One Health Lens on Rabies: Human–Bat Interactions and Genomic Insights of Rabies Virus in Rural Lilongwe, Malawi
Abstract
1. Introduction
2. Materials and Methods
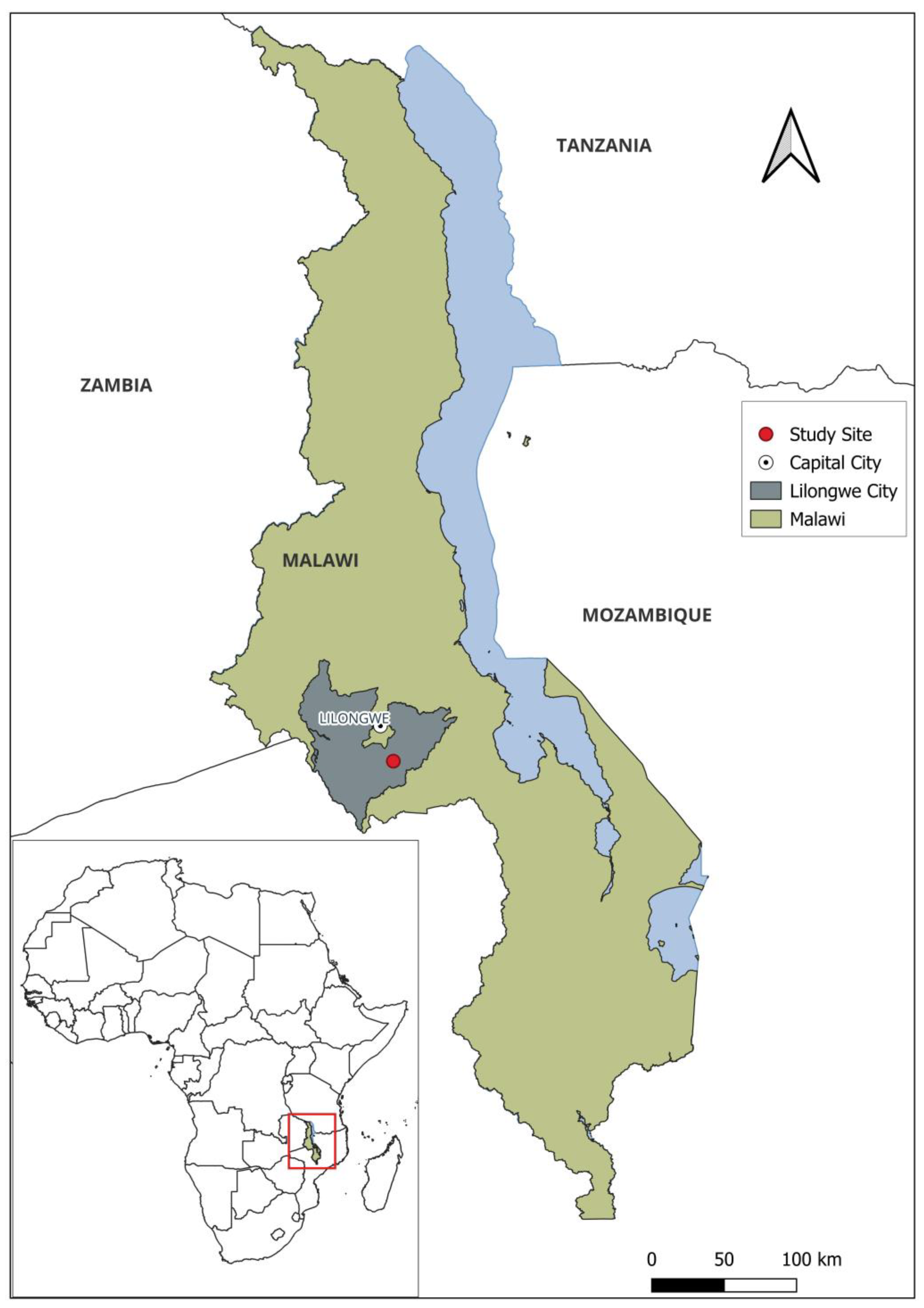
2.1. Study Site and Population
2.2. Study Design and Sampling
2.3. Data Collection Procedure
2.4. DNA Extraction and RT-PCR Screening of Bat Samples
2.5. Whole-Genome Sequencing of Rabies Virus
3. Data Analysis
3.1. Epidemiological Data Analysis
3.2. Sequence Alignment and Phylogenetic Analysis
4. Results
4.1. Sociodemographic Characteristics of Study Participants
4.2. Reported Human–Bat Interactions
4.3. Knowledge Related to Human–Bat Interactions
4.4. Practices Related to Human–Bat Interactions
4.5. Analysis of Risk Factors Associated with Human–Bat Interactions
4.6. Amplification of Lyssavirus RNA in Wildlife Hosts
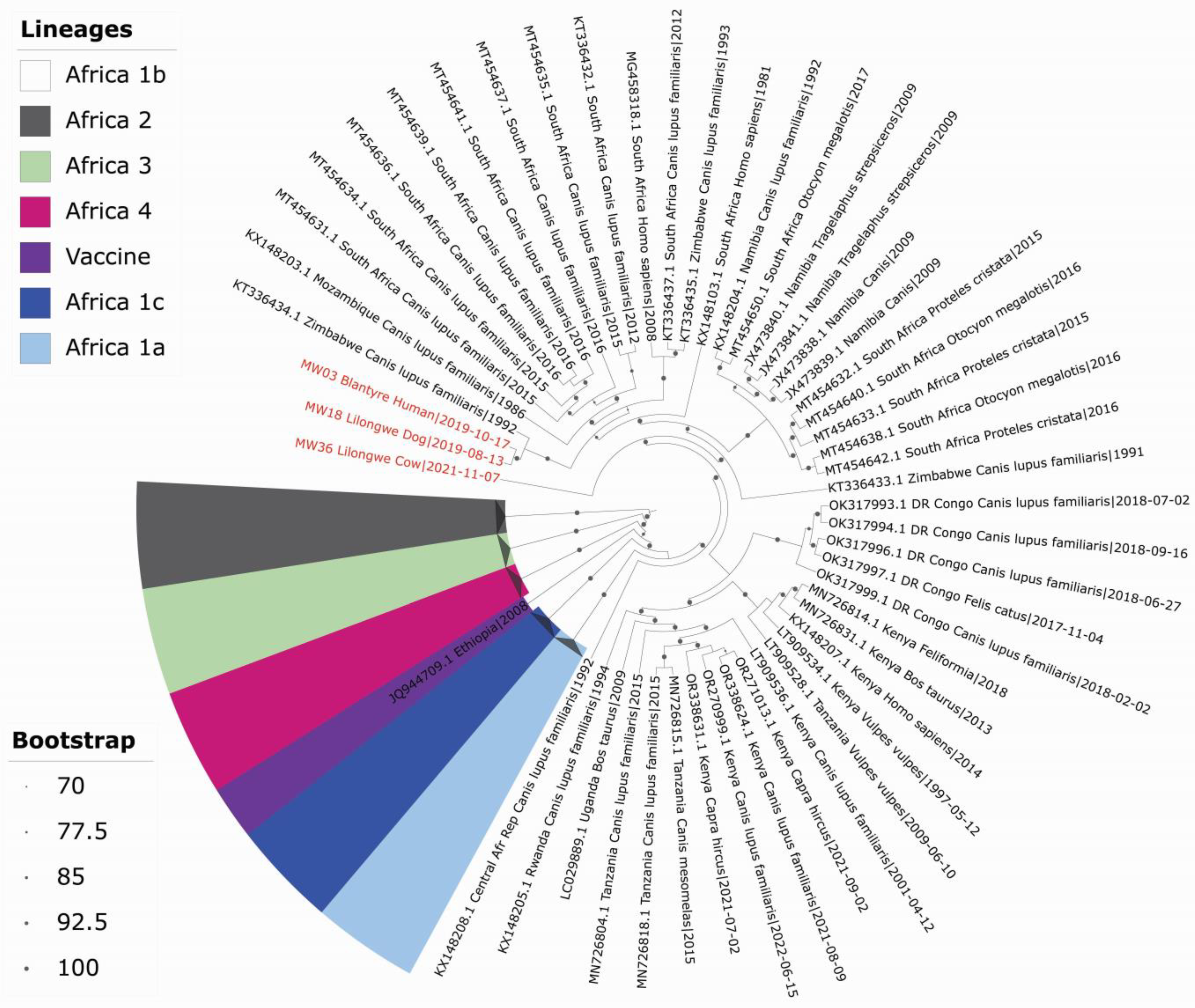
4.7. Phylogenetic Analysis of Whole-Genome Sequences of Lyssavirus
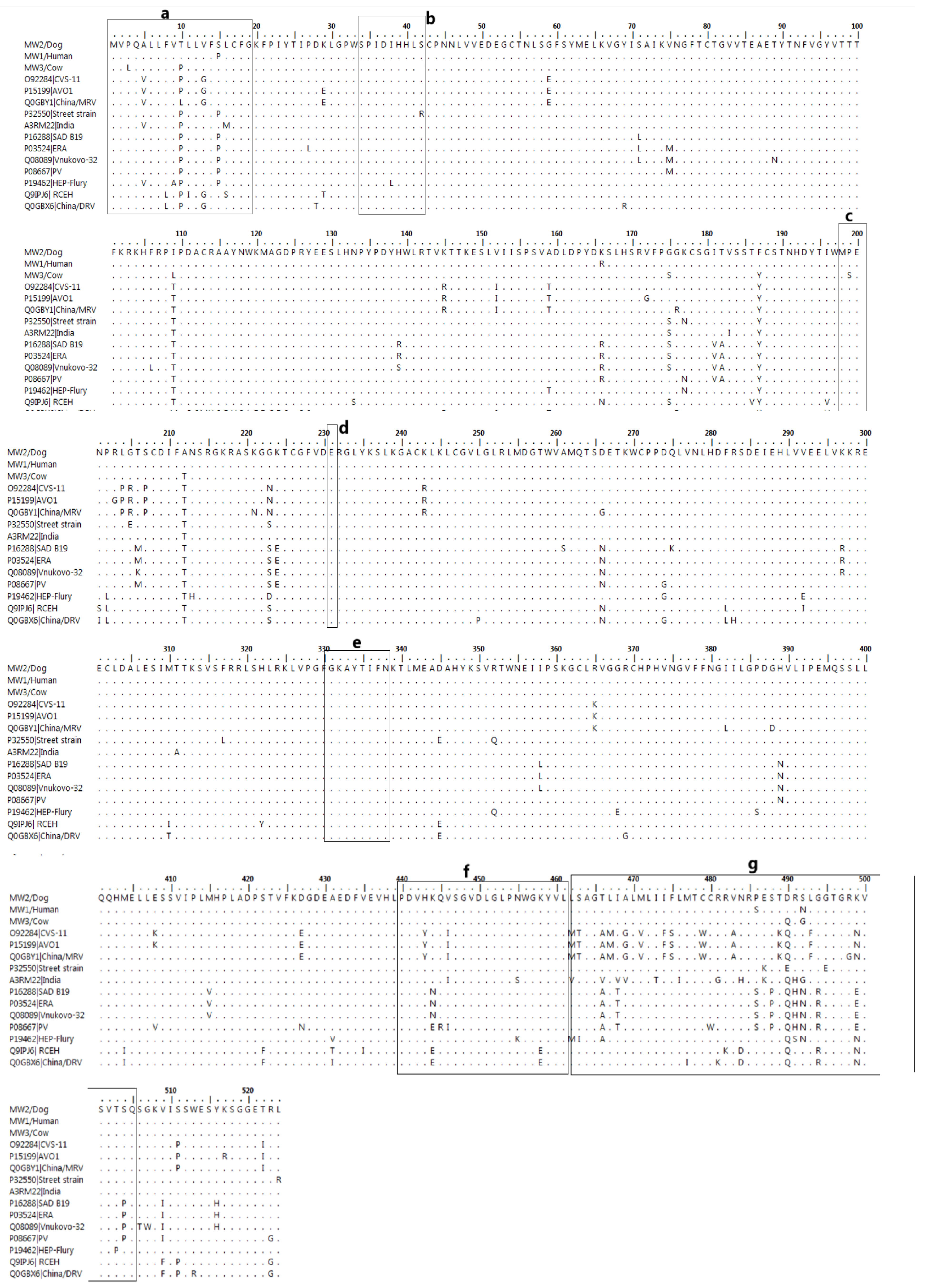
4.8. Characterisation of RABV from a Human, Cow, and Domestic Dog
Features of the Structural Proteins of RABV from Human, Cow, and Domestic Dog
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
| RRVs | Rabies-related lyssaviruses |
| RT-PCR | Reverse Transcriptase Polymerase Chain Reaction |
| PCR | Polymerase Chain Reaction |
| RABV | Rabies virus |
| OR | Odds Ratio |
| aOR | Adjusted Odds Ratio |
| RNA | Ribonucleic acid |
| cDNA | Complimentary deoxyribonucleic acid |
| DNA | Deoxyribonucleic acid |
| WGS | Whole-genome sequencing |
| MW | Malawi |
| MSA | Multiple sequence alignment |
| Aa | Amino acid |
References
