Knowledge Discovery from Bioactive Peptide Data in the PepLab Database Through Quantitative Analysis and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Description and Preprocessing
2.2. Methodology of Descriptive Statistical Analysis
2.3. Structural Modeling of Descriptor Interdependencies Using DEMATEL
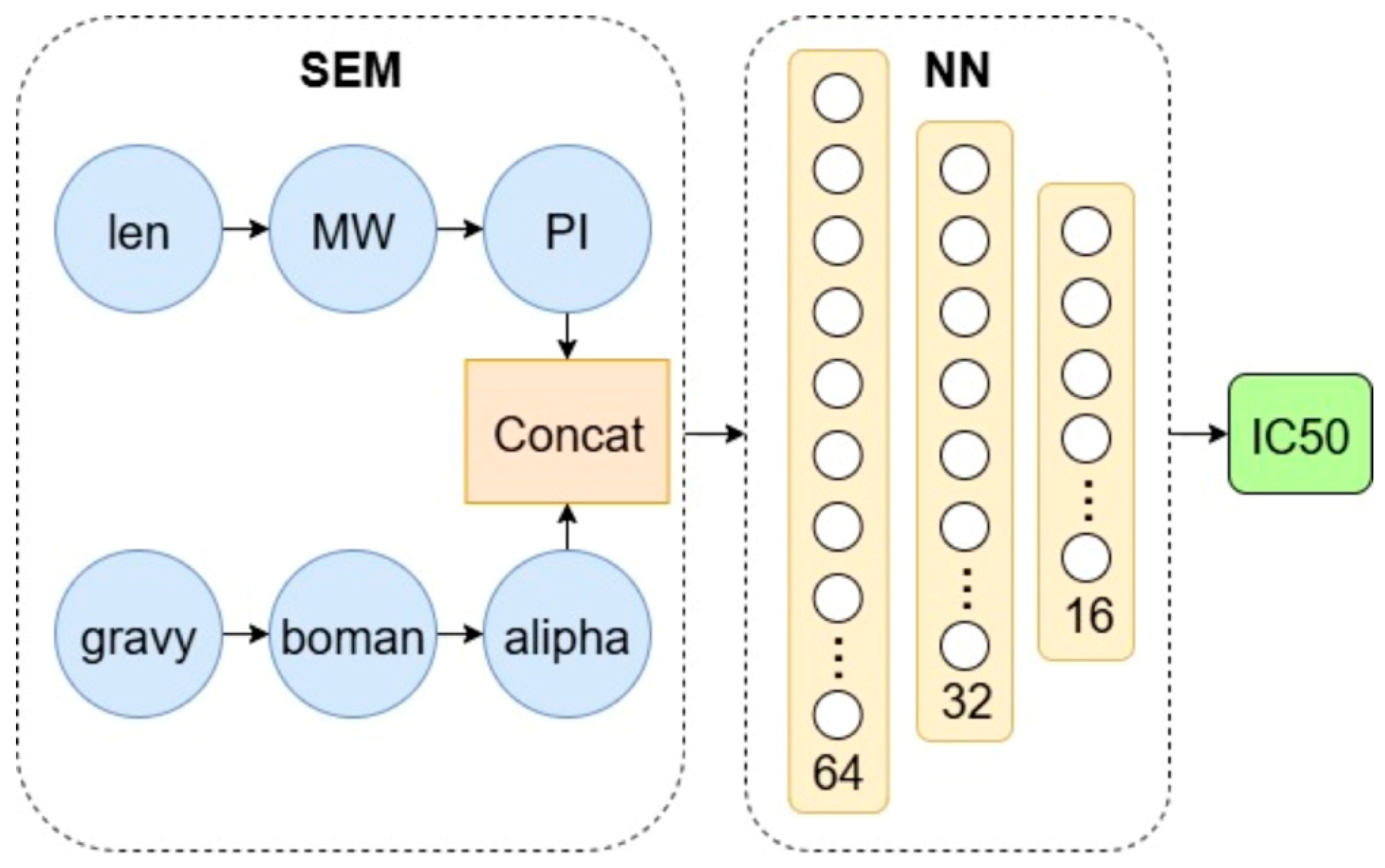
2.4. SEM and SEM-NN Architecture
2.5. Domain-Specific Data Augmentation
2.6. Multiclass Classification Model
- Interpretability is facilitated through the visualization of feature attribution masks;
- The robustness of the system is enhanced through the implementation of flexible loss weighting;
- The efficiency of the system is optimized on small training sets through the utilization of sparsity and adaptive attention mechanisms;
- The system can automatically select relevant descriptors, thus negating the necessity for prior reduction or normalization.
