Is It Human or Animal? The Origin of Pathogenic E. coli in the Drinking Water of a Low-Income Urban Community in Bangladesh
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Sample Collection and the Culture of Bacterial Strains
2.3. Extraction of Bacterial DNA
2.4. Detection of Virulence Genes and Phylogenetic Groups
2.5. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troeger, C.; Blacker, B.F.; Khalil, I.A.; Rao, P.C.; Cao, S.; Zimsen, S.R.; Albertson, S.B.; Stanaway, J.D.; Deshpande, A.; Abebe, Z. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef] [Green Version]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Paton, J.C.; Paton, A.W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 1998, 11, 450–479. [Google Scholar] [CrossRef] [Green Version]
- Welinder-Olsson, C.; Kaijser, B. Enterohemorrhagic Escherichia coli (EHEC). Scand. J. Infect. Dis. 2005, 37, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.; Lanoix, J.N.; World Health Organization. Excreta disposal for rural areas and small communities; World Health Organization: Geneva, Swtizerland, 1958. [Google Scholar]
- Prendergast, A.J.; Gharpure, R.; Mor, S.; Viney, M.; Dube, K.; Lello, J.; Berger, C.; Siwila, J.; Joyeux, M.; Hodobo, T. Putting the “A” into WaSH: A call for integrated management of water, animals, sanitation, and hygiene. Lancet Planet. Health 2019, 3, e336–e337. [Google Scholar] [CrossRef] [Green Version]
- Schriewer, A.; Odagiri, M.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Clasen, T.; Jenkins, M.W. Human and animal fecal contamination of community water sources, stored drinking water and hands in rural India measured with validated microbial source tracking assays. Am. J. Trop. Med. Hyg. 2015, 93, 509–516. [Google Scholar] [CrossRef]
- Scott, T.M.; Rose, J.B.; Jenkins, T.M.; Farrah, S.R.; Lukasik, J. Microbial source tracking: Current methodology and future directions. Appl. Environ. Microbiol. 2002, 68, 5796–5803. [Google Scholar] [CrossRef] [Green Version]
- Dick, L.K.; Bernhard, A.E.; Brodeur, T.J.; Santo Domingo, J.W.; Simpson, J.M.; Walters, S.P.; Field, K.G. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 2005, 71, 3184–3191. [Google Scholar] [CrossRef] [Green Version]
- Witty, M.; Nickels, J.; Lisa, J.; Tiedemann, J. Ecology, DNA, and the future of microbial source tracking. Water air soil Pollut. 2009, 201, 219–232. [Google Scholar] [CrossRef]
- Odagiri, M.; Schriewer, A.; Hanley, K.; Wuertz, S.; Misra, P.R.; Panigrahi, P.; Jenkins, M.W. Validation of Bacteroidales quantitative PCR assays targeting human and animal fecal contamination in the public and domestic domains in India. Sci. Total. Environ. 2015, 502, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Baldy-Chudzik, K.; Mackiewicz, P.; Stosik, M. Phylogenetic background, virulence gene profiles, and genomic diversity in commensal Escherichia coli isolated from ten mammal species living in one zoo. Vet. Microbiol. 2008, 131, 173–184. [Google Scholar] [CrossRef]
- Carlos, C.; Pires, M.M.; Stoppe, N.C.; Hachich, E.M.; Sato, M.I.; Gomes, T.A.; Amaral, L.A.; Ottoboni, L.M. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Escobar-Páramo, P.; Le Menac’h, A.; Le Gall, T.; Amorin, C.; Gouriou, S.; Picard, B.; Skurnik, D.; Denamur, E. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 2006, 8, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Stoppe, N.d.C.; Silva, J.S.; Torres, T.T.; Carlos, C.; Hachich, E.M.; Sato, M.I.Z.; Saraiva, A.M.; Ottoboni, L.M.M. Clustering of water bodies in unpolluted and polluted environments based on Escherichia coli phylogroup abundance using a simple interaction database. Genet. Mol. Biol. 2014, 37, 694–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiology 2003, 149, 3575–3586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, C.L.; Gordon, D.M. Effect of diet and gut dynamics on the establishment and persistence of Escherichia coli. Microbiology 2011, 157, 1375–1384. [Google Scholar] [CrossRef] [Green Version]
