Enzymes Degrading Fungal Cell Wall Components vs. Those Exhibiting Lactonase Activity as Participants of Antifungals
Abstract
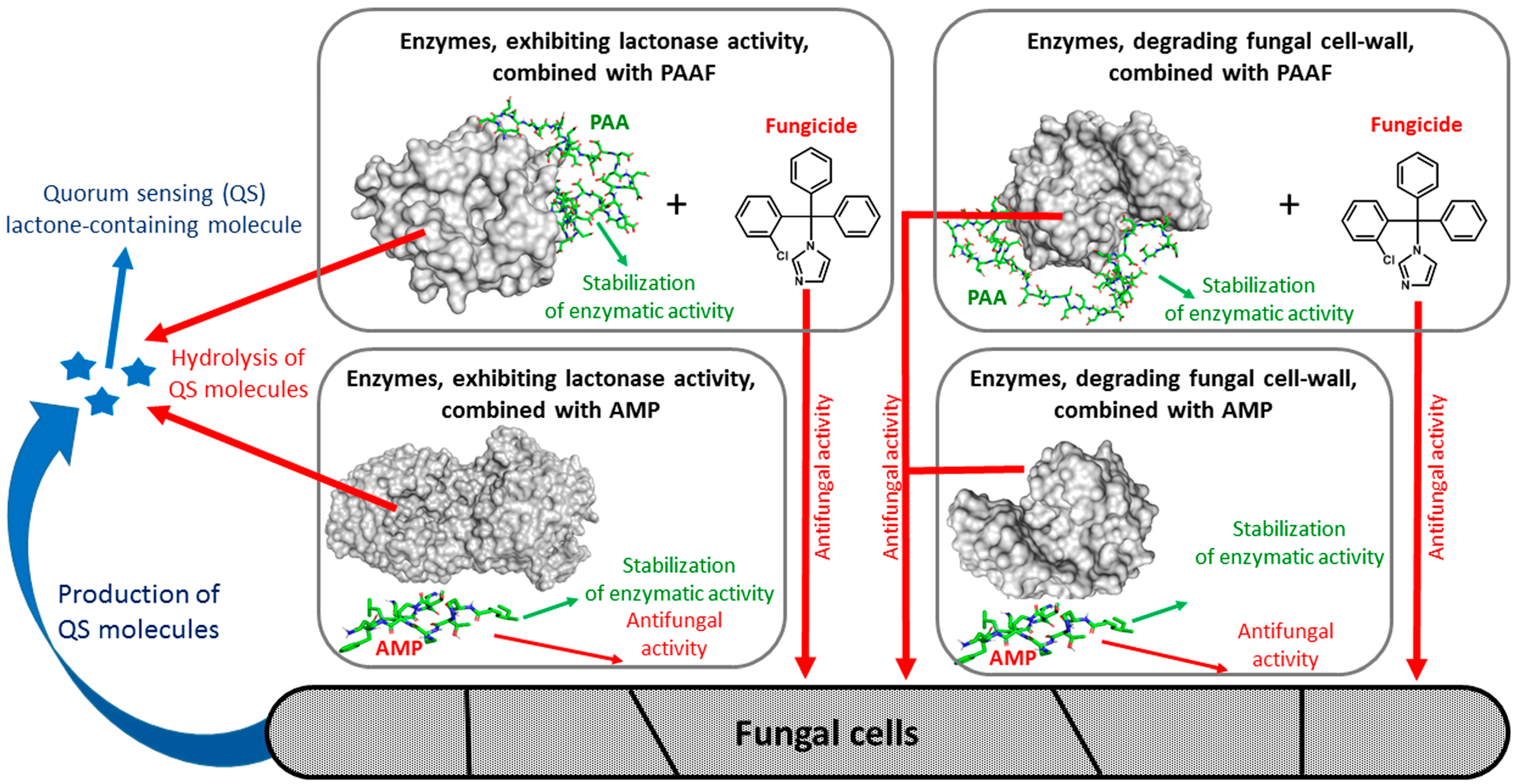
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Molecular Docking
2.3. Enzymatic Activity and Physical-Chemical Characteristics
2.4. Antifungal Activity
3. Results
3.1. Computational Modeling of the Interactions of Hydrolytic Enzymes with the Fungal Cell Wall Components
3.2. Computational Modeling of the Interactions of Enzymes Degrading the Fungal Cell Wall Components with AMPs or PAAs
3.2.1. Interaction of AMPs with the Enzymes Degrading Fungal Cell Wall Components
3.2.2. Interaction of PAAs with the Molecules of Enzymes Degrading Fungal Cell Wall Components
3.3. Computational Modeling of the Interactions of PAAs with Enzymes Exhibiting Lactonase Activity
3.4. Catalytic and Physical-Chemical Characteristics of Antifungal Enzyme Combinations
3.5. The Effect of Antifungal Enzyme Combinations on Fungi
3.5.1. Modeling of Interactions of Fungicides with Enzyme–PAA Combinations
3.5.2. Antifungal Activity of the Combinations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3OHC6-HSL | N-(3-hydroxy-hexanoyl)-l-homoserine lactone |
| AMP | Antimicrobial polypeptide |
| ANOVA | Analysis of variance |
| APBS | Adaptive Poisson-Boltzmann solver |
| ATP | Adenosine triphosphate |
| Bgy6-like | 1,3–1,4-β-d-glucanase |
| BMAP-18 | Bovine Myeloid Antimicrobial Peptide-18 |
| DMSO | Dimethyl sulfoxide |
| Ega3 | Endo-α-1,4-galactosaminidase |
| His6-OPH | Hexahistidine containing organophosphate hydrolase |
| hLF | Human lactoferrin |
| I-TASSER | Iterative Threading ASSEmbly Refinement |
| Lfampin B | Lactoferrampin B |
| MUF-3-NAG | 4-Methylumbelliferyl β-d-N,N′,N″-triacetylchitotrioside |
| NDM-1 | New Delhi metallo-beta-lactamase 1 |
| NTA | Nanoparticle tracking analysis |
| PAA | Polyamino acid |
| PAAF | Polyamino acids supplemented with fungicides |
| PEG | Polyethylene glycol |
| PLD | Poly-l-aspartic acid |
| PLE | Poly-l-glutamic acid |
| RcAlb-PepII | Peptide based on the structure of 2S albumin from Ricinus communis |
References
- WHO. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action. 2022. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 22 September 2025).