2.7. Computational Resources and Methods
3. Results and Discussion
3.1. Descriptive Statistical Analysis
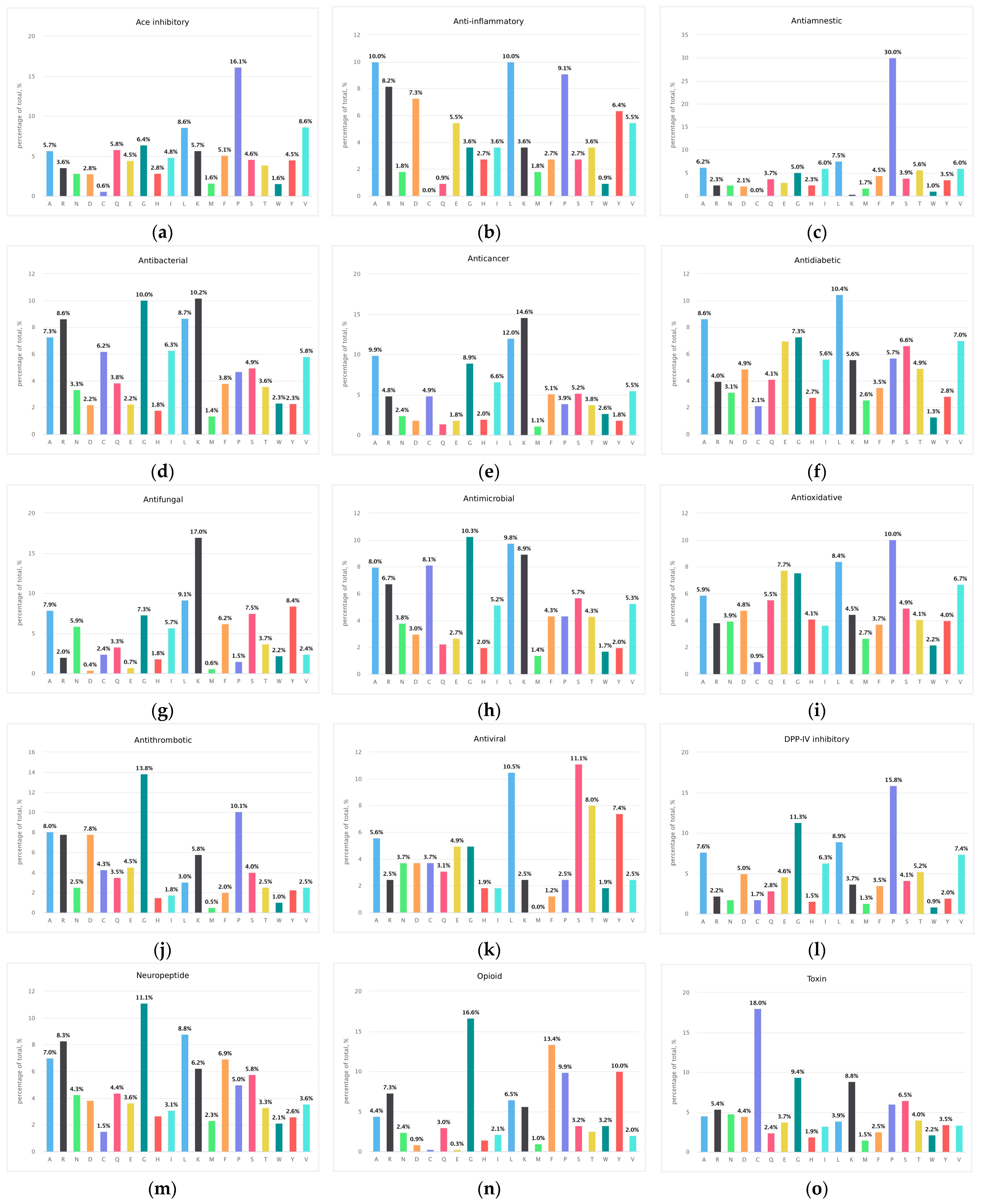
3.1.1. ACE Inhibitory Peptides
3.1.2. Anti-Inflammatory Peptides
3.1.3. Antiamnestic Peptides
3.1.4. Antibacterial Peptides
3.1.5. Anticancer Peptides
3.1.6. Antidiabetic Peptides
3.1.7. Antifungal Peptides
3.1.8. Antimicrobial Peptides
3.1.9. Antioxidative Peptides
3.1.10. Antithrombotic Peptides
3.1.11. Antiviral Peptides
3.1.12. DPP-IV Inhibitor Peptides
3.1.13. Neuropeptides
3.1.14. Opioid Peptides
3.1.15. Toxin Peptides
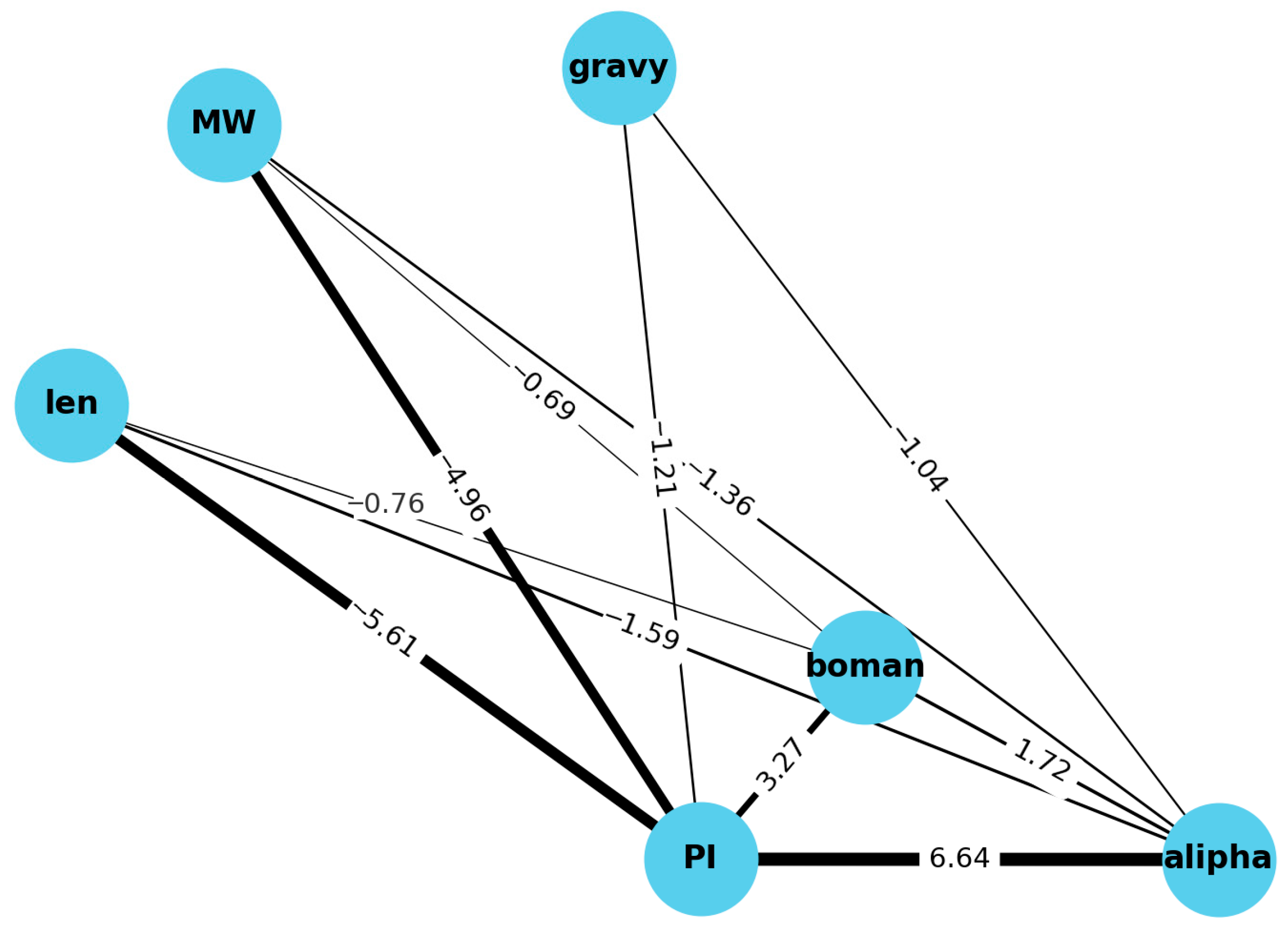
3.2. DEMATEL Analysis
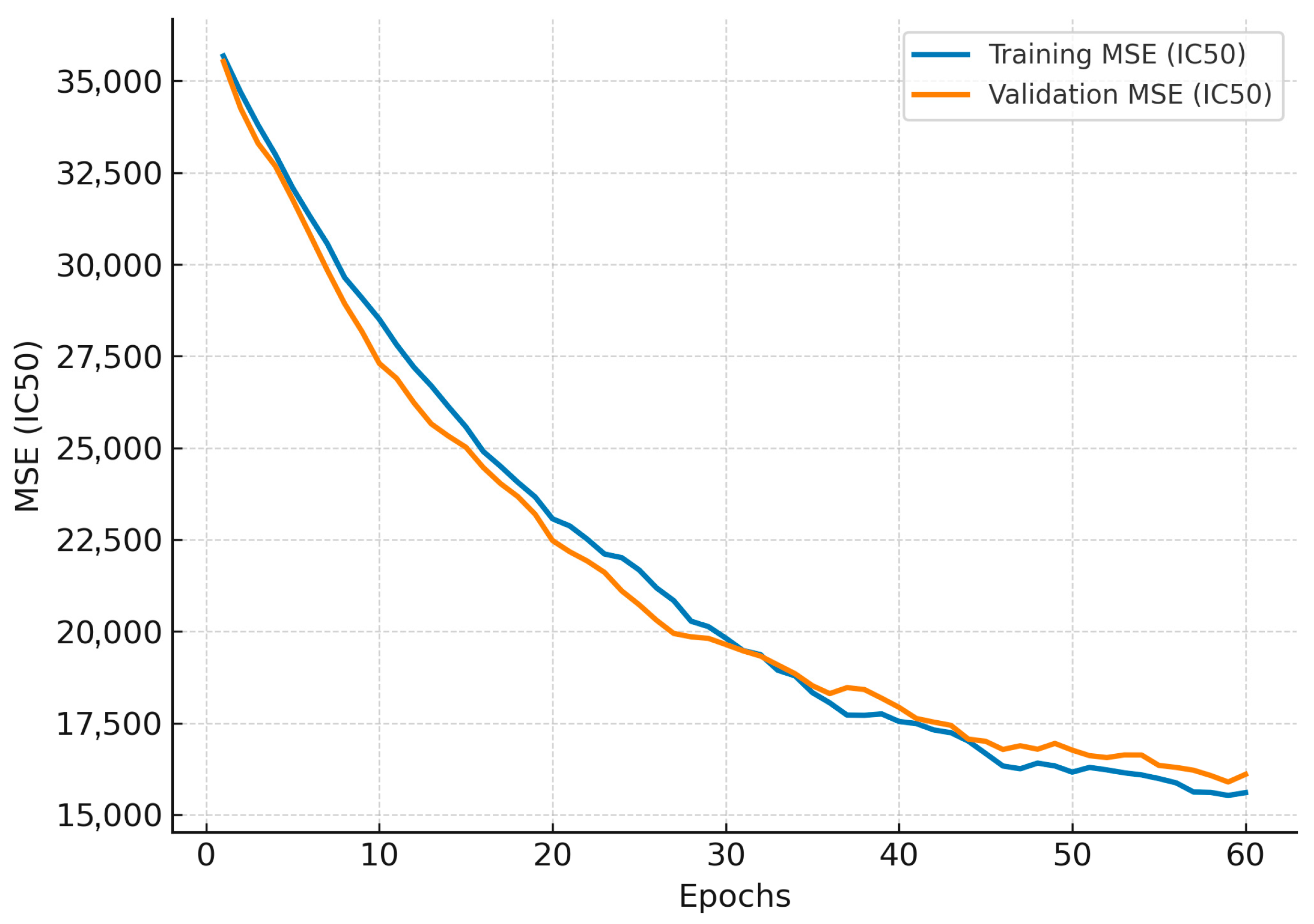
3.3. Structural Equation Modeling (SEM) and Its Neural Extension (SEM-NN)
3.4. AI-Based Multiclass Classification of Peptide Bioactivity
| Algorithm 1. Pseudo code of the augmentation process. |
| def augmentation (D, T, CI): |
| FOR each class c in D: |
| Determine deficit = max(T,|c|) − |c| |
| IF deficit NOT NULL |
| FOR i = 1 to deficit: |
| Generate new sequence |
| Compute descriptors of sequence |
| IF descriptors within CI bounds of c THEN |
| Add sequence to class c as synthetic |
| ENDIF |
| ENDFOR |
| ENDIF |
| ENDFOR |
| Return augmented dataset D* |
4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAC | Amino Acid Composition |
| ACE | Angiotensin-Converting Enzyme |
| AMP | Antimicrobial Peptide |
| BAP | Bioactive Peptide |
| D | Outward Influence (DEMATEL) |
| D+R | Centrality (Sum of Outward and Inward Influence) |
| D–R | Causality (Difference between Outward and Inward Influence) |
| Da | Dalton (Molecular Weight Unit) |
| DEMATEL | Decision Making Trial and Evaluation Laboratory |
| DPP-IV | Dipeptidyl Peptidase IV |
| GRAVY | Grand Average of Hydropathy |
| kDa | Kilodalton |
| MLP | Multi-Layer Perceptron |
| MW | Molecular Weight |
| PCA | Principal Component Analysis |
| pI | Isoelectric Point |
| QSAR | Quantitative Structure–Activity Relationship |
| R | Inward Influence (DEMATEL) |
| SEM | Structural Equation Modeling |
| SEM-NN | Structural Equation Modeling with Neural Networks |
| SHAP | SHapley Additive exPlanations |
| TabNet | Attentive Interpretable Tabular Learning |
Appendix A
| Molecular Weight (DA) | ||||||||||
| Interval | 468.51 to 1008.61 | 1008.61 to 1548.7 | 1548.7 to 2088.8 | 2088.8 to 2628.89 | 2628.89 to 3168.99 | 3168.99 to 3709.08 | 3709.08 to 4249.18 | 4249.18 to 4789.27 | 4789.27 to 5329.37 | 5329.37 to 5869.46 |
| Data (%) | 64.44 | 22.94 | 9.37 | 0.96 | 1.53 | 0.57 | 0.00 | 0.00 | 0.00 | 0.19 |
| Isoelectric Point (pH) | ||||||||||
| Interval | 2.63 to 3.73 | 3.73 to 4.82 | 4.82 to 5.92 | 5.92 to 7.02 | 7.02 to 8.12 | 8.12 to 9.21 | 9.21 to 10.31 | 10.31 to 11.41 | 11.41 to 12.5 | 12.5 to 13.6 |