- Valat, C.; Auvray, F.; Forest, K.; Métayer, V.; Gay, E.; de Garam, C.P.; Madec, J.-Y.; Haenni, M. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl. Environ. Microbiol. 2012, 78, 4677–4682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, D.M. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 2001, 147, 1079–1085. [Google Scholar] [CrossRef] [Green Version]
- Ferdous, J.; Sultana, R.; Rashid, R.B.; Tasnimuzzaman, M.; Nordland, A.; Begum, A.; Jensen, P.K. A comparative analysis of Vibrio cholerae contamination in point-of-drinking and source water in a low-income urban community, Bangladesh. Front. Microbiol. 2018, 9, 489. [Google Scholar] [CrossRef] [Green Version]
- Sultana, R.; Tamason, C.C.; Carstensen, L.S.; Ferdous, J.; Hossain, Z.Z.; Begum, A.; Jensen, P.K.M. Water usage, hygiene and diarrhea in low-income urban communities—A mixed method prospective longitudinal study. MethodsX 2019, 6, 2822–2837. [Google Scholar] [CrossRef]
- Ferdous, J.; Sultana, R.; Rashid, R.B.; Saima, S.; Begum, A.; Jensen, P.K.M. Comparative assessment of fecal contamination in piped-to-plot communal source and point-of-drinking water. Water 2021, 13, 1139. [Google Scholar] [CrossRef]
- Azman, A.S.; Lessler, J.; Satter, S.M.; Mckay, M.V.; Khan, A.; Ahmed, D.; Gurley, E.S. Tracking cholera through surveillance of oral rehydration solution sales at pharmacies: Insights from urban Bangladesh. PLoS Negl. Trop. Dis. 2015, 9, e0004230. [Google Scholar] [CrossRef] [PubMed]
- Gurley, E.S.; Hossain, M.J.; Paul, R.C.; Sazzad, H.M.; Islam, M.S.; Parveen, S.; Faruque, L.I.; Husain, M.; Ara, K.; Jahan, Y. Outbreak of hepatitis E in urban Bangladesh resulting in maternal and perinatal mortality. Clin. Infect. Dis. 2014, 59, 658–665. [Google Scholar] [CrossRef] [Green Version]
- Bangladesh Bureau of Statistics. Population and Housing Census 2011, Community Report Gazipur. Dhaka, Bangladesh. Available online: https://www.google.com/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&cad=rja&uact=8&ved=2ahUKEwiKn4Lyh5rxAhVaOisKHQlOCKMQFjAAegQIBBAD&url=http%3A%2F%2F203.112.218.65%3A8008%2FWebTestApplica-tion%2Fuserfiles%2FImage%2FPopCen2011%2FCom_Gazipur.pdf&usg=AOvVaw3hE1VoMntAchUYITX0G1Of (accessed on 14 September 2021).
- Hossain, Z.Z.; Ferdous, J.; Tulsiani, S.; Jensen, P.; Begum, A. Quantitative analysis of nucleic acid extraction methods for vibrio cholerae using real-time PCR and conventional PCR. Mymensingh Med J. MMJ 2018, 27, 327–335. [Google Scholar] [PubMed]
- Ferdous, J.; Hossain, Z.; Tulsiani, S.; Rashid, R.; Jensen, P.; Begum, A. Optimization and validation of real time PCR assays for absolute quantification of toxigenic Vibrio cholerae and Escherichia coli. Trop. Biomed. 2016, 33, 641–651. [Google Scholar] [PubMed]
- Nguyen, T.V.; Le Van, P.; Le Huy, C.; Gia, K.N.; Weintraub, A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J. Clin. Microbiol. 2005, 43, 755–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anastasi, E.M.; Matthews, B.; Stratton, H.; Katouli, M. Pathogenic Escherichia coli found in sewage treatment plants and environmental waters. Appl. Environ. Microbiol. 2012, 78, 5536–5541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamelin, K.; Bruant, G.; El-Shaarawi, A.; Hill, S.; Edge, T.A.; Fairbrother, J.; Harel, J.; Maynard, C.; Masson, L.; Brousseau, R. Occurrence of virulence and antimicrobial resistance genes in Escherichia coli isolates from different aquatic ecosystems within the St. Clair River and Detroit River areas. Appl. Environ. Microbiol. 2007, 73, 477–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, M.J.; Hadi, A.Z.; Griffith, J.F.; Ishii, S.; Sadowsky, M.J. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 2010, 44, 5463–5473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Power, M.L.; Littlefield-Wyer, J.; Gordon, D.M.; Veal, D.A.; Slade, M.B. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes. Environ. Microbiol. 