- Nel, L.H.; Markotter, W. Lyssaviruses. Crit. Rev. Microbiol. 2007, 33, 301–324. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Ahmed, K.; Wimalaratne, O.; Yamada, K.; Nanayakkara, S.; Perera, D.; Karunanayake, D.; Nishizono, A. Whole-genome analysis of a human rabies virus from Sri Lanka. Arch. Virol. 2011, 156, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Nagaraja, T.; Madhusudana, S.; Desai, A. Molecular characterization of the full-length genome of a rabies virus isolate from India. Virus Genes 2008, 36, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Raux, H.; Flamand, A.; Blondel, D. Interaction of the Rabies Virus P Protein with the LC8 Dynein Light Chain. J. Virol. 2000, 74, 10212–10216. [Google Scholar] [CrossRef]
- Finke, S.; Mueller-Waldeck, R.; Conzelmann, K.-K. Rabies virus matrix protein regulates the balance of virus transcription and replication. J. Gen. Virol. 2003, 84 Pt 6, 1613–1621. [Google Scholar] [CrossRef]
- Morimoto, K.; Hooper, D.C.; Spitsin, S.; Koprowski, H.; Dietzschold, B. Pathogenicity of Different Rabies Virus Variants Inversely Correlates with Apoptosis and Rabies Virus Glycoprotein Expression in Infected Primary Neuron Cultures. J. Virol. 1999, 73, 510–518. [Google Scholar] [CrossRef]
- Muleya, W.; Chambaro, H.M.; Sasaki, M.; Gwenhure, L.F.; Mwenechanya, R.; Kajihara, M.; Saasa, N.; Mupila, Z.; Mori-Kajihara, A.; Qiu, Y.; et al. Genetic diversity of rabies virus in different host species and geographic regions of Zambia and Zimbabwe. Virus Genes 2019, 55, 713–719. [Google Scholar] [CrossRef]
- Hampson, K.; Coudeville, L.; Lembo, T.; Sambo, M.; Kieffer, A.; Attlan, M.; Barrat, J.; Blanton, J.D.; Briggs, D.J.; Cleaveland, S.; et al. Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 2015, 9, e0003709. [Google Scholar] [CrossRef]