| Data (%) | 11.85 | 4.97 | 17.02 | 19.31 | 11.66 | 2.49 | 18.74 | 7.65 | 4.97 | 1.34 |
| GRAVY | ||||||||||
| Interval | −3.9 to −3.26 | −3.26 to −2.62 | −2.62 to −1.98 | −1.98 to −1.33 | −1.33 to −0.69 | −0.69 to −0.05 | −0.05 to 0.59 | 0.59 to 1.23 | 1.23 to 1.87 | 1.87 to 2.51 |
| Data (%) | 0.76 | 0.76 | 2.68 | 6.50 | 16.63 | 28.49 | 26.77 | 11.85 | 3.44 | 2.10 |
| Aliphatic index | ||||||||||
| Interval | 0 to 29.29 | 29.29 to 58.57 | 58.57 to 87.86 | 87.86 to 117.14 | 117.14 to 146.43 | 146.43 to 175.72 | 175.72 to 205 | 205 to 234.29 | 234.29 to 263.57 | 263.57 to 292.86 |
| Data (%) | 11.28 | 19.69 | 24.86 | 23.14 | 11.09 | 4.97 | 3.82 | 0.57 | 0.38 | 0.19 |
| Boman index | ||||||||||
| Interval | −3.11 to −1.73 | −1.73 to −0.36 | −0.36 to 1.01 | 1.01 to 2.38 | 2.38 to 3.76 | 3.76 to 5.13 | 5.13 to 6.5 | 6.5 to 7.88 | 7.88 to 9.25 | 9.25 to 10.62 |
| Data (%) | 4.59 | 19.31 | 36.52 | 23.52 | 10.33 | 3.82 | 0.96 | 0.19 | 0.57 | 0.19 |
| Molecular Weight (DA) | |||||
| Interval | 581.71 to 803.56 | 803.56 to 1025.41 | 1025.41 to 1247.27 | 1247.27 to 1469.12 | 1469.12 to 1690.97 |
| Data (%) | 42.86 | 28.57 | 7.14 | 7.14 | 14.29 |
| Isoelectric Point (pH) | |||||
| Interval | 3.08 to 4.76 | 4.76 to 6.45 | 6.45 to 8.13 | 8.13 to 9.82 | 9.82 to 11.5 |
| Data (%) | 35.71 | 21.43 | 0.00 | 35.71 | 7.14 |
| GRAVY | |||||
| Interval | −2.78 to −1.95 | −1.95 to −1.11 | −1.11 to −0.28 | −0.28 to 0.55 | 0.55 to 1.39 |
| Data (%) | 14.29 | 14.29 | 28.57 | 21.43 | 21.43 |
| Aliphatic index | |||||
| Interval | 0 to 39 | 39 to 78 | 78 to 117 | 117 to 156 | 156 to 195 |
| Data (%) | 14.29 | 28.57 | 21.43 | 28.57 | 7.14 |
| Boman index | |||||
| Interval | −1.51 to 0.22 | 0.22 to 1.95 | 1.95 to 3.69 | 3.69 to 5.42 | 5.42 to 7.15 |
| Data (%) | 35.71 | 14.29 | 35.71 | 0.00 | 14.29 |
| Molecular Weight (DA) | ||||||
| Interval | 519.64 to 818.63 | 818.63 to 1117.61 | 1117.61 to 1416.6 | 1416.6 to 1715.59 | 1715.59 to 2014.57 | 2014.57 to 2313.56 |
| Data (%) | 21.2766 | 42.5532 | 8.5106 | 2.1277 | 17.0213 | 8.5106 |
| Isoelectric Point (pH) | ||||||
| Interval | 2.76 to 4.28 | 4.28 to 5.81 | 5.81 to 7.33 | 7.33 to 8.85 | 8.85 to 10.38 | 10.38 to 11.9 |
| Data (%) | 21.2766 | 17.0213 | 36.1702 | 17.0213 | 0.0000 | 8.5106 |
| GRAVY | ||||||
| Interval | −2.5 to −1.83 | −1.83 to −1.16 | −1.16 to −0.49 | −0.49 to 0.18 | 0.18 to 0.84 | 0.84 to 1.51 |
| Data (%) | 2.1277 | 2.1277 | 17.0213 | 31.9149 | 42.5532 | 4.2553 |
| Aliphatic index | ||||||
| Interval | 0 to 23.52 | 23.52 to 47.04 | 47.04 to 70.55 | 70.56 to 94.07 | 94.07 to 117.59 | 117.59 to 141.11 |
| Data (%) | 6.3830 | 6.3830 | 17.0213 | 38.2979 | 17.0213 | 14.8936 |
| Boman index | ||||||