- Denning, D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 24, e428–e438. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Chowdhary, A.; Gold, J.A. The rapid emergence of antifungal-resistant human-pathogenic fungi. Nat. Rev. Microbiol. 2023, 21, 818–832. [Google Scholar] [CrossRef]
- Logan, A.; Wolfe, A.; Williamson, J.C. Antifungal resistance and the role of new therapeutic agents. Curr. Infect. Dis. Rep. 2022, 24, 105–116. [Google Scholar] [CrossRef]
- Rhijn, N.v.; Rhodes, J. Evolution of antifungal resistance in the environment. Nat. Microbiol. 2025, 10, 1804–1815. [Google Scholar] [CrossRef]
- Bédard, C.; Pageau, A.; Fijarczyk, A.; Mendoza-Salido, D.; Alcañiz, A.J.; Després, P.C.; Durand, R.; Plante, S.; Alexander, E.M.M.; Rouleau, F.D.; et al. FungAMR: A comprehensive database for investigating fungal mutations associated with antimicrobial resistance. Nat. Microbiol. 2025, 10, 2338–2352. [Google Scholar] [CrossRef]
- Aslanli, A.; Domnin, M.; Stepanov, N.; Senko, O.; Efremenko, E. Action enhancement of antimicrobial peptides by their combination with enzymes hydrolyzing fungal quorum molecules. Int. J. Biol. Macromol. 2024, 280, 136066. [Google Scholar] [CrossRef]
- Efremenko, E.; Aslanli, A.; Domnin, M.; Stepanov, N.; Senko, O. Enzymes with Lactonase Activity against Fungal Quorum Molecules as Effective Antifungals. Biomolecules 2024, 14, 383. [Google Scholar] [CrossRef]
- Naga, N.G.; El-Badan, D.; Ghanem, K.M.; Shaaban, M.I. It is the time for quorum sensing inhibition as alternative strategy of antimicrobial therapy. Cell Commun. Signal. 2023, 21, 133. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Q.; Ren, K.; Xu, T.; Zhang, Z.; Xu, M.; Rao, Z.; Zhang, X. A Review of Antimicrobial Peptides: Structure, Mechanism of Action, and Molecular Optimization Strategies. Fermentation 2024, 10, 540. [Google Scholar] [CrossRef]
- Mufida, D.R.A.; Putra, I.P.; Nawangsih, A.A.; Krishanti, N.P.R.A.; Wahyudi, A.T. Glucanase enzyme activity from rhizospheric Streptomyces spp. inhibit growth and damage the cell wall of Fusarium oxysporum. Rhizosphere 2024, 32, 100991. [Google Scholar] [CrossRef]