2005, 7, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, S.; de Laurent, Z.R.; de Villiers, E.P.; Githinji, G.; Charles, K.J. The utility of Escherichia coli as a contamination indicator for rural drinking water: Evidence from whole genome sequencing. PLoS ONE 2021, 16, e0245910. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Bergholz, P.W.; Noar, J.D.; Buckley, D.H. Environmental patterns are imposed on the population structure of Escherichia coli after fecal deposition. Appl. Environ. Microbiol. 2011, 77, 211–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walk, S.T.; Alm, E.W.; Calhoun, L.M.; Mladonicky, J.M.; Whittam, T.S. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 2007, 9, 2274–2288. [Google Scholar] [CrossRef]
- Donnenberg, M. Escherichia Coli: Pathotypes and Principles of Pathogenesis; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Devane, M.; Moriarty, E.; Weaver, L.; Cookson, A.; Gilpin, B. Fecal indicator bacteria from environmental sources; strategies for identification to improve water quality monitoring. Water Res. 2020, 185, 116204. [Google Scholar] [CrossRef]
- Begum, Y.A.; Talukder, K.A.; Nair, G.B.; Qadri, F.; Sack, R.B.; Svennerholm, A.-M. Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. J. Clin. Microbiol. 2005, 43, 3582–3583. [Google Scholar] [CrossRef] [Green Version]
- Harada, H.; Fujimori, Y.; Gomi, R.; Ahsan, M.N.; Fujii, S.; Sakai, A.; Matsuda, T. Pathotyping of Escherichia coli isolated from community toilet wastewater and stored drinking water in a slum in Bangladesh. Lett. Appl. Microbiol. 2018, 66, 542–548. [Google Scholar] [CrossRef] [Green Version]
- Talukdar, P.K.; Rahman, M.; Rahman, M.; Nabi, A.; Islam, Z.; Hoque, M.M.; Endtz, H.P.; Islam, M.A. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE 2013, 8, e61090. [Google Scholar] [CrossRef] [Green Version]
- Lothigius, Å.; Janzon, A.; Begum, Y.; Sjöling, Å.; Qadri, F.; Svennerholm, A.M.; Bölin, I. Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area by real-time PCR. J. Appl. Microbiol. 2008, 104, 1128–1136. [Google Scholar] [CrossRef]
- Ahmed, D.; Islam, M.S.; Begum, Y.A.; Janzon, A.; Qadri, F.; Sjöling, Å. Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. J. Appl. Microbiol. 2013, 114, 1223–1229. [Google Scholar] [CrossRef]
- Pfaff-McDonough, S.J.; Horne, S.M.; Giddings, C.W.; Ebert, J.O.; Doetkott, C.; Smith, M.H.; Nolan, L.K. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Dis. 2000, 44, 23–33. [Google Scholar] [CrossRef]
- Subedi, M.; Luitel, H.; Devkota, B.; Bhattarai, R.K.; Phuyal, S.; Panthi, P.; Shrestha, A.; Chaudhary, D.K. Antibiotic resistance pattern and virulence genes content in avian pathogenic Escherichia coli (APEC) from broiler chickens in Chitwan, Nepal. BMC Vet. Res. 2018, 14, 1–6. [Google Scholar] [CrossRef]
- Delicato, E.R.; de Brito, B.G.; Gaziri, L.C.J.; Vidotto, M.C. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 2003, 94, 97–103. [Google Scholar] [CrossRef]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Nolan, L.K. Characterizing the APEC pathotype. Vet. Res. 2005, 36, 241–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Fakhr, M.K.; Nolan, L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005, 151, 2097–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zambrano, L.D.; Levy, K.; Menezes, N.P.; Freeman, M.C. Human diarrhea infections associated with domestic animal husbandry: A systematic review and meta-analysis. Trans. R. Soc. Trop. Med. Hyg. 2014, 108, 313–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ercumen, A.; Pickering, A.J.; Kwong, L.H.; Arnold, B.F.; Parvez, S.M.; Alam, M.; Sen, D.; Islam, S.; Kullmann, C.; Chase, C. Animal feces contribute to domestic fecal contamination: Evidence from E. coli measured in water, hands, food, flies, and soil in Bangladesh. Environ. Sci. Technol. 2017, 51, 8725–8734. [Google Scholar] [CrossRef] [Green Version]