- Lembo, T.; Hampson, K.; Kaare, M.T.; Ernest, E.; Knobel, D.; Kazwala, R.R.; Haydon, D.T.; Cleaveland, S. The Feasibility of Canine Rabies Elimination in Africa: Dispelling Doubts with Data. PLoS Negl. Trop. Dis. 2010, 4, e626. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Genet. 2020, 18, 461–471. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; López-Roig, M.; Lavenir, R.; Abdelatif, E.; Boucekkine, W.; Elharrak, M.; Harif, B.; El Ayachi, S.; Salama, A.A.; Nayel, M.A.; et al. Active sero-survey for European bat lyssavirus type-1 circulation in North African insectivorous bats. Emerg. Microbes Infect. 2018, 7, 213. [Google Scholar] [CrossRef] [PubMed]
- Bourhy, H.; Reynes, J.-M.; Dunham, E.J.; Dacheux, L.; Larrous, F.; Huong, V.T.Q.; Xu, G.; Yan, J.; Miranda, M.E.G.; Holmes, E.C. The origin and phylogeography of dog rabies virus. J. Gen. Virol. 2008, 89, 2673–2681. [Google Scholar] [CrossRef] [PubMed]
- Talbi, C.; Holmes, E.C.; de Benedictis, P.; Faye, O.; Nakouné, E.; Gamatié, D.; Diarra, A.; Elmamy, B.O.; Sow, A.; Adjogoua, E.V.; et al. Evolutionary history and dynamics of dog rabies virus in western and central Africa. J. Gen. Virol. 2009, 90, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kainga, H.; Chatanga, E.; Phonera, M.C.; Kothowa, J.P.; Dzimbiri, P.; Kamwendo, G.; Mulavu, M.; Khumalo, C.S.; Changula, K.; Chambaro, H.; et al. Current status and molecular epidemiology of rabies virus from different hosts and regions in Malawi. Arch. Virol. 2023, 168, 61. [Google Scholar] [CrossRef]