- Zhang, L.; Li, W.; Tao, Y.; Zhao, S.; Yao, L.; Cai, Y.; Niu, Q. Overexpression of the key virulence factor 1,3–1,4-β-D-glucanase in the endophytic bacterium Bacillus halotolerans Y6 to improve Verticillium resistance in cotton. J. Agric. Food Chem. 2019, 67, 6828–6836. [Google Scholar] [CrossRef] [PubMed]
- Kezuka, Y.; Ohishi, M.; Itoh, Y.; Watanabe, J.; Mitsutomi, M.; Watanabe, T.; Nonaka, T. Structural studies of a two-domain chitinase from Streptomyces griseus HUT6037. J. Mol. Biol. 2006, 358, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Kanai, R.; Kawase, T.; Tanabe, T.; Mitsutomi, M.; Sakuda, S.; Miyashita, K. Family 19 chitinases of Streptomyces species: Characterization and distribution. Microbiology 1999, 145, 3353–3363. [Google Scholar] [CrossRef]
- Bamford, N.C.; Le Mauff, F.; Subramanian, A.S.; Yip, P.; Millán, C.; Zhang, Y.; Zacharias, C.; Forman, A.; Nitz, M.; Codée, J.D.C.; et al. Ega3 from the fungal pathogen Aspergillus fumigatus is an endo-α-1,4-galactosaminidase that disrupts microbial biofilms. J. Biol. Chem. 2019, 294, 13833–13849. [Google Scholar] [CrossRef]
- Wang, A.; Paul, J.; Weldrick, P.J.; Madden, L.A.; Paunov, V.N. Enhanced clearing of Candida biofilms on a 3D urothelial cell in vitro model using lysozyme-functionalized fluconazole-loaded shellac nanoparticles. Biomater. Sci. 2021, 9, 6927–6939. [Google Scholar] [CrossRef]
- Hernández-Téllez, C.N.; Rodríguez-Córdova, F.J.; Rosas-Burgos, E.C.; Cortez-Rocha, M.O.; Burgos-Hernández, A.; Lizardi-Mendoza, J.; Torres-Arreola, W.; Martínez-Higuera, A.; Plascencia-Jatomea, M. Activity of chitosan-lysozyme nanoparticles on the growth, membrane integrity, and β-1,3-glucanase production by Aspergillus parasiticus. 3 Biotech 2017, 7, e279. [Google Scholar] [CrossRef]
- Numata, K. Poly(amino acid)s/polypeptides as potential functional and structural materials. Polym. J. 2015, 47, 537–545. [Google Scholar] [CrossRef]