| Interval | −1.79 to −0.96 | −0.96 to −0.12 | −0.12 to 0.71 | 0.71 to 1.54 | 1.54 to 2.38 | 2.38 to 3.21 |
| Data (%) | 31.9149 | 19.1489 | 21.2766 | 14.8936 | 8.5106 | 4.2553 |
| Molecular Weight (DA) | |||||||||
| Interval | 514.63 to 1149.25 | 1149.25 to 1783.88 | 1783.88 to 2418.5 | 2418.5 to 3053.13 | 3053.13 to 3687.75 | 3687.75 to 4322.38 | 4322.38 to 4957 | 4957 to 5591.62 | 5591.62 to 6226.25 |
| Data (%) | 9.6591 | 17.3295 | 14.4886 | 12.7841 | 11.3636 | 19.8864 | 10.2273 | 3.6932 | 0.5682 |
| Isoelectric Point (pH) | |||||||||
| Interval | 2.74 to 4.01 | 4.01 to 5.28 | 5.28 to 6.54 | 6.54 to 7.81 | 7.81 to 9.08 | 9.08 to 10.35 | 10.35 to 11.61 | 11.61 to 12.88 | 12.88 to 14.15 |
| Data (%) | 4.2614 | 2.2727 | 4.2614 | 5.9659 | 9.6591 | 25.5682 | 25.2841 | 11.3636 | 11.3636 |
| GRAVY | |||||||||
| Interval | −3.59 to −3 | −3 to −2.42 | −2.42 to −1.83 | −1.83 to −1.25 | −1.25 to −0.66 | −0.66 to −0.08 | −0.08 to 0.51 | 0.51 to 1.1 | 1.1 to 1.68 |
| Data (%) | 1.1364 | 0.8523 | 2.5568 | 9.6591 | 13.9205 | 30.1136 | 21.3068 | 11.0795 | 9.3750 |
| Aliphatic index | |||||||||
| Interval | 0 to 25.28 | 25.28 to 50.56 | 50.56 to 75.83 | 75.83 to 101.11 | 101.11 to 126.39 | 126.39 to 151.67 | 151.67 to 176.94 | 176.94 to 202.22 | 202.22 to 227.5 |
| Data (%) | 10.2273 | 18.1818 | 16.7614 | 18.1818 | 17.8977 | 11.3636 | 3.6932 | 2.2727 | 1.4205 |
| Boman index | |||||||||
| Interval | −2.48 to −1.45 | −1.45 to −0.42 | −0.42 to 0.61 | 0.61 to 1.64 | 1.64 to 2.67 | 2.67 to 3.7 | 3.7 to 4.73 | 4.73 to 5.77 | 5.77 to 6.8 |
| Data (%) | 3.1250 | 10.7955 | 16.4773 | 20.4545 | 22.7273 | 17.3295 | 5.9659 | 2.8409 | 0.2841 |
| Molecular Weight (DA) | |||||||||
| Interval | 573.67 to 1097.51 | 1097.51 to 1621.34 | 1621.34 to 2145.18 | 2145.18 to 2669.01 | 2669.01 to 3192.85 | 3192.85 to 3716.68 | 3716.68 to 4240.52 | 4240.52 to 4764.35 | 4764.35 to 5288.19 |
| Data (%) | 5.3004 | 21.5548 | 27.2085 | 18.0212 | 16.6078 | 6.3604 | 3.1802 | 0.7067 | 1.0601 |
| Isoelectric Point (pH) | |||||||||
| Interval | 2.94 to 4.17 | 4.17 to 5.39 | 5.39 to 6.62 | 6.62 to 7.85 | 7.85 to 9.07 | 9.07 to 10.3 | 10.3 to 11.53 | 11.53 to 12.75 | 12.75 to 13.98 |
| Data (%) | 3.8869 | 7.4205 | 4.5936 | 9.5406 | 7.0671 | 16.6078 | 33.9223 | 6.7138 | 10.2473 |
| GRAVY | |||||||||
| Interval | −2.74 to −2.22 | −2.22 to −1.7 | −1.7 to −1.18 | −1.18 to −0.66 | −0.66 to −0.14 | −0.14 to 0.39 | 0.39 to 0.91 | 0.91 to 1.43 | 1.43 to 1.95 |
| Data (%) | 0.7067 | 1.0601 | 3.5336 | 10.2473 | 19.4346 | 23.3216 | 25.4417 | 12.7208 | 3.5336 |
| Aliphatic index | |||||||||
| Interval | 0 to 25.49 | 25.49 to 50.98 | 50.98 to 76.47 | 76.47 to 101.96 | 101.96 to 127.45 | 127.45 to 152.94 | 152.94 to 178.43 | 178.43 to 203.92 | 203.92 to 229.41 |