- Sultana, R.; Nahar, N.; Rimi, N.A.; Azad, S.; Islam, M.S.; Gurley, E.S.; Luby, S.P. Backyard poultry raising in Bangladesh: A valued resource for the villagers and a setting for zoonotic transmission of avian influenza. A qualitative study. Rural Remote Health 2012, 12. [Google Scholar] [CrossRef]
- Rimi, N.A.; Sultana, R.; Ishtiak-Ahmed, K.; Khan, S.U.; Sharker, M.Y.; Zaman, R.U.; Azziz-Baumgartner, E.; Gurley, E.S.; Nahar, N.; Luby, S.P. Poultry slaughtering practices in rural communities of Bangladesh and risk of avian influenza transmission: A qualitative study. EcoHealth 2014, 11, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Wardrop, N.A.; Hill, A.G.; Dzodzomenyo, M.; Aryeetey, G.; Wright, J.A. Livestock ownership and microbial contamination of drinking-water: Evidence from nationally representative household surveys in Ghana, Nepal and Bangladesh. Int. J. Hyg. Environ. Health 2018, 221, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Acharjee, M.; Rahman, F.; Beauty, S.A.; Feroz, F.; Rahman, M.M.; Noor, R. Microbiological study on supply water and treated water in Dhaka city. Stamford J. Microbiol. 2012, 1, 42–45. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.M.M.; Mohamed, Z.K.; Klena, J.D.; Ahmed, S.F.; Moussa, T.A.; Ghenghesh, K.S. Molecular characterization of diarrheagenic Escherichia coli from Libya. Am. J. Trop. Med. Hyg. 2012, 86, 866–871. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Singh, A.K.; Mukherjee, S.; Rajendran, K.; Saha, D.R.; Koley, H.; Basu, S. Composition of Escherichia coli population in the neonatal gut: Phylogroups and virulence determinants. J. Med Microbiol. 2013, 62, 1680–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rúgeles, L.C.; Bai, J.; Martínez, A.J.; Vanegas, M.C.; Gómez-Duarte, O.G. Molecular characterization of diarrheagenic Escherichia coli strains from stools samples and food products in Colombia. Int. J. food Microbiol. 2010, 138, 282–286. [Google Scholar] [CrossRef] [Green Version]
- Unno, T.; Han, D.; Jang, J.; Lee, S.-N.; Ko, G.; Choi, H.Y.; Kim, J.H.; Sadowsky, M.J.; Hur, H.-G. Absence of Escherichia coli phylogenetic group B2 strains in humans and domesticated animals from Jeonnam Province, Republic of Korea. Appl. Environ. Microbiol. 2009, 75, 5659–5666. [Google Scholar] [CrossRef] [Green Version]
- Saha, O.; Hoque, M.N.; Islam, O.K.; Rahaman, M.; Sultana, M.; Hossain, M.A. Multidrug-resistant avian pathogenic Escherichia coli strains and association of their virulence genes in Bangladesh. Microorganisms 2020, 8, 1135. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.J.; Blau, K.; Jechalke, S.; Smalla, K.; Djordjevic, S.P. Whole genome sequencing of Escherichia coli from store-bought produce. Front. Microbiol. 2020, 10, 3050. [Google Scholar] [CrossRef]
- Gomi, R.; Matsuda, T.; Matsui, Y.; Yoneda, M. Fecal source tracking in water by next-generation sequencing technologies using host-specific Escherichia coli genetic markers. Environ. Sci. Technol. 2014, 48, 9616–9623. [Google Scholar] [CrossRef]
- Li, X.; Harwood, V.J.; Nayak, B.; Staley, C.; Sadowsky, M.J.; Weidhaas, J. A novel microbial source tracking microarray for pathogen detection and fecal source identification in environmental systems. Environ. Sci. Technol. 2015, 49, 7319–7329. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Buddenborg, S.; Yoder-Himes, D.R.; Tiedje, J.M.; Konstantinidis, K.T. Genomic diversity of Escherichia isolates from diverse habitats. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warish, A.; Triplett, C.; Gomi, R.; Gyawali, P.; Hodgers, L.; Toze, S. Assessment of genetic markers for tracking the sources of human wastewater associated Escherichia coli in environmental waters. Environ. Sci. Technol. 2015, 49, 9341–9346. [Google Scholar] [CrossRef] [PubMed]
- WHO and Unicef. WASH in the 2030 Agenda: New Global Indicators for Drinking Water, Sanitation and Hygiene. Available online: https://washdata.org/sites/default/files/documents/reports/2017-07/JMP-2017-WASH-in-the-2030-agenda.pdf (accessed on 10 December 2018).