- Lyagin, I.V.; Efremenko, E.N. Biomolecular engineering of biocatalysts hydrolyzing neurotoxic organophosphates. Biochimie 2018, 144, 115–121. [Google Scholar] [CrossRef]
- Sivasankaran, R.P.; Snell, K.; Kunkel, G.; Georgiou, P.; Puente, E.G.; Maynard, H.D. Polymer-mediated protein/peptide therapeutic stabilization: Current progress and future directions. Prog. Polym. Sci. 2024, 156, 101867. [Google Scholar] [CrossRef]
- Efremenko, E.; Votchitseva, Y.; Plieva, F.; Galaev, I.; Mattiasson, B. Purification of His6-organophosphate hydrolase using monolithic supermacroporous polyacrylamide cryogels developed for immobilized metal affinity chromatography. Appl. Microbiol. Biotechnol. 2006, 70, 558–563. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35 (Suppl. 2), W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Voevodin, V.; Antonov, A.; Nikitenko, D.; Shvets, P.; Sobolev, S.; Sidorov, I.; Stefanof, K.; Voevodin, V.; Zhumatiy, S. Supercomputer Lomonosov-2: Large scale, deep monitoring and fine analytics for the user community. Supercomput. Front. Innov. 2019, 6, 4–11. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Grigorenko, V.G.; Khrenova, M.G.; Andreeva, I.P.; Rubtsova, M.Y.; Lev, A.I.; Novikova, T.S.; Detusheva, E.V.; Fursova, N.K.; Dyatlov, I.A.; Egorov, A.M. Drug repurposing of the unithiol: Inhibition of metallo-β-lactamases for the treatment of carbapenem-resistant Gram-negative bacterial infections. Int. J. Mol. Sci. 2022, 23, 1834. [Google Scholar] [CrossRef]
- Bergonzi, C.; Schwab, M.; Elias, M. The quorum-quenching lactonase from Geobacillus caldoxylosilyticus: Purification, characterization, crystallization and crystallographic analysis. Acta Crystallogr. Sect. Struct. Biol. Commun. 2016, 72, 681–686. [Google Scholar] [CrossRef]
- Shen, C.R.; Chen, Y.S.; Yang, C.J.; Chen, J.K.; Liu, C.L. Colloid chitin azure is a dispersible, low-cost substrate for chitinase measurements in a sensitive, fast, reproducible assay. SLAS Discov. Adv. Sci. Drug Discov. 2010, 15, 213–217. [Google Scholar] [CrossRef]