- Zimmer, B.; Gamble, L.; Foster, R.; Kennedy, N.; Mayer, D.; Bailey, J.B.; Lemon, J.; Langton, J. Assessment of the impact on paediatric rabies at Queen Elizabeth Central Hospital, Blantyre, Malawi, following a mass canine rabies vaccination programme. Int. J. Infect. Dis. 2019, 79, 64. [Google Scholar] [CrossRef]
- Suu-Ire, R.; Obodai, E.; Bel-Nono, S.O.; Ampofo, W.K.; Mazet, J.A.K.; Goldstein, T.; Johnson, C.K.; Smith, B.; Boaatema, L.; Asigbee, T.W.; et al. Surveillance for potentially zoonotic viruses in rodent and bat populations and behavioral risk in an agricultural settlement in Ghana. One Health Outlook 2022, 4, 6. [Google Scholar] [CrossRef]
- O’malley, K.D.; Kunin, W.E.; Town, M.; Mgoola, W.O.; Stone, E.L. Roost selection by Mauritian tomb bats (Taphozus mauritianus) in Lilongwe city, Malawi—Importance of woodland for sustainable urban planning. PLoS ONE 2020, 15, e0240434. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Jeggo, M. The One Health Approach—Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- Stärk, K.D.; Kuribreña, M.A.; Dauphin, G.; Vokaty, S.; Ward, M.P.; Wieland, B.; Lindberg, A. One Health surveillance—More than a buzz word? Prev. Veter. Med. 2015, 120, 124–130. [Google Scholar] [CrossRef]
- Otu, A.; Effa, E.; Meseko, C.; Cadmus, S.; Ochu, C.; Athingo, R.; Namisango, E.; Ogoina, D.; Okonofua, F.; Ebenso, B. Africa needs to prioritize One Health approaches that focus on the environment, animal health and human health. Nat. Med. 2021, 27, 943–946. [Google Scholar] [CrossRef]
- Israel, G.D. Determining Sample Size. Available online: https://www.psycholosphere.com/Determining%20sample%20size%20by%20Glen%20Israel.pdf (accessed on 3 March 2025).