| Phylogenetic Subgroup by Carlos et al., 2010 [14] | Prevalent Host Species | Carlos et al. [14] (Ssolates, N = 241) | Phylogenetic Subgroup by Escobar et al., 2006 [15] | Escobar-Páramo et al. [15] (Isolates, N = 2658) | ||
| Prevalent | Less prevalent | Prevalent | Less prevalent | |||
| A0 | Animal (Carlos et al., 2010, Escobar-Páramo et al., 2006) [14,15] | Animal: 100% (28) [Cow 43% (12/28), chicken 25% (7/28), pig 14% (4/28), sheep 14% (4/28), goat 4% (1/28)] | A | Human 43% | Animal 34% [mammals (39%), bird (16%)] | |
| A1 | Humans (Carlos et al., 2010, Escobar-Páramo et al. 2006) [14,15] | Human 61% (38/62) | Pig 27% (17/62), cow 3% (2/62), goat 3% (2/62), chicken 5% (3/62) | |||
| B1 | Animals, birds (Carlos et al., 2010, Escobar-Páramo et al., 2006) [14,15] | Animal: 88% (71/81) [Cow 36% (29/81), sheep 25% (20/81), goat 16% (13/81), pig 11% (9/81)] | Human 10% (8/81), chicken 2% (2/81) | B1 | Animal 32% [bird (30%), mammals (32%)] | Human 22% |
| B22 | Humans (Carlos et al., 2010, Escobar-Páramo et al., 2006) [14,15] | Human 62% (5/8), | Pig 25% (2/8), chicken 13% (1/8) | B2 | Human 24% | Animal 13% [bird (20%), mammals (12%)] |
| B23 | Humans (Carlos et al., 2010, Escobar-Páramo et al., 2006) [14,15] | Human 100% (7) | ||||
| D1 | Animals, birds (Escobar-Páramo et al., 2006) [14,15] | Human 68% (26/38), | Pig 13% (5/38), cow 11% (4/38), sheep 8% (3/38) | D | Animal 21% [bird (38%), mammals (18%)] | Human 11% |
| D2 | Animals, birds (Escobar-Páramo et al., 2006) [14,15] | Human 59% (10/17), | Cow 18% (3/17), pig 12% (2/17), sheep 12% (2/17) | |||
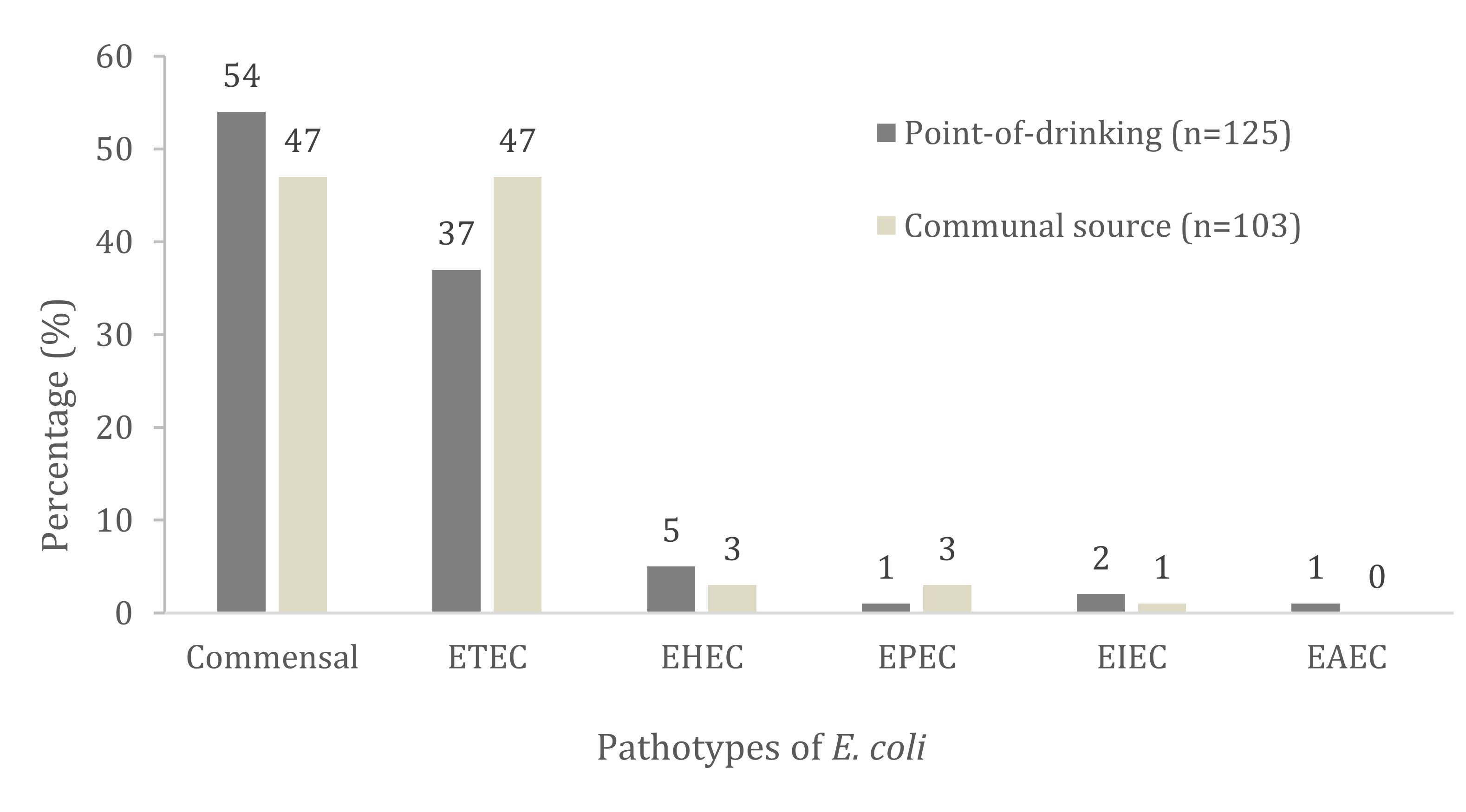
| Virulence Genes and Pathotypes Assignment | Total no. with Trait n = 228 (%) | Point-of-Drinking Water n = 125 (%) | Source Water n = 103 (%) | p Value |