- Ismayilov, I.T.; Stepanov, N.A.; Efremenko, E.N.; Abbasov, V.M. Evaluation of biocidal properties of vegetable oil-based corrosion inhibitors using bioluminescent enzymatic method. Mosc. Univ. Chem. Bull. 2015, 70, 197–201. [Google Scholar] [CrossRef]
- Efremenko, E.; Lyagin, I.; Aslanli, A.; Stepanov, N.; Maslova, O.; Senko, O. Carrier variety used in immobilization of His6-OPH extends its application areas. Polymers 2023, 15, 591. [Google Scholar] [CrossRef]
- Kaul, L.; Süss, R.; Zannettino, A.; Richter, K. The revival of dithiocarbamates: From pesticides to innovative medical treatments. iScience 2021, 24, 102092. [Google Scholar] [CrossRef]
- Haq, M.; Deshmukh, P. Review of recurrent otomycosis and clotrimazole in its treatment. Cureus 2022, 14, e30098. [Google Scholar] [CrossRef]
- Ladetto, M.F.; Ancarola, M.E.; Lamas, D.G.; Kollrich, B.A.; Cucher, M.A.; Álvarez, V.A.; Glisoni, R.J.; Castro, G.R.; Islan, G.A.; Cuestas, M.L. Enhanced antifungal efficacy of clotrimazole and itraconazole delivered via solid lipid nanoparticles for potential otomycosis therapy. J. Drug Deliv. Sci. Technol. 2025, 113, 107401. [Google Scholar] [CrossRef]
- Karpe, S.C.; Kiran, M.; Majhi, S.; Meena, J.; Kumar, R.; Chander, H.; Anvikar, A.R.; Kasana, H. Clotrimazole as a new frontier: Drug repurposing and its efficacy in cancer therapy. Cancer Pathog. Ther. 2025, 3, E14–E23. [Google Scholar] [CrossRef]
- Bucataru, C.; Ciobanasu, C. Antimicrobial peptides: Opportunities and challenges in overcoming resistance. Microbiol. Res. 2024, 286, 127822. [Google Scholar] [CrossRef] [PubMed]
- Oliveira Junior, N.G.; Souza, C.M.; Buccini, D.F.; Cardoso, M.H.; Franco, O.L. Antimicrobial peptides: Structure, functions and translational applications. Nat. Rev. Microbiol. 2025, 23, 687–700. [Google Scholar] [CrossRef]
- London, N.; Raveh, B.; Schueler-Furman, O. Druggable protein–protein interactions–from hot spot to hot segments. Curr. Opin. Chem. Biol. 2013, 17, 952–959. [Google Scholar] [CrossRef]