- De Benedictis, P.; De Battisti, C.; Dacheux, L.; Marciano, S.; Ormelli, S.; Salomoni, A.; Caenazzo, S.T.; Lepelletier, A.; Bourhy, H.; Capua, I.; et al. Lyssavirus Detection and Typing Using Pyrosequencing. J. Clin. Microbiol. 2011, 49, 1932–1938. [Google Scholar] [CrossRef] [PubMed]
- Brunker, K.; Jaswant, G.; Thumbi, S.; Lushasi, K.; Lugelo, A.; Czupryna, A.M.; Ade, F.; Wambura, G.; Chuchu, V.; Steenson, R.; et al. Rapid in-country sequencing of whole virus genomes to inform rabies elimination programmes. Wellcome Open Res. 2020, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Yang, J.; Koprowski, H.; Dietzschold, B.; Fu, Z.F. Phosphorylation of Rabies Virus Nucleoprotein Regulates Viral RNA Transcription and Replication by Modulating Leader RNA Encapsidation. J. Virol. 1999, 73, 1661–1664. [Google Scholar] [CrossRef]
- Anzai, J.; Takamatsu, F.; Takeuchi, K.; Kohno, T.; Morimoto, K.; Goto, H.; Minamoto, N.; Kawai, A. Identification of a Phosphatase-Sensitive Epitope of Rabies Virus Nucleoprotein Which Is Recognized by a Monoclonal Antibody 5-2-26. Microbiol. Immunol. 1997, 41, 229–240. [Google Scholar] [CrossRef]
- Goto, H.; Minamoto, N.; Ito, H.; Ito, N.; Sugiyama, M.; Kinjo, T.; Kawai, A. Mapping of epitopes and structural analysis of antigenic sites in the nucleoprotein of rabies virus. Microbiology 2000, 81 Pt 1, 119–127. [Google Scholar] [CrossRef]
- Lo, K.W.-H.; Naisbitt, S.; Fan, J.-S.; Sheng, M.; Zhang, M. The 8-kDa Dynein Light Chain Binds to Its Targets via a Conserved (K/R)XTQT Motif. J. Biol. Chem. 2001, 276, 14059–14066. [Google Scholar] [CrossRef]
- Gould, A.R.; Kattenbelt, J.A.; Gumley, S.G.; Lunt, R.A. Characterisation of an Australian bat lyssavirus variant isolated from an insectivorous bat. Virus Res. 2002, 89, 1–28. [Google Scholar] [CrossRef]
- Finke, S.; Conzelmann, K.-K. Dissociation of Rabies Virus Matrix Protein Functions in Regulation of Viral RNA Synthesis and Virus Assembly. J. Virol. 2003, 77, 12074–12082. [Google Scholar] [CrossRef]
- Fooks, A.R.; Banyard, A.C.; Horton, D.L.; Johnson, N.; McElhinney, L.M.; Jackson, A.C. Current status of rabies and prospects for elimination. Lancet 2014, 384, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Banyard, A.C.; Evans, J.S.; Luo, T.R.; Fooks, A.R. Lyssaviruses and Bats: Emergence and Zoonotic Threat. Viruses 2014, 6, 2974–2990. [Google Scholar] [CrossRef] [PubMed]
- Bunuma, E.K.; Keyyu, J.; Maziku, J.; Bitanyi, S.; Fyumagwa, R.; Changula, K.; Mubemba, B.; Simulundu, E.; Chitanga, S.; Horton, D.L.; et al. Risk Factors for Human Contact with Bats in Northern Tanzania. Zoonotic Dis. 2024, 4, 293–309. [Google Scholar] [CrossRef]
- van Thiel, P.-P.A.M.; de Bie, R.M.A.; Eftimov, F.; Tepaske, R.; Zaaijer, H.L.; van Doornum, G.J.J.; Schutten, M.; Osterhaus, A.D.M.E.; Majoie, C.B.L.M.; Aronica, E.; et al. Fatal Human Rabies due to Duvenhage Virus from a Bat in Kenya: Failure of Treatment with Coma-Induction, Ketamine, and Antiviral Drugs. PLoS Negl. Trop. Dis. 2009, 3, e428. [Google Scholar] [CrossRef]
- Moran, D.; Juliao, P.; Álvarez, D.; Lindblade, K.A.; Ellison, J.A.; Gilbert, A.T.; Petersen, B.; Rupprecht, C.; Recuenco, S. Knowledge, attitudes and practices regarding rabies and exposure to bats in two rural communities in Guatemala. BMC Res. Notes 2015, 8, 955. [Google Scholar] [CrossRef]
- Nathwani, D.; McIntyre, P.G.; White, K.; Shearer, A.J.; Reynolds, N.; Walker, D.; Orange, G.V.; Fooks, A.R. Fatal Human Rabies Caused by European Bat Lyssavirus Type 2a Infection in Scotland. Clin. Infect. Dis. 2003, 37, 598–601. [Google Scholar] [CrossRef]
- Paweska, J.T.; Blumberg, L.H.; Liebenberg, C.; Hewlett, R.H.; Grobbelaar, A.A.; Leman, P.A.; Croft, J.E.; Nel, L.H.; Nutt, L.; Swanepoel, R. Fatal Human Infection with Rabies-related Duvenhage Virus, South Africa. Emerg. Infect. Dis. 2006, 12, 1965–1967. [Google Scholar] [CrossRef]