| Data (%) | 7.0671 | 12.0141 | 13.7809 | 15.5477 | 15.9011 | 17.3145 | 14.1343 | 3.1802 | 1.0601 |
| Boman index | |||||||||
| Interval | −2.64 to −1.56 | −1.56 to −0.47 | −0.47 to 0.61 | 0.61 to 1.7 | 1.7 to 2.79 | 2.79 to 3.87 | 3.87 to 4.96 | 4.96 to 6.04 | 6.04 to 7.13 |
| Data (%) | 3.8869 | 20.4947 | 30.3887 | 23.3216 | 14.4876 | 3.8869 | 1.7668 | 1.0601 | 0.7067 |
| Molecular Weight (DA) | ||||||||
| Interval | 1028.09 to 1491.66 | 1491.66 to 1955.24 | 1955.24 to 2418.81 | 2418.81 to 2882.38 | 2882.38 to 3345.95 | 3345.95 to 3809.53 | 3809.53 to 4273.1 | 4273.1 to 4736.67 |
| Data (%) | 11.6379 | 19.3966 | 42.6724 | 12.5000 | 11.6379 | 0.0000 | 1.2931 | 0.8621 |
| Isoelectric Point (pH) | ||||||||
| Interval | 2.73 to 4.1 | 4.1 to 5.47 | 5.47 to 6.84 | 6.84 to 8.21 | 8.21 to 9.57 | 9.57 to 10.94 | 10.94 to 12.31 | 12.31 to 13.68 |
| Data (%) | 20.6897 | 25.4310 | 11.6379 | 14.2241 | 7.7586 | 16.3793 | 3.4483 | 0.4310 |
| GRAVY | ||||||||
| Interval | −2.15 to −1.64 | −1.64 to −1.13 | −1.13 to −0.62 | −0.62 to −0.11 | −0.11 to 0.4 | 0.4 to 0.9 | 0.9 to 1.41 | 1.41 to 1.92 |
| Data (%) | 2.1552 | 6.8966 | 16.3793 | 27.1552 | 24.5690 | 12.9310 | 7.7586 | 2.1552 |
| Aliphatic index | ||||||||
| Interval | 0 to 25.29 | 25.29 to 50.58 | 50.58 to 75.87 | 75.87 to 101.15 | 101.15 to 126.44 | 126.44 to 151.73 | 151.73 to 177.02 | 177.02 to 202.31 |
| Data (%) | 3.4483 | 10.7759 | 20.6897 | 26.7241 | 22.4138 | 11.6379 | 3.4483 | 0.8621 |
| Boman index | ||||||||
| Interval | −1.54 to −0.73 | −0.73 to 0.08 | 0.08 to 0.89 | 0.89 to 1.71 | 1.71 to 2.52 | 2.52 to 3.33 | 3.33 to 4.14 | 4.14 to 4.95 |
| Data (%) | 5.1724 | 14.2241 | 22.8448 | 22.4138 | 21.5517 | 6.4655 | 5.6034 | 1.7241 |
| Molecular Weight (DA) | ||||||
| Interval | 653.69 to 1070.02 | 1070.02 to 1486.36 | 1486.36 to 1902.69 | 1902.69 to 2319.02 | 2319.02 to 2735.36 | 2735.36 to 3151.69 |
| Data (%) | 9.7561 | 26.8293 | 56.0976 | 0.0000 | 0.0000 | 7.3171 |
| Isoelectric Point (pH) | ||||||
| Interval | 3.29 to 4.97 | 4.97 to 6.65 | 6.65 to 8.34 | 8.34 to 10.02 | 10.02 to 11.7 | 11.7 to 13.38 |
| Data (%) | 2.4390 | 0.0000 | 9.7561 | 41.4634 | 43.9024 | 2.4390 |
| GRAVY | ||||||
| Interval | −1.59 to −1.18 | −1.18 to −0.76 | −0.76 to −0.35 | −0.35 to 0.06 | 0.06 to 0.48 | 0.48 to 0.89 |
| Data (%) | 7.3171 | 14.6341 | 41.4634 | 9.7561 | 14.6341 | 12.1951 |
| Aliphatic index | ||||||
| Interval | 0 to 31.11 | 31.11 to 62.22 | 62.22 to 93.34 | 93.34 to 124.45 | 124.45 to 155.56 | 155.56 to 186.67 |
| Data (%) | 12.1951 | 41.4634 | 12.1951 | 0.0000 | 26.8293 | 7.3171 |
| Boman index | ||||||
| Interval | −0.89 to 0.09 | 0.09 to 1.07 | 1.07 to 2.04 | 2.04 to 3.02 | 3.02 to 4 | 4 to 4.98 |
| Data (%) | 24.3902 | 17.0732 | 43.9024 | 7.3171 | 2.4390 | 4.8780 |
