. Drug-Resistant TB notifications | District | quarterly 2009–2018 |
| 6. Master list of TB facilities | Health facility | 2019 |
| 7. Sputum smear testing data | District | quarterly 2009–2017 |
| 8. Private sector notifications | District | Yearly 2017–2018 |
| 9. HIV registrations | Province | 2001–2018 |
| 10. HIV testing rates among TB cases | District | quarterly 2009–2018 |
| 11. Census Population estimates | District | 2017 |
| 12. Shape files | District | 2019 |
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |
|---|---|---|---|---|---|
| Modelling framework | Binomial-logistic regression | Binomial-logistic regression | Binomial-logistic regression | Small Area Estimation (SAE) and Latent Markov (LM) modelling as linking model for SAE | Self-Organising Maps (SOM) on binomial |
| Inference | Bayesian inference with Markov Chain Monte Carlo with No-U-Turn-Sampler (NUTS) | Approximate Bayesian inference with integrated nested Laplace Approximations (INLA) | Approximate Bayesian inference with Broyden–Fletcher–Goldfarb–Shanno algorithm | Bayesian inference with Data Augmentation Markov Chain Monte Carlo and Gibbs sampler | Bayesian Artificial Neural Network |
| Covariance structure | Spatially explicit hierarchical model with fixed and random effects. | Spatially explicit hierarchical model with fixed and random effects | Spatially explicit hierarchical model with fixed and random effects | Hierarchical Discrete latent state model depending on a Gaussian linking model | N/A |
| Outcome variable | Bacteriologically-confirmed TB cases from TB prevalence survey at cluster-level by age and sex | Bacteriologically-confirmed TB cases from TB prevalence survey at cluster-level | Bacteriologically-confirmed TB cases from TB prevalence survey at cluster-level | Bacteriologically-confirmed TB cases from TB prevalence survey at district level | Bacteriologically-confirmed TB cases from TB prevalence survey at district level |
| Final set of predictors 1 | SES, HH size, Indoor smoke, BMI, WAZ, Vaccination coverage, Prevalence of cough, Distance to health facility | Age 15–24 Female Age 15–24 * female Ag 65+ Age_65+ * Sindh Underweight Underweight * KPH | Population density Access to cities [10] Density of TB facilities Poverty Urban extents Locations of protests Locations of violent acts Aridity | Urban households Rural households Urban male pop Rural male pop Urban female pop Rural female pop Pop growth overall Bac+ notifications Bac- notifications EP notifications | All-forms TB notifications Bac+ TB notifications SS+ rate among tested Population density Average household size Percentage rural population Growth rate (urban, rural) Sex ratio (urban, rural) Log gross national income Life expectancy Expected years of schooling Mean years of schooling Human development index |
| Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |
|---|---|---|---|---|---|
| Summary statistics 1 | Min = 104 Max = 7425 Mean = 754 Median = 378 | Min = 276 Max = 2050 Mean = 508 Median = 430 | Min = 51 Max = 456 Mean = 192 Median = 162 | Min = 0 Max = 1000 Mean = 362 Median = 382 | Min = 44 Max = 906 Mean = 366 Median = 289 |
| Completenes 2 | 131 | 143 | 142 | 94 | 139 |
| Pseudo-accuracy by LOOCV for 2010 3 | R2 = 0.404 | R2 = 0.320 | R2 = 0.733 4 | R2 = 0.115 | |
| Cross-validation 5 | Model 2: r = −0.0882 Model 3: r = 0.2305 Model 4: r = −0.0041 Model 5: r = 0.0001 | Model 1: r = −0.0882 Model 3: r = 0.4029 Model 4: r = 0.2492 Model 5: r = 0.1495 | Model 1: r = 0.2305 Model 2: r = 0.4029 Model 4: r = 0.2402 Model 5: r = 0.1583 | Model 1: r = −0.0041 Model 2: r = 0.2492 Model 3: r = 0.2402 Model 5: r = 0.0778 | Model 1: r = 0.0001 Model 2: r = 0.1495 Model 3: r = 0.1583 Model 4: r = 0.0778 |
| Precision 6 | Ratio = 2.69 | Ratio = 0.78 | Ratio = 5.30 | Ratio = 0.63 | Ratio = 2.06 |
| Credibility score 7 | Rater 1: 3 Rater 2: 4 Rater 3: 3 Rater 4: 5 Mean score = 3.75 | Rater 1: 5 Rater 2: 3 Rater 3: 3 Rater 4: 5 Mean score = 4 | Rater 1: 7 Rater 2: 7 Rater 3: 7 Rater 4: 6 Mean score = 6.75 | Rater 1: 4 Rater 2: 4 Rater 3: 3 Rater 4: 5 Mean score = 4 | Rater 1: 7 Rater 2: 8 Rater 3: 6 Rater 4: 6 Mean score = 6.75 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alba, S.; Rood, E.; Mecatti, F.; Ross, J.M.; Dodd, P.J.; Chang, S.; Potgieter, M.; Bertarelli, G.; Henry, N.J.; LeGrand, K.E.; et al. TB Hackathon: Development and Comparison of Five Models to Predict Subnational Tuberculosis Prevalence in Pakistan. Trop. Med. Infect. Dis. 2022, 7, 13. https://doi.org/10.3390/tropicalmed7010013
Alba S, Rood E, Mecatti F, Ross JM, Dodd PJ, Chang S, Potgieter M, Bertarelli G, Henry NJ, LeGrand KE, et al. TB Hackathon: Development and Comparison of Five Models to Predict Subnational Tuberculosis Prevalence in Pakistan. Tropical Medicine and Infectious Disease. 2022; 7(1):13. https://doi.org/10.3390/tropicalmed7010013
Chicago/Turabian StyleAlba, Sandra, Ente Rood, Fulvia Mecatti, Jennifer M. Ross, Peter J. Dodd, Stewart Chang, Matthys Potgieter, Gaia Bertarelli, Nathaniel J. Henry, Kate E. LeGrand, and et al. 2022. "TB Hackathon: Development and Comparison of Five Models to Predict Subnational Tuberculosis Prevalence in Pakistan" Tropical Medicine and Infectious Disease 7, no. 1: 13. https://doi.org/10.3390/tropicalmed7010013
APA StyleAlba, S., Rood, E., Mecatti, F., Ross, J. M., Dodd, P. J., Chang, S., Potgieter, M., Bertarelli, G., Henry, N. J., LeGrand, K. E., Trouleau, W., Shaweno, D., MacPherson, P., Qin, Z. Z., Mergenthaler, C., Giardina, F., Augustijn, E.-W., Baloch, A. Q., & Latif, A. (2022). TB Hackathon: Development and Comparison of Five Models to Predict Subnational Tuberculosis Prevalence in Pakistan. Tropical Medicine and Infectious Disease, 7(1), 13. https://doi.org/10.3390/tropicalmed7010013