- Dah, I.; Poueme Namegni, R.S.; Mohamed, M.M.M.; Jumbo, S.D.; Noumedem, R.N.G.; Conclois, I.; Florian, L.; God-Yang, L.; Kameni, J.M.F.; Wade, A.; et al. Prevalence and public health significance of Lyssavirus in bats in North region of Cameroon. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kamins, A.O.; Rowcliffe, J.M.; Ntiamoa-Baidu, Y.; Cunningham, A.A.; Wood, J.L.N.; Restif, O. Characteristics and Risk Perceptions of Ghanaians Potentially Exposed to Bat-Borne Zoonoses through Bushmeat. Ecohealth 2015, 12, 104–120. [Google Scholar] [CrossRef]
- Kamins, A.; Restif, O.; Ntiamoa-Baidu, Y.; Suu-Ire, R.; Hayman, D.; Cunningham, A.; Wood, J.; Rowcliffe, J. Uncovering the fruit bat bushmeat commodity chain and the true extent of fruit bat hunting in Ghana, West Africa. Biol. Conserv. 2011, 144, 3000–3008. [Google Scholar] [CrossRef]
- Rocha, R.; Fernández-Llamazares, Á.; López-Baucells, A.; Andriamitandrina, S.F.M.; Andriatafika, Z.E.; Temba, E.M.; Torrent, L.; Burgas, D.; Cabeza, M. Human-Bat Interactions in Rural Southwestern Madagascar through a Biocultural Lens. J. Ethnobiol. 2021, 41, 53–69. [Google Scholar] [CrossRef]
- Wright, E.; Anuradha, S.; Richards, R.; Reid, S. A review of the circumstances and health-seeking behaviours associated with bat exposures in high-income countries. Zoonoses Public Health 2022, 69, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.L.B.; Gamble, L.; Gibson, A.D.; Bronsvoort, B.M.D.; Handel, I.G.; Mellanby, R.J.; Mazeri, S. A rabies lesson improves rabies knowledge amongst primary school children in Zomba, Malawi. PLoS Negl. Trop. Dis. 2018, 12, e0006293. [Google Scholar] [CrossRef]
- Fatema, K.; Hossain, S.; Natasha, K.; Chowdhury, H.A.; Akter, J.; Khan, T.; Ali, L. Knowledge attitude and practice regarding diabetes mellitus among Nondiabetic and diabetic study participants in Bangladesh. BMC Public Healh 2017, 17, 364. [Google Scholar] [CrossRef]
- Lee, M.; Kang, B.A.; You, M. Knowledge, attitudes, and practices (KAP) toward COVID-19: A cross-sectional study in South Korea. BMC Public Health 2021, 21, 295. [Google Scholar] [CrossRef]
- Suwannarong, K.; Chanabun, S.; Kanthawee, P.; Khiewkhern, S.; Boonyakawee, P.; Suwannarong, K.; Saengkul, C.; Bubpa, N.; Amonsin, A. Risk factors for bat contact and consumption behaviors in Thailand; a quantitative study. BMC Public Healh 2020, 20, 841. [Google Scholar] [CrossRef]
- Anti, P.; Owusu, M.; Agbenyega, O.; Annan, A.; Badu, E.K.; Nkrumah, E.E.; Tschapka, M.; Oppong, S.; Adu-Sarkodie, Y.; Drosten, C. Human–Bat Interactions in Rural West Africa. Emerg. Infect. Dis. 2015, 21, 1418–1421. [Google Scholar] [CrossRef]
- Jackson, R.T.; Lunn, T.J.; DeAnglis, I.K.; Ogola, J.G.; Webala, P.W.; Forbes, K.M. Frequent and intense human-bat interactions occur in buildings of rural Kenya. PLoS Negl. Trop. Dis. 2024, 18, e001198. [Google Scholar] [CrossRef]
- Euren, J.; Bangura, J.; Gbakima, A.; Sinah, M.; Yonda, S.; Lange, C.E.; McIver, D.J.; LeBreton, M.; Wolking, D.; Monagin, C.G.; et al. Human Interactions with Bat Populations in Bombali, Sierra Leone. Ecohealth 2020, 17, 292–301. [Google Scholar] [CrossRef]
- Nauwelaers, I.; Eynde, C.V.D.; Terryn, S.; Vandendriessche, B.; Willems, W.; Dekeukeleire, D.; Van Gucht, S. Detection and Serological Evidence of European Bat Lyssavirus 1 in Belgian Bats between 2016 and 2018. Trop. Med. Infect. Dis. 2024, 9, 151. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; Amengual, B.; Abellan, C.; Bourhy, H. European bat lyssavirus infection in Spanish bat populations. Emerg. Infect. Dis. 2002, 8, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Constantine, D. Bat Rabies and Other Lyssavirus Infections’, Reston, Virgina. 2009. Available online: https://pubs.usgs.gov/circ/circ1329/ (accessed on 16 November 2023).