| Molecular Weight (DA) | |||||||||
| Interval | 658.8 to 1249.26 | 1249.26 to 1839.72 | 1839.72 to 2430.18 | 2430.18 to 3020.64 | 3020.64 to 3611.09 | 3611.09 to 4201.55 | 4201.55 to 4792.01 | 4792.01 to 5382.47 | 5382.47 to 5972.93 |
| Data (%) | 4.2857 | 14.8980 | 21.8367 | 17.3469 | 10.8163 | 10.2041 | 12.4490 | 5.7143 | 2.4490 |
| Isoelectric Point (pH) | |||||||||
| Interval | 2.27 to 3.63 | 3.63 to 4.98 | 4.98 to 6.34 | 6.34 to 7.7 | 7.7 to 9.05 | 9.05 to 10.41 | 10.41 to 11.77 | 11.77 to 13.12 | 13.12 to 14.48 |
| Data (%) | 3.0612 | 5.5102 | 3.4694 | 5.7143 | 20.2041 | 28.9796 | 19.5918 | 6.1224 | 7.3469 |
| GRAVY | |||||||||
| Interval | −2.88 to −2.25 | −2.25 to −1.62 | −1.61 to −0.98 | −0.98 to −0.35 | −0.35 to 0.28 | 0.28 to 0.92 | 0.92 to 1.55 | 1.55 to 2.18 | 2.18 to 2.81 |
| Data (%) | 0.8163 | 2.2449 | 4.8980 | 25.9184 | 27.5510 | 23.4694 | 12.2449 | 2.4490 | 0.4082 |
| Aliphatic index | |||||||||
| Interval | 0 to 24.65 | 24.65 to 49.29 | 49.29 to 73.94 | 73.94 to 98.59 | 98.59 to 123.23 | 123.23 to 147.88 | 147.88 to 172.53 | 172.53 to 197.17 | 197.17 to 221.82 |
| Data (%) | 6.3265 | 14.8980 | 21.6327 | 18.9796 | 16.9388 | 12.4490 | 5.3061 | 2.6531 | 0.8163 |
| Boman index | |||||||||
| Interval | −3.58 to −2.09 | −2.09 to −0.6 | −0.6 to 0.89 | 0.89 to 2.37 | 2.38 to 3.86 | 3.86 to 5.35 | 5.35 to 6.84 | 6.84 to 8.33 | 8.33 to 9.81 |
| Data (%) | 1.8367 | 14.0816 | 34.8980 | 28.9796 | 15.5102 | 2.6531 | 1.6327 | 0.2041 | 0.2041 |
| Molecular Weight (DA) | |||||||||
| Interval | 389.41 to 671.04 | 671.04 to 952.67 | 952.67 to 1234.29 | 1234.29 to 1515.92 | 1515.92 to 1797.55 | 1797.55 to 2079.18 | 2079.18 to 2360.8 | 2360.8 to 2642.43 | 2642.43 to 2924.06 |
| Data (%) | 29.3893 | 32.0611 | 11.4504 | 11.4504 | 11.4504 | 3.0534 | 0.3817 | 0.3817 | 0.3817 |
| Isoelectric Point (pH) | |||||||||
| Interval | 2.71 to 3.84 | 3.84 to 4.97 | 4.97 to 6.11 | 6.11 to 7.24 | 7.24 to 8.37 | 8.37 to 9.5 | 9.5 to 10.64 | 10.64 to 11.77 | 11.77 to 12.9 |
| Data (%) | 21.3740 | 9.5420 | 24.0458 | 10.3053 | 10.6870 | 3.0534 | 12.2137 | 7.6336 | 1.1450 |
| GRAVY | |||||||||
| Interval | −3.38 to −2.63 | −2.63 to −1.88 | −1.88 to −1.12 | −1.12 to −0.37 | −0.37 to 0.38 | 0.38 to 1.13 | 1.13 to 1.88 | 1.88 to 2.63 | 2.63 to 3.38 |
| Data (%) | 1.9084 | 7.6336 | 15.6489 | 27.4809 | 26.3359 | 14.8855 | 4.1985 | 1.5267 | 0.3817 |
| Aliphatic index | |||||||||
| Interval | 0 to 27.04 | 27.04 to 54.07 | 54.07 to 81.11 | 81.11 to 108.15 | 108.15 to 135.18 | 135.18 to 162.22 | 162.22 to 189.26 | 189.26 to 216.29 | 216.29 to 243.33 |
| Data (%) | 17.5573 | 14.1221 | 27.8626 | 16.7939 | 14.8855 | 6.4885 | 0.7634 | 1.1450 | 0.3817 |
| Boman index | |||||||||