- Hashemi, Z.S.; Zarei, M.; Fath, M.K.; Ganji, M.; Farahani, M.S.; Afsharnouri, F.; Pourzardosht, N.; Khalesi, B.; Jahangiri, A.; Rahbar, M.R.; et al. In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions. Front. Mol. Biosci. 2021, 8, 669431. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, C.; Chen, X.; Wang, B.; Sun, F.; Li, W.; Liu, D.; Jin, H. Possible adverse events of imidazole antifungal drugs during treatment of vulvovaginal candidiasis: Analysis of the FDA Adverse Event Reporting System. Sci. Rep. 2024, 14, 14560. [Google Scholar] [CrossRef]
- Dalal, A.; Kumar, P.; Bansal, R.; Tushir, R.; Chauhan, A. A descriptive review on pharmacokinetics and pharmacodynamics profile of an antifungal agent: Clotrimazole. Eur. J. Pharm. Med. Res. 2022, 9, 204–216. [Google Scholar] [CrossRef]
- Malik, N.A.; Nazir, N.; Manzoor, M.; Gull, F. Fungicide-albumin interactions: Unraveling the complex relationship—A comprehensive review. Biophys. Rev. 2024, 16, 417–439. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.; Latge, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P.; Wang, T. Modern biophysics redefines our understanding of fungal cell wall structure, complexity, and dynamics. mBio 2022, 13, e01145-22. [Google Scholar] [CrossRef]

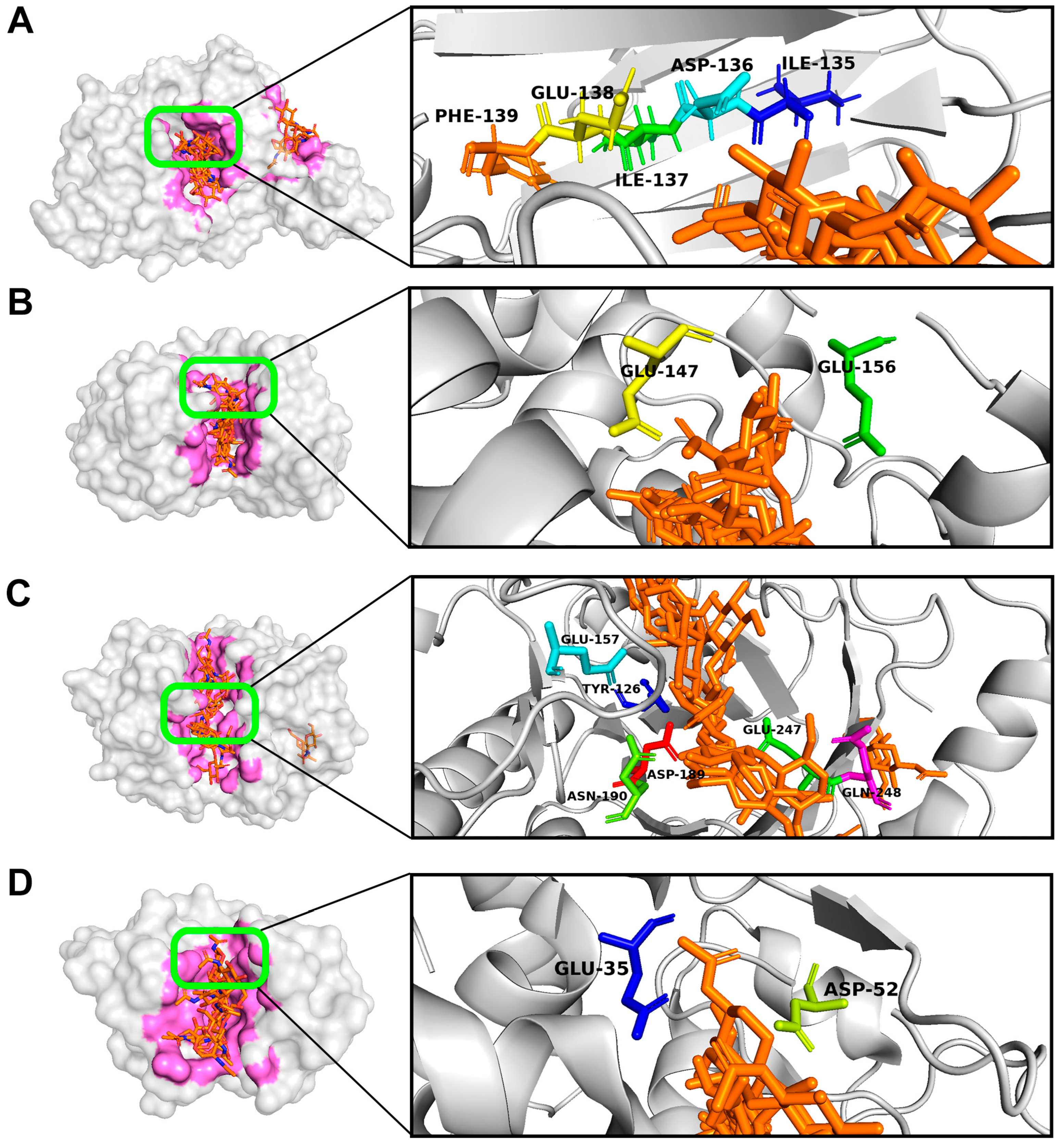
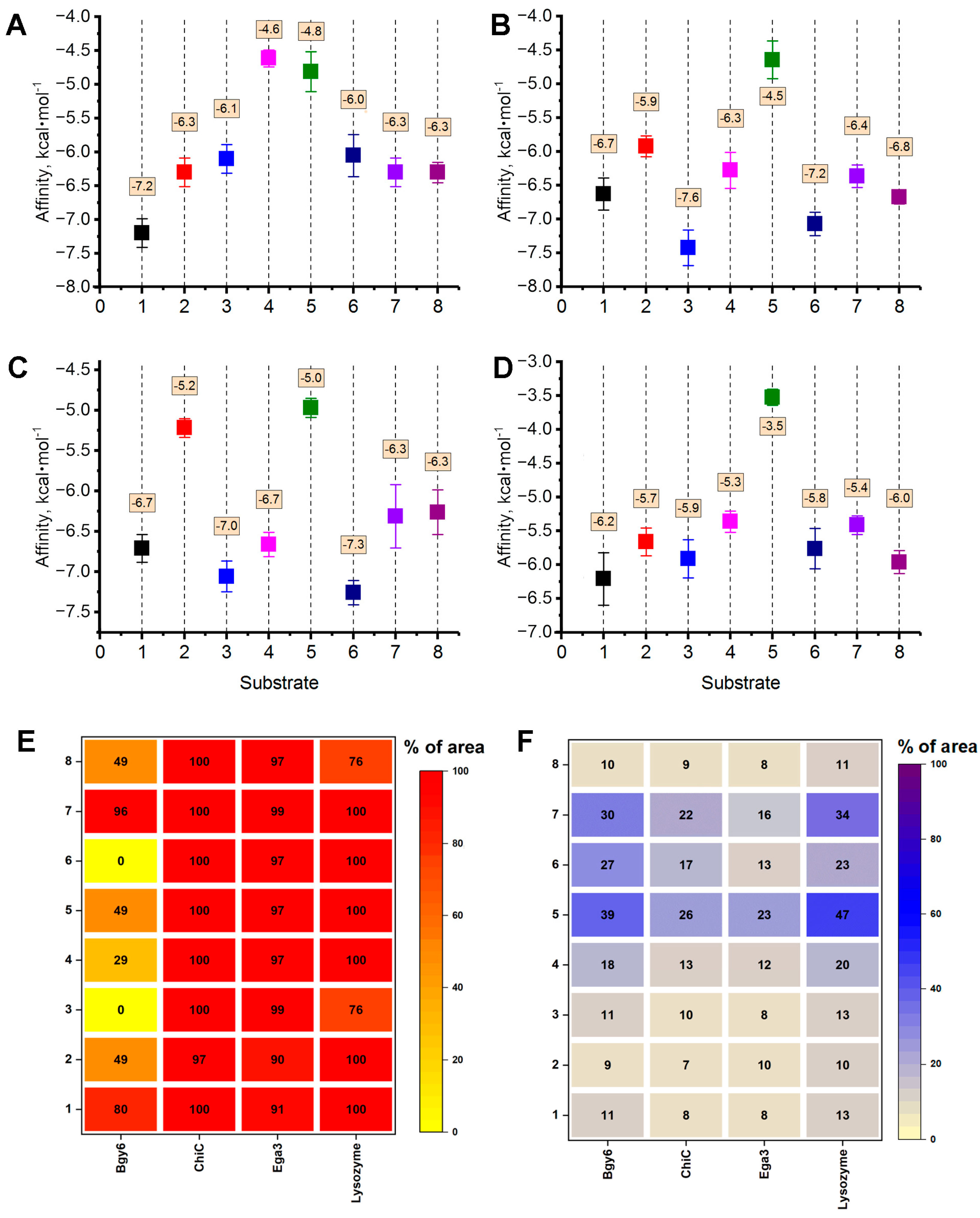

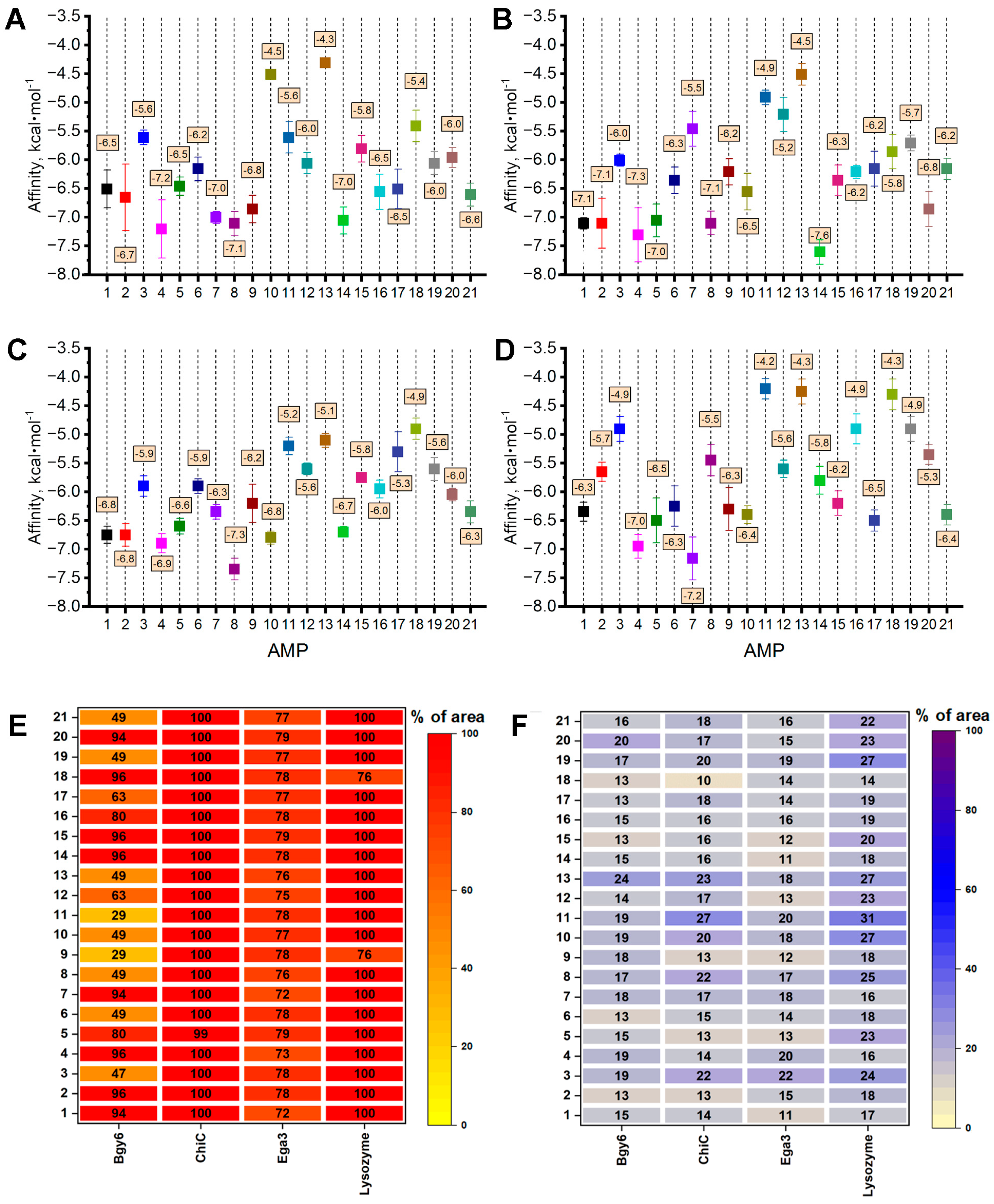
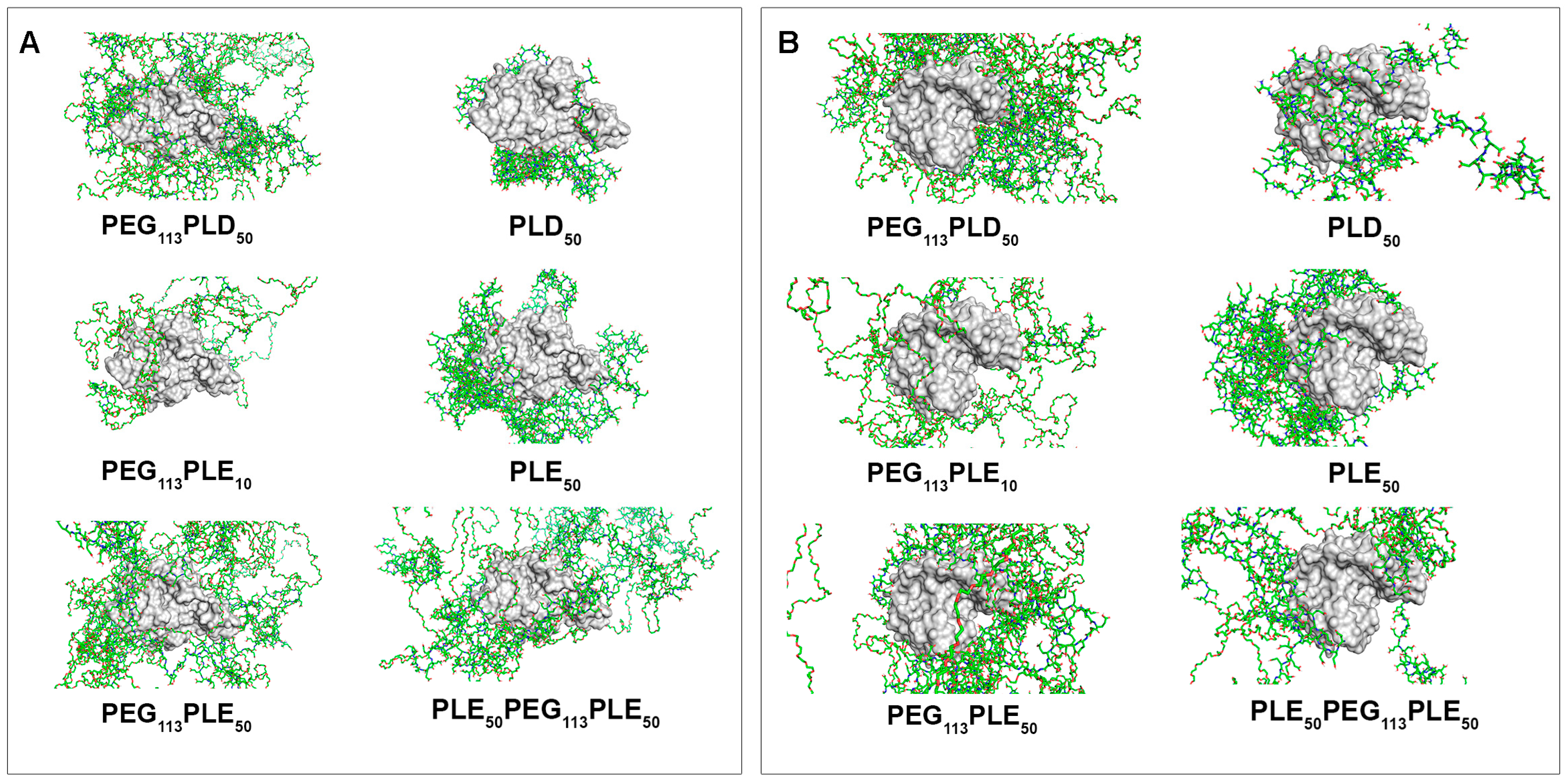

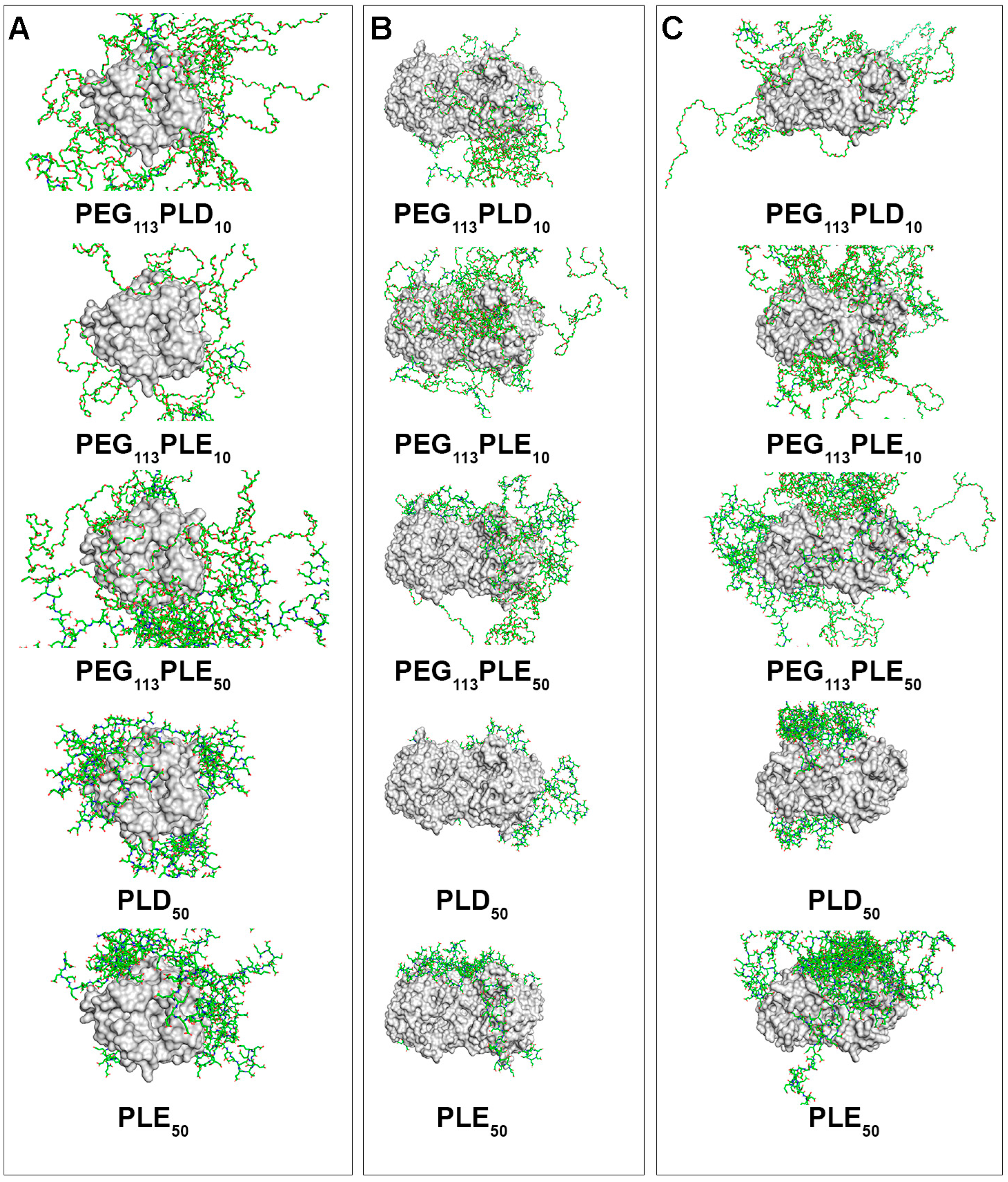
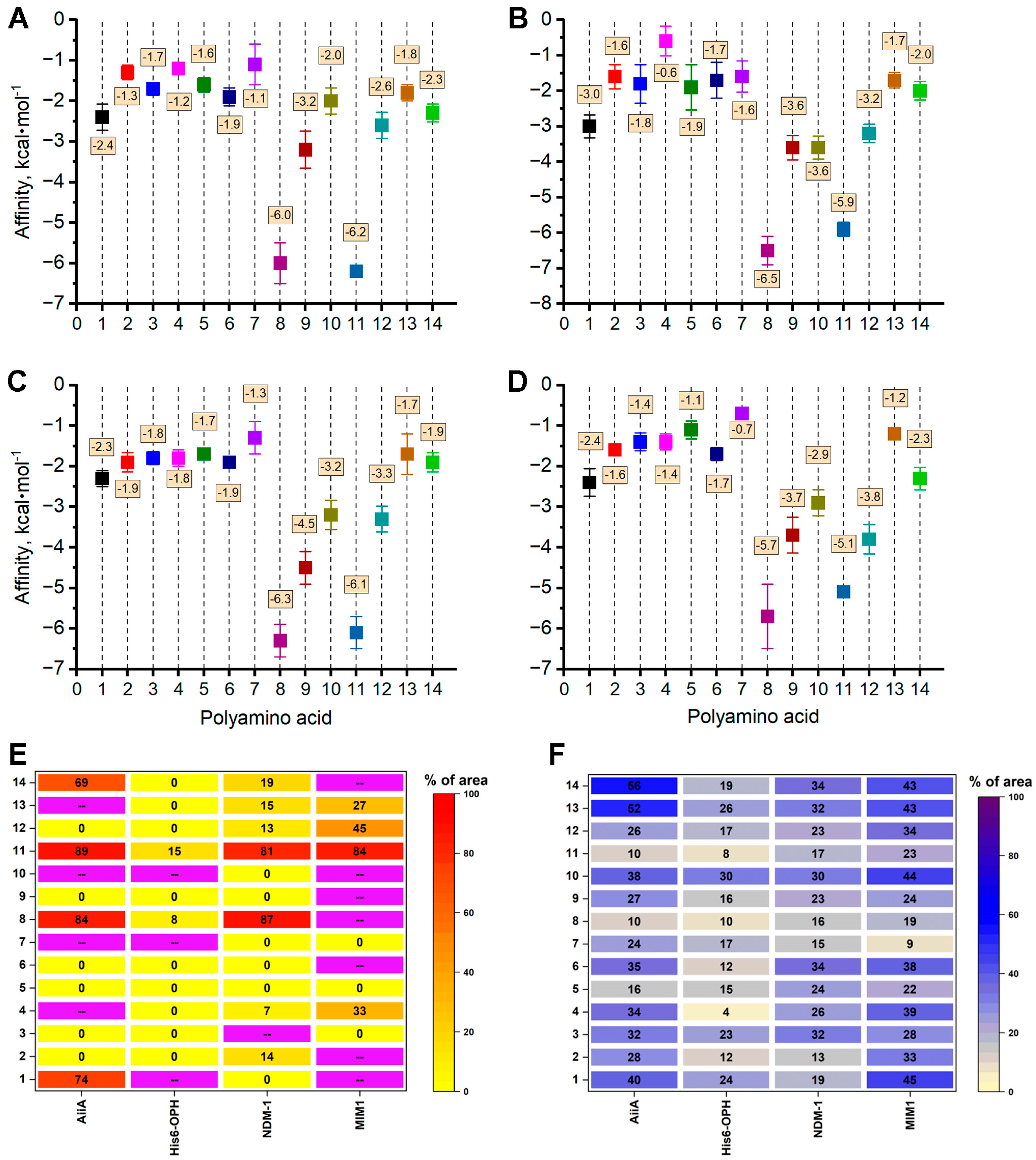
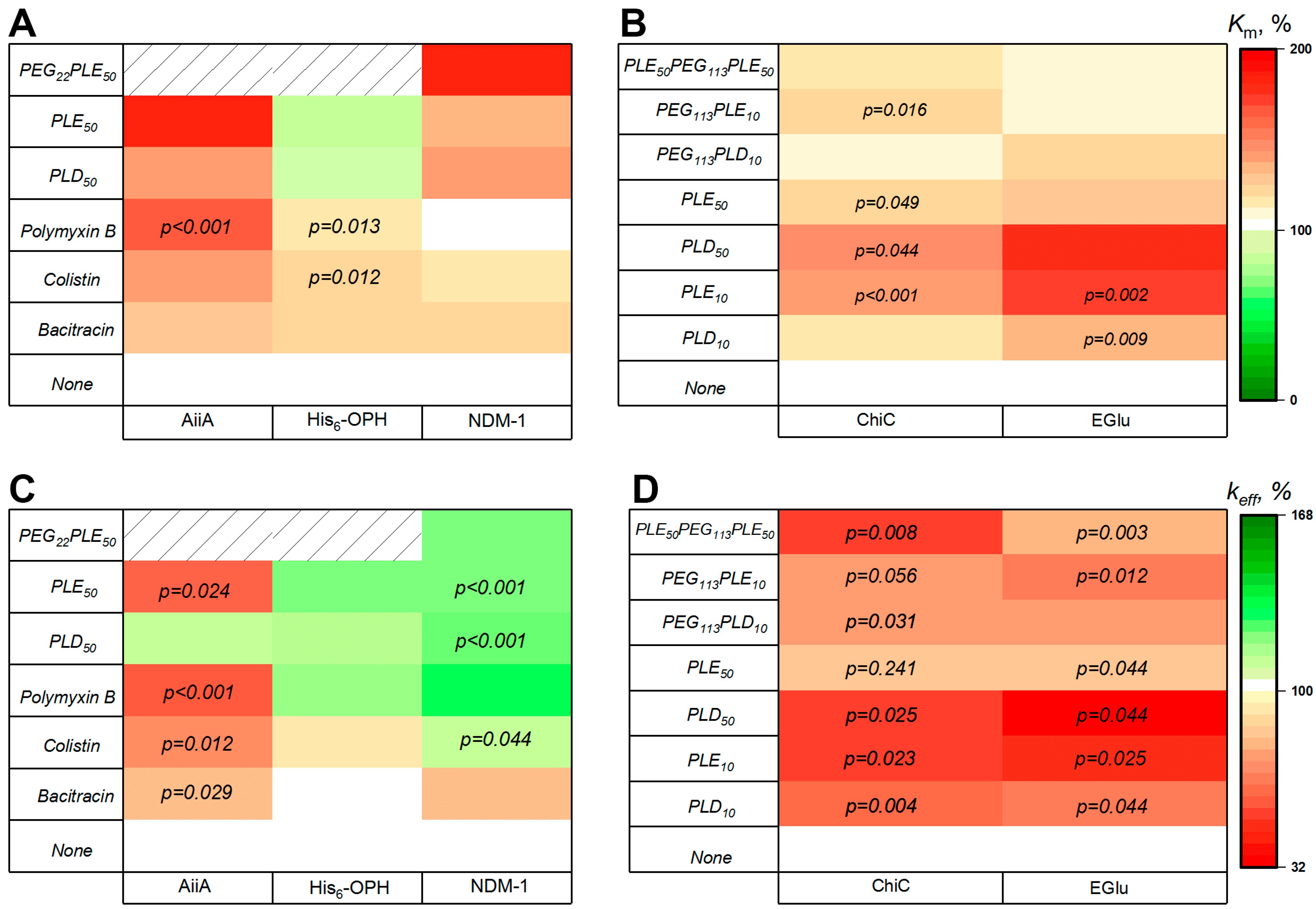

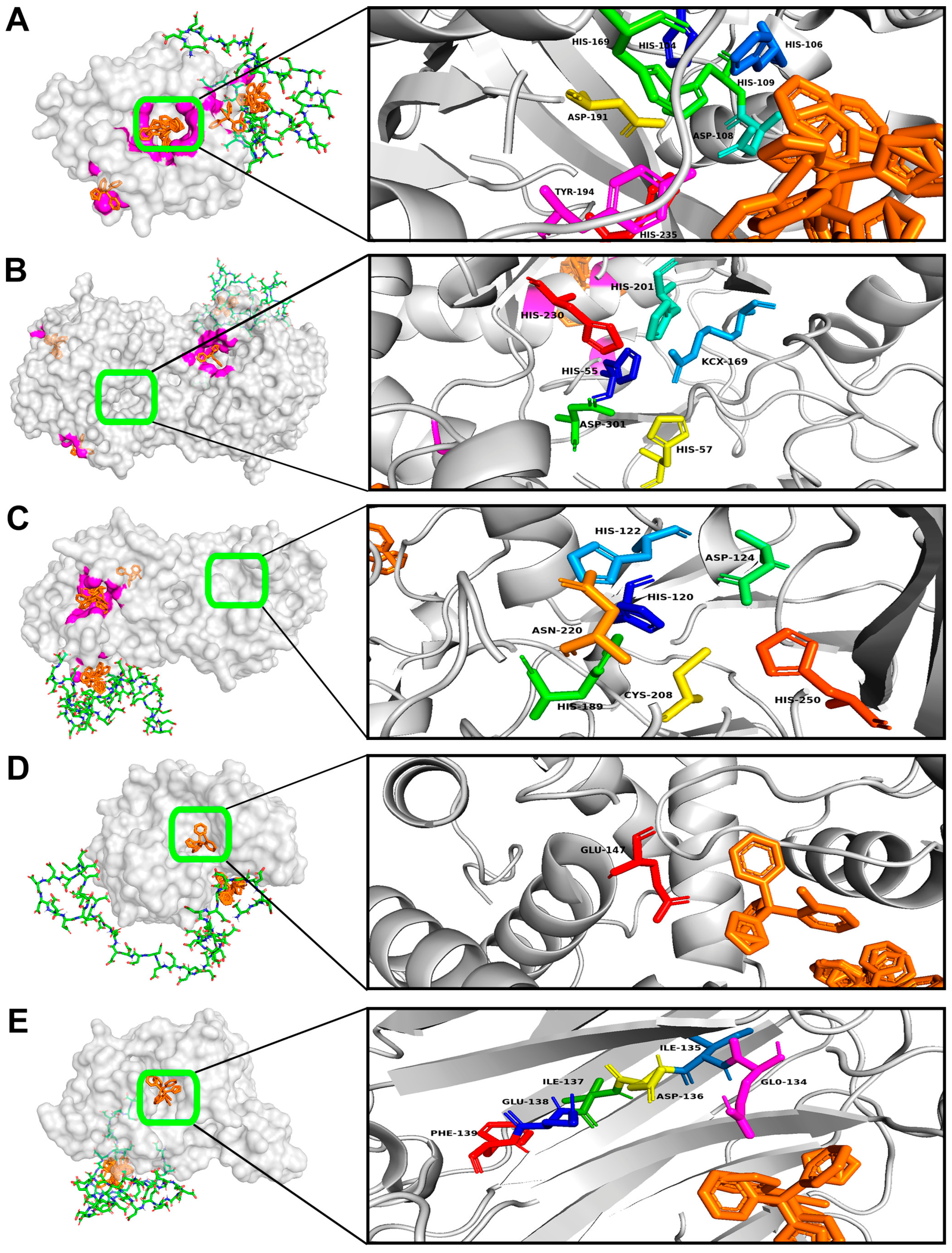
| Enzyme, Origin [Reference] | Object of Action | Mechanism of Action |
|---|---|---|
| β-(1-3)-glucanase Bgy6 from Bacillus halotolerans [12] | Verticillium dahliae | Pronounced inhibition of both spore germination and mycelial growth in fungal cultures. |
| Chitinase ChiC from Streptomyces griseus [13,14] | Trichoderma reesei | Ability to inhibit hyphal extension of T. reesei. |
| Endo-α-1,4-galactosaminidase Ega3 from Aspergillus fumigatus [15] | Aspergillus fumigatus | The enzyme catalyzes the breakdown of the exopolysaccharide galactosaminogalactan, which is a major structural component of A. fumigatus matrix. |