- Garcia, A.B.; de Carvalho, C.; Casagrande, D.; Picinato, M.A.D.C.; Pedro, W.A.; Marinho, M.; Queiroz, L.H. Rabies in Bats (Chiroptera, Mammalia) in Brazil: Prevalence and Potential Risk Factors Based on Twenty Years of Research in the Northwestern Region of São Paulo, Brazil. Veter. Sci. 2023, 10, 34. [Google Scholar] [CrossRef]
- Jones, B.D.; Kaufman, E.J.; Peel, A.J. Viral Co-Infection in Bats: A Systematic Review. Viruses 2023, 15, 1860. [Google Scholar] [CrossRef] [PubMed]
- Kia, G.; Huang, Y.; Zhou, M.; Zhou, Z.; Gnanadurai, C.; Leysona, C.; Umoh, J.; Kazeem, H.; Ehizibolo, D.; Kwaga, J.; et al. Molecular characterization of a rabies virus isolated from trade dogs in Plateau State, Nigeria. Sokoto J. Veter. Sci. 2018, 16, 54. [Google Scholar] [CrossRef]
- Fisher, C.R.; Streicker, D.G.; Schnell, M.J. The spread and evolution of rabies virus: Conquering new frontiers. Nat. Rev. Microbiol. 2018, 16, 241–255. [Google Scholar] [CrossRef]
- Streicker, D.G.; Altizer, S.M.; Velasco-Villa, A.; Rupprecht, C.E. Variable evolutionary routes to host establishment across repeated rabies virus host shifts among bats. Proc. Natl. Acad. Sci. USA 2012, 109, 19715–19720. [Google Scholar] [CrossRef]
- Tuffereau, C.; Leblois, H.; Bénéjean, J.; Coulon, P.; Lafay, F.; Flamand, A. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence in adult mice. Virology 1989, 172, 206–212. [Google Scholar] [CrossRef]
- Xie, T.; Yu, H.; Wu, J.; Ming, P.; Huang, S.; Shen, Z.; Xu, G.; Yan, J.; Yu, B.; Zhou, D. Molecular characterization of the complete genome of a street rabies virus WH11 isolated from donkey in China. Virus Genes 2012, 45, 452–462. [Google Scholar] [CrossRef]
- Kissi, B.; Tordo, N.; Bourhy, H. Genetic Polymorphism in the Rabies Virus Nucleoprotein Gene. Virology 1995, 209, 526–537. [Google Scholar] [CrossRef]
- Al-Eitan, L.N.; Wu, G.; Golding, M.; Tang, Y.; Goharriz, H.; Marston, D.A.; Fooks, A.R.; McElhinney, L.M. Whole-genome sequencing and phylogenetic analysis of rabies viruses from Jordan. PLoS Negl. Trop. Dis. 2021, 15, e0009431. [Google Scholar] [CrossRef]
| Variable | Category | Number | Proportion (%) |
|---|---|---|---|
| Sex | Female Male | 58 14 | 81 19 |
| Age | <36 ≥36 | 39 32 | 54 46 |
| Education | None Primary Secondary Tertiary | 7 43 16 5 | 10 60 23 7 |
| Marital status | Married Single/divorced | 18 54 | 25 75 |
| Family size | <5 members ≥5 members | 57 15 | 79 21 |
| Occupation | Farmers Govt/private | 62 10 | 86 14 |
| Daily income | <MWK 2500 (USD 1.50) ≥MWK 2500 (USD 1.50) | 50 8 | 86 14 |
| Length of residence | <5 years ≥5 years | 9 63 | 13 87 |
| Variable | Category | Number | Proportion (%) |
|---|---|---|---|
| Touched bats | Yes No | 11 56 | 16 84 |
| Bat consumption | Yes No | 36 36 | 50 50 |
| Bat caves for traditional medicine | Yes No | 4 67 | 6 94 |
| Bats coming inside dwellings | Yes No | 32 38 | 46 54 |
| Bats are seen in the ceiling | Yes No | 15 57 | 21 79 |
| Bats are seen in trees near my dwelling | Yes No | 19 53 | 26 74 |
| Equipment for handling bats | None Gloves | 31 23 | 57 43 |
| Knowledge Variables | Category | Number | Proportion (%) |
|---|---|---|---|
| Bats as a source of infectious diseases | Yes No | 20 51 | 28 72 |
| Knew that sick/dead bats are dangerous to come in contact with | Yes No Don’t know | 36 16 19 | 51 22 27 |
| Knew that bats get sick | Yes No Don’t know | 17 13 40 | 24 19 57 |
| Heard about bat conservation | Yes No | 3 66 | 4 96 |
| Knew that some bats eat fruits | Yes No | 31 41 | 43 57 |
| Knowledge on vaccines | Yes No | 25 46 | 35 65 |
| Knew that educating people can prevent bat-related diseases | Yes No | 59 11 | 84 16 |
| Knowledge | |||||
|---|---|---|---|---|---|
| Demographic Variables | Low | High | χ2 | p Value | |
| N (%) | N (%) | ||||
| Overall | 41 (57) | 31 (43) | |||
| Gender | Female Male | 36 (88) 5 (12) | 22 (71) 9 (29) | 3.195 | 0.074 |
| Age | <36 years ≥36 years | 20 (50) 20 (50) | 17 (59) 12 (41) | 0.502 | 0.478 |
| Educational level | <Secondary ≥Secondary | 30 (73) 11 (27) | 20 (67) 10 (33) | 0.352 | 0.553 |
| Marital status | Single | 12 (29) | 6 (19) | 0.925 | 0.336 |
| Married | 29 (71) | 25 (81) | |||
| Duration of residence in area | <5 years | 5 (12) | 4 (13) | n/a | 0.928 |
| ≥5 years | 36 (88) | 27 (87) | |||