| Interval | −3.5 to −2.39 | −2.39 to −1.28 | −1.28 to −0.17 | −0.17 to 0.94 | 0.94 to 2.06 | 2.06 to 3.17 | 3.17 to 4.28 | 4.28 to 5.39 | 5.39 to 6.5 |
| Data (%) | 1.9084 | 5.7252 | 18.7023 | 15.2672 | 27.0992 | 14.5038 | 10.3053 | 3.4351 | 3.0534 |
| Molecular Weight (DA) | ||||||
| Interval | 482.54 to 1306.31 | 1306.31 to 2130.07 | 2130.07 to 2953.84 | 2953.84 to 3777.61 | 3777.61 to 4601.37 | 4601.37 to 5425.14 |
| Data (%) | 77.1429 | 14.2857 | 0.0000 | 2.8571 | 0.0000 | 5.7143 |
| Isoelectric Point (pH) | ||||||
| Interval | 2.77 to 4.59 | 4.59 to 6.41 | 6.41 to 8.22 | 8.23 to 10.04 | 10.04 to 11.86 | 11.86 to 13.68 |
| Data (%) | 22.8571 | 2.8571 | 34.2857 | 14.2857 | 22.8571 | 2.8571 |
| GRAVY | ||||||
| Interval | −3.66 to −3.02 | −3.02 to −2.37 | −2.37 to −1.73 | −1.73 to −1.08 | −1.08 to −0.44 | −0.44 to 0.21 |
| Data (%) | 8.5714 | 0.0000 | 14.2857 | 34.2857 | 22.8571 | 20 |
| Aliphatic index | ||||||
| Interval | 0 to 14.67 | 14.67 to 29.33 | 29.33 to 44 | 44 to 58.67 | 58.67 to 73.33 | 73.33 to 88 |
| Data (%) | 40.0000 | 17.1429 | 5.7143 | 14.2857 | 8.5714 | 14.2857 |
| Boman index | ||||||
| Interval | −0.59 to 0.88 | 0.88 to 2.34 | 2.34 to 3.8 | 3.81 to 5.27 | 5.27 to 6.73 | 6.73 to 8.2 |
| Data (%) | 8.5714 | 40.0000 | 31.4286 | 11.4286 | 2.8571 | 5.7143 |
| Molecular Weight (DA) | |||||
| Interval | 623.71 to 1128.72 | 1128.72 to 1633.73 | 1633.73 to 2138.74 | 2138.74 to 2643.75 | 2643.75 to 3148.76 |
| Data (%) | 52.9412 | 11.7647 | 23.5294 | 0.0000 | 11.7647 |
| Isoelectric Point (pH) | |||||
| Interval | 3.29 to 4.65 | 4.65 to 6.01 | 6.01 to 7.38 | 7.38 to 8.74 | 8.74 to 10.1 |
| Data (%) | 47.0588 | 5.8824 | 11.7647 | 11.7647 | 23.5294 |
| GRAVY | |||||
| Interval | −1.34 to −0.86 | −0.86 to −0.37 | −0.37 to 0.11 | 0.11 to 0.6 | 0.6 to 1.08 |
| Data (%) | 23.5294 | 35.2941 | 11.7647 | 17.6471 | 11.7647 |
| Aliphatic index | |||||
| Interval | 0 to 39 | 39 to 78 | 78 to 117 | 117 to 156 | 156 to 195 |
| Data (%) | 35.2941 | 23.5294 | 11.7647 | 17.6471 | 11.7647 |
| Boman index | |||||
| Interval | −0.73 to 0.18 | 0.18 to 1.1 | 1.1 to 2.02 | 2.02 to 2.93 | 2.93 to 3.85 |
| Data (%) | 23.5294 | 11.7647 | 29.4118 | 23.5294 | 11.7647 |
| Molecular Weight (DA) | ||||||
| Interval | 359.38 to 613.86 | 613.86 to 868.34 | 868.34 to 1122.81 | 1122.82 to 1377.29 | 1377.29 to 1631.77 | 1631.77 to 1886.25 |
| Data (%) | 14.5455 | 43.6364 | 16.3636 | 18.1818 | 3.6364 | 3.6364 |
| Isoelectric Point (pH) | ||||||
| Interval | 2.89 to 4.37 | 4.37 to 5.86 | 5.86 to 7.34 | 7.34 to 8.82 | 8.82 to 10.31 | 10.31 to 11.79 |
| Data (%) | 40.0000 | 16.3636 | 23.6364 | 0.0000 | 12.7273 | 7.2727 |
| GRAVY | ||||||
| Interval | −2.19 to −1.44 | −1.43 to −0.68 | −0.68 to 0.08 | 0.08 to 0.83 | 0.83 to 1.59 | 1.59 to 2.34 |