| Lysozyme from hen egg white [16,17] | Biofilm of C. albicans; Aspergillus parasiticus | Biofilm clearing effect was observed. The decrease in fungal cell viability was 100%; an inhibitory effect on the germination of spores was confirmed. |
| Microorganism | Polymyxin B | Colistin | Clotrimazole | |
| no enzyme | ||||
| Aspergillus niger | 23.4 ± 1.4 | 60.6 ± 4.1 | 100 ± 4.1 | |
| Fusarium solani | 1.8 ± 0.2 | 3.4 ± 0.5 | 48.1 ± 1.2 | |
| Trichoderma atroviride | 100 ± 4.3 | 100 ± 5.4 | 2.3 ± 0.3 | |
| Rhizopus oryzae | 100 ± 4.6 | 56.1 ± 4.3 | 22.7 ± 3.1 | |
| Saccharomyces cerevisiae | 84.0 ± 3.2 | 88.5 ± 1.4 | 39.7 ± 1.2 | |
| Candida tropicalis | 93.0 ± 2.8 | 92.2 ± 4.9 | 6.6 ± 1.3 | |
| AiiA | +PLD50 | +PLE50 | ||
| Aspergillus niger | 15.7 ± 2.7 | 25.2 ± 2.3 | 0.15 ± 0.04 | 0.27 ± 0.04 |
| Fusarium solani | 4.1 ± 0.9 | 4 ± 0.2 | 2.3 ± 0.1 | 3.5 ± 0.05 |
| Trichoderma atroviride | 4.0 ± 0.3 | 3.9 ± 1.0 | 0.11 ± 0.03 | 0.13 ± 0.02 |
| Rhizopus oryzae | 0.9 ± 0.1 | 0.8 ± 0.1 | 15.2 ± 1.4 | 14.1 ± 0.12 |
| Saccharomyces cerevisiae | 78.3 ± 3.9 | 82.9 ± 4.1 | 70.1 ± 3.1 | 96.4 ± 2.3 |
| Candida tropicalis | 91.2 ± 2.8 | 91.2 ± 5.7 | 33.8 ± 1.3 | 5.5 ± 1.2 |
| His6-OPH | +PLD50 | +PLE50 | ||
| Aspergillus niger | 5.0 ± 0.8 | 3.4 ± 0.5 | 6.2 ± 0.7 | 10.9 ± 0.8 |
| Fusarium solani | 21.7 ± 1.7 | 15.2 ± 1.6 | 100 ± 2.5 | 100 ± 3.6 |
| Trichoderma atroviride | 0.02 ± 0.001 | 0.05 ± 0.001 | 38.3 ± 1.2 | 100 ± 4.1 |
| Rhizopus oryzae | 0.05 ± 0.001 | 0.6 ± 0.02 | 100 ± 4.5 | 96.5 ± 1.4 |
| Saccharomyces cerevisiae | 70.0 ± 1.4 | 73.8 ± 5.7 | 79.4 ± 2.2 | 100 ± 3.4 |
| Candida tropicalis | 67.9 ± 4.3 | 67.3 ± 1.3 | 28.8 ± 1.3 | 32.3 ± 0.7 |
| NDM-1 | +PLD50 | +PLE50 | ||
| Aspergillus niger | 100 ± 5.5 | 35.7 ± 1.8 | 2.9 ± 0.4 | 0.72 ± 0.08 |
| Fusarium solani | 0.2 ± 0.07 | 16.6 ± 0.7 | 34 ± 1.1 | 11.4 ± 0.4 |
| Trichoderma atroviride | 100 ± 6.5 | 100 ± 4.4 | 1.9 ± 0.09 | 18.9 ± 1.7 |
| Rhizopus oryzae | 0.6 ± 0.02 | 100 ± 3.1 | 0.67 ± 0.03 | 1.5 ± 0.08 |
| Saccharomyces cerevisiae | 100 ± 4.1 | 96.1 ± 4.1 | 100 ± 4.4 | 45.5 ± 2.1 |