| Occupation | Farmers | 36 (88) | 26 (84) | 0.228 | 0.633 |
| Others | 5 (12) | 5 (16) | |||
| Daily income | <MWK 2500 (USD 1.50) | 30 (66) | 20 (74) | 0.970 | 0.325 |
| ≥MWK 2500 (USD 1.50) | 15 (33) | 7 (26) | |||
| Practice Variables | Category | Number | Proportion (%) |
|---|---|---|---|
| Wash fruits before eating | Yes No | 66 6 | 92 8 |
| Wash using soap after bat scratch or bite | Yes No | 36 26 | 58 42 |
| If you get scratched, where would you go? | Hospital Nowhere | 51 11 | 82 18 |
| Wash hands using soap after handling bats | Yes No | 41 26 | 61 39 |
| Method of disposing of a dead bat | Proper Improper | 28 23 | 55 45 |
| Disinfecting environment after bat disposal | Yes No | 33 35 | 49 51 |
| Dog vaccination against rabies | Yes No | 23 36 | 39 61 |
| Practice | |||||
|---|---|---|---|---|---|
| Demographic Variables | Poor | Good | χ2 | p Value | |
| N (%) | N (%) | ||||
| Overall | 33 (46) | 39 (54) | |||
| Gender | Female | 27 (82) | 31 (80) | 0.062 | 0.803 |
| Male | 6 (18) | 8 (20) | |||
| Age | <36 years | 21 (66) | 16 (43) | 3.457 | 0.063 |
| ≥36 years | 11 (34) | 43 (57) | |||
| Educational level | <Secondary | 21 (64) | 29 (76) | 1.363 | 0.243 |
| ≥Secondary | 12 (36) | 9 (24) | |||
| Marital status | Single | 10 (30) | 8 (20) | 0.914 | 0.339 |
| Married | 23 (70) | 31 (80) | |||
| Duration of residence in area | <5 years | 5 (15) | 4 (10) | n/a | 0.531 |
| ≥5 years | 28 (85) | 35 (90) | |||
| Occupation | Farmers | 28 (85) | 34 (87) | n/a | 0.776 |
| Others | 5 (15) | 5 (13) | |||
| Daily income | <MWK 2500 (USD 1.50) | 22 (52) | 28 (80) | 5.240 | 0.022 |
| ≥MWK 2500 (USD 1.50) | 20 (48) | 7 (20) | |||
| Variable | Number | Test Static | p-Value |
|---|---|---|---|
| Education | |||
| <Secondary level | 50 | 1.497 a | 0.3 |
| ≥Secondary level | 21 | ||
| Age | |||
| <36 years old | 37 | 4.176 a | 0.55 |
| ≥36 years old | 32 | ||
| Duration of residence | * | ||
| <5 years | 9 | 6.222 b | 0.028 |
| ≥5 years | 63 | ||
| Occupation | * | ||
| Farmer | 62 | 7.432 a | 0.014 |
| Other | 10 | ||
| Daily income | |||
| <MWK 2500 | 50 | 1.424 b | 0.272 |
| ≥MWK 2500 | 8 | ||
| Location of bat roosts | |||
| Ceiling/trees nearby | 31 | 2.837 a | 0.148 |
| Other places | 39 | ||
| Bats coming inside houses | |||
| Yes | 38 | 0.230 a | 0.811 |
| No | 32 | ||
| Do you dislike bats? | * | ||
| Yes | 42 | 4.799 a | 0.041 |
| No | 24 |
| Unadjusted | Adjusted | |||||
|---|---|---|---|---|---|---|
| Odds Ratio | 95% CI | p-Value | Odds Ratio | 95% CI | p-Value | |
| Age | ||||||
| <36 years | Ref | Ref | ||||
| ≥36 years | 0.40 | 0.047–1.239 | 0.092 | 0.19 | 0.127–1.979 | 0.075 |
| Location of bat roosts | ||||||
| Ceiling | Ref | |||||
| Other places | 0.35 | 0.092–1.353 | 0.129 | 0.35 | 0.08–1.53 | 0.067 |
| Bats coming inside dwellings | ||||||
| No | Ref | |||||
| Yes | 0.40 | 0.275–0.605 | 0.002 * | 0.075 | 0.008–0.68 | 0.021 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singano, N.; Kainga, H.; Chatanga, E.; Nkhoma, J.; Njunga, G.; Chulu, J.; Tembo, R.; Sawa, H.; Muleya, W. One Health Lens on Rabies: Human–Bat Interactions and Genomic Insights of Rabies Virus in Rural Lilongwe, Malawi. Trop. Med. Infect. Dis. 2025, 10, 95. https://doi.org/10.3390/tropicalmed10040095
Singano N, Kainga H, Chatanga E, Nkhoma J, Njunga G, Chulu J, Tembo R, Sawa H, Muleya W. One Health Lens on Rabies: Human–Bat Interactions and Genomic Insights of Rabies Virus in Rural Lilongwe, Malawi. Tropical Medicine and Infectious Disease. 2025; 10(4):95. https://doi.org/10.3390/tropicalmed10040095
Chicago/Turabian StyleSingano, Nathan, Henson Kainga, Elisha Chatanga, Joseph Nkhoma, Gilson Njunga, Julius Chulu, Rabecca Tembo, Hirofumi Sawa, and Walter Muleya. 2025. "One Health Lens on Rabies: Human–Bat Interactions and Genomic Insights of Rabies Virus in Rural Lilongwe, Malawi" Tropical Medicine and Infectious Disease 10, no. 4: 95. https://doi.org/10.3390/tropicalmed10040095
APA StyleSingano, N., Kainga, H., Chatanga, E., Nkhoma, J., Njunga, G., Chulu, J., Tembo, R., Sawa, H., & Muleya, W. (2025). One Health Lens on Rabies: Human–Bat Interactions and Genomic Insights of Rabies Virus in Rural Lilongwe, Malawi. Tropical Medicine and Infectious Disease, 10(4), 95. https://doi.org/10.3390/tropicalmed10040095