|---|---|---|---|---|
| Pathotype assignment (DEC) | ||||
| eltB | 27 (12) | 14 (11) | 13 (13) | 0.761 |
| estA | 46 (20) | 22 (18) | 24 (23) | 0.303 |
| eltB+estA | 21 (9) | 10 (8) | 11 (11) | 0.501 |
| ETEC | 94 (41) | 46 (37) | 48 (47) | 0.152 |
| vt1 | 2 (1) | 2 (2) | 1 (1) | 0.896 |
| vt2 | 3 (1) | 4 (3) | 2(2) | 0.112 |
| vt1+eaeA | 1 (0.4) | 1 (1) | - | 0.361 |
| vt2+eaeA | - | - | - | - |
| EHEC | 10 (4) | 7 (5) | 3 (3) | 0.152 |
| eaeA | 4 (2) | 1 (1) | 3 (3) | 0.231 |
| eaeA+bfp | - | - | - | - |
| EPEC | 4 (2) | 1 (1) | 3 (3) | 0.231 |
| ipaH | 3 (1) | 2 (2) | 2 (1) | 0.672 |
| EIEC | 3 (1) | 2 (2) | 1 (1) | 0.672 |
| pCVD | 1 (0.4) | 1 (1) | - | 0.361 |
| EAEC | 1 (0.4) | 1 (1) | - | 0.361 |
| Commensal | 116 (51) | 68 (54) | 48 (47) | 0.067 |
| Categories | Prevalence of Pathotypes by Phylogenetic Group, no. (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Point-of-Drinking Water (n = 125) | Source Water (n = 103) | |||||||||
| A1 (n = 2) | B1 (n = 91) | B22 (n = 2) | B23 (n = 13) | D1 (n = 4) | D2 (n = 13) | B1 (n = 89) | B22 (n = 2) | B23 (n = 7) | D2 (n = 5) | |
| ETEC (n = 94) | - | 36 (38) | - | 2 (2) | 3 (3) | 5 (5) | 46 (49) | - | 1 (1) | 1 (1) |
| EHEC (n = 10) | - | 6 (67) | - | 1 (17) | - | - | 2 (17) | - | - | 1 |
| EPEC (n = 4) | - | 1 (25) | - | - | - | - | 3 (75) | - | - | - |
| EIEC (n = 3) | - | 2 (67) | - | - | - | - | 1 (33) | - | - | - |
| EAEC (n = 1) | - | 1 (100) | - | - | - | - | - | - | - | - |
| Commensal (n = 116) | 2 (1) | 45 (40) | 2 (2) | 10 (8) | 1 (1) | 8 (5) | 37 (34) | 2 (2) | 6 (5) | 3 (3) |
| APEC Associated Virulence Genes | Total no. with Trait n = 228 (%) | Point-of-Drinking Water, n = 125 (%) | Source Water, n = 103 (%) | p Value |