| Candida tropicalis | 100 ± 5.9 | 100 ± 5.3 | 100 ± 3.4 | 100 ± 2.8 |
| ChiC | +PLD50 | +PLE50 | ||
| Aspergillus niger | 32.4 ±1.8 | 1.3 ± 0.1 | 100 ± 2.7 | 41.4 ± 3.1 |
| Fusarium solani | 3.4 ± 0.3 | 38.8 ± 1.5 | 100 ± 2.6 | 8.4 ± 0.4 |
| Trichoderma atroviride | 0.6 ± 0.06 | 3.7 ± 0.4 | 10.5 ± 2.5 | 32.8 ± 1.6 |
| Rhizopus oryzae | 3.9 ± 0.5 | 0.2 ± 0.07 | 1.2 ± 0.2 | 67.1 ± 1.2 |
| Saccharomyces cerevisiae | 23.6 ± 0.8 | 1.4 ± 0.03 | 6.6 ± 0.3 | 1.5 ± 0.02 |
| Candida tropicalis | 51.2 ± 1.7 | 2.9 ± 0.02 | 6.7 ± 0.3 | 1.2 ± 0.06 |
| Bgy6-like | +PLD50 | +PLE50 | ||
| Aspergillus niger | 43.8 ± 1.1 | 16.4 ± 2.3 | 0.7 ± 0.1 | 56.7 ± 2.8 |
| Fusarium solani | 4.2 ± 0.2 | 3.9 ± 1.2 | 100 ± 3.3 | 83.1 ± 3.2 |
| Trichoderma atroviride | 100 ± 2.5 | 40.7 ± 2.1 | 74.2 ± 3.7 | 10.6 ± 2.5 |
| Rhizopus oryzae | 0.1 ± 0.01 | 15.9 ± 1.1 | 100 ± 2.7 | 8.8 ± 0.9 |
| Saccharomyces cerevisiae | 100 ± 1.4 | 7.6 ± 0.8 | 5.6 ± 0.2 | 15.8 ± 0.7 |
| Candida tropicalis | 94.2 ± 4.7 | 10.5 ± 0.5 | 10.5 ± 0.5 | 93.9 ± 3.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domnin, M.; Aslanli, A.; Senko, O.; Stepanov, N.; Efremenko, E. Enzymes Degrading Fungal Cell Wall Components vs. Those Exhibiting Lactonase Activity as Participants of Antifungals. Sci 2025, 7, 169. https://doi.org/10.3390/sci7040169
Domnin M, Aslanli A, Senko O, Stepanov N, Efremenko E. Enzymes Degrading Fungal Cell Wall Components vs. Those Exhibiting Lactonase Activity as Participants of Antifungals. Sci. 2025; 7(4):169. https://doi.org/10.3390/sci7040169
Chicago/Turabian StyleDomnin, Maksim, Aysel Aslanli, Olga Senko, Nikolay Stepanov, and Elena Efremenko. 2025. "Enzymes Degrading Fungal Cell Wall Components vs. Those Exhibiting Lactonase Activity as Participants of Antifungals" Sci 7, no. 4: 169. https://doi.org/10.3390/sci7040169
APA StyleDomnin, M., Aslanli, A., Senko, O., Stepanov, N., & Efremenko, E. (2025). Enzymes Degrading Fungal Cell Wall Components vs. Those Exhibiting Lactonase Activity as Participants of Antifungals. Sci, 7(4), 169. https://doi.org/10.3390/sci7040169