|---|---|---|---|---|
| iutA | 31 (14) | 20 (16) | 11 (11) | 0.232 |
| fyuA | 12 (5) | 8 (6) | 4 (4) | 0.388 |
| cnf1 | 30 (13) | 24 (19) | 6 (6) | 0.003 † |
| cvaC | 13 (6) | 9 (7) | 4 (4) | 0.275 |
| Iss | 32 (14) | 27 (22) | 5 (5) | 0.000 † |
| ompT | 18 (8) | 8 (6) | 10 (10) | 0.368 |
| ibe10 | 66 (29) | 49 (39) | 17 (16) | 0.000 † |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferdous, J.; Rashid, R.B.; Sultana, R.; Saima, S.; Jahan Prima, M.; Begum, A.; Mackie Jensen, P.K. Is It Human or Animal? The Origin of Pathogenic E. coli in the Drinking Water of a Low-Income Urban Community in Bangladesh. Trop. Med. Infect. Dis. 2021, 6, 181. https://doi.org/10.3390/tropicalmed6040181
Ferdous J, Rashid RB, Sultana R, Saima S, Jahan Prima M, Begum A, Mackie Jensen PK. Is It Human or Animal? The Origin of Pathogenic E. coli in the Drinking Water of a Low-Income Urban Community in Bangladesh. Tropical Medicine and Infectious Disease. 2021; 6(4):181. https://doi.org/10.3390/tropicalmed6040181
Chicago/Turabian StyleFerdous, Jannatul, Ridwan Bin Rashid, Rebeca Sultana, Sabera Saima, Musharrat Jahan Prima, Anowara Begum, and Peter Kjær Mackie Jensen. 2021. "Is It Human or Animal? The Origin of Pathogenic E. coli in the Drinking Water of a Low-Income Urban Community in Bangladesh" Tropical Medicine and Infectious Disease 6, no. 4: 181. https://doi.org/10.3390/tropicalmed6040181
APA StyleFerdous, J., Rashid, R. B., Sultana, R., Saima, S., Jahan Prima, M., Begum, A., & Mackie Jensen, P. K. (2021). Is It Human or Animal? The Origin of Pathogenic E. coli in the Drinking Water of a Low-Income Urban Community in Bangladesh. Tropical Medicine and Infectious Disease, 6(4), 181. https://doi.org/10.3390/tropicalmed6040181