| Data (%) | 5.4545 | 12.7273 | 30.9091 | 29.0909 | 9.0909 | 12.7273 |
| Aliphatic index | ||||||
| Interval | 0 to 43.33 | 43.33 to 86.67 | 86.67 to 130 | 130 to 173.33 | 173.33 to 216.67 | 216.67 to 260 |
| Data (%) | 16.3636 | 34.5455 | 27.2727 | 14.5455 | 3.6364 | 3.6364 |
| Boman index | ||||||
| Interval | −2.95 to −1.74 | −1.74 to −0.52 | −0.52 to 0.69 | 0.69 to 1.91 | 1.91 to 3.12 | 3.12 to 4.34 |
| Data (%) | 10.9091 | 21.8182 | 38.1818 | 14.5455 | 5.4545 | 9.0909 |
| Molecular Weight (DA) | |||||||
| Interval | 573.67 to 1304.24 | 1304.24 to 2034.81 | 2034.81 to 2765.38 | 2765.38 to 3495.95 | 3495.95 to 4226.52 | 4226.52 to 4957.09 | 4957.09 to 5687.66 |
| Data (%) | 46.4789 | 30.9859 | 4.2254 | 7.0423 | 4.2254 | 2.8169 | 4.2254 |
| Isoelectric Point (pH) | |||||||
| Interval | 3.07 to 4.56 | 4.56 to 6.05 | 6.05 to 7.54 | 7.54 to 9.03 | 9.03 to 10.52 | 10.52 to 12.01 | 12.01 to 13.5 |
| Data (%) | 7.0423 | 9.8592 | 18.3099 | 9.8592 | 18.3099 | 22.5352 | 14.0845 |
| GRAVY | |||||||
| Interval | −3.19 to −2.48 | −2.48 to −1.78 | −1.78 to −1.07 | −1.07 to −0.37 | −0.37 to 0.34 | 0.34 to 1.04 | 1.05 to 1.75 |
| Data (%) | 2.8169 | 2.8169 | 11.2676 | 35.2113 | 33.8028 | 11.2676 | 2.8169 |
| Aliphatic index | |||||||
| Interval | 0 to 21.84 | 21.84 to 43.67 | 43.67 to 65.51 | 65.51 to 87.35 | 87.35 to 109.19 | 109.19 to 131.02 | 131.02 to 152.86 |
| Data (%) | 14.0845 | 16.9014 | 21.1268 | 23.9437 | 15.4930 | 5.6338 | 2.8169 |
| Boman index | |||||||
| Interval | −2.52 to −1.26 | −1.26 to −0.01 | −0.01 to 1.25 | 1.25 to 2.51 | 2.51 to 3.76 | 3.76 to 5.02 | 5.02 to 6.28 |
| Data (%) | 8.4507 | 8.4507 | 18.3099 | 32.3944 | 23.9437 | 7.0423 | 1.4085 |
| Molecular Weight (DA) | |||||||
| Interval | 549.63 to 966.12 | 966.12 to 1382.6 | 1382.6 to 1799.09 | 1799.09 to 2215.57 | 2215.57 to 2632.06 | 2632.06 to 3048.54 | 3048.54 to 3465.03 |
| Data (%) | 67.50 | 11.25 | 5.00 | 15.00 | 0.00 | 0.00 | 1.25 |
| Isoelectric Point (pH) | |||||||
| Interval | 3.1 to 4.54 | 4.54 to 5.99 | 5.99 to 7.43 | 7.43 to 8.87 | 8.87 to 10.31 | 10.31 to 11.76 | 11.76 to 13.2 |
| Data (%) | 3.75 | 42.5 | 1.25 | 3.75 | 21.25 | 10 | 17.5 |
| GRAVY | |||||||
| Interval | −2.11 to −1.58 | −1.58 to −1.04 | −1.04 to −0.5 | −0.5 to 0.04 | 0.04 to 0.58 | 0.58 to 1.12 | 1.12 to 1.66 |
| Data (%) | 8.75 | 10 | 16.25 | 25 | 23.75 | 15 | 1.25 |
| Aliphatic index | |||||||
| Interval | 0 to 22.29 | 22.29 to 44.57 | 44.57 to 66.86 | 66.86 to 89.14 | 89.14 to 111.43 | 111.43 to 133.71 | 133.71 to 156 |
| Data (%) | 50 | 5 | 16.25 | 12.5 | 8.75 | 3.75 | 3.75 |
| Boman index | |||||||
| Interval | −2.18 to −1.35 | −1.35 to −0.51 | −0.51 to 0.32 | 0.32 to 1.16 | 1.16 to 2 | 2 to 2.83 | 2.83 to 3.67 |
| Data (%) | 15 | 22.5 | 17.5 | 8.75 | 16.25 | 7.5 | 12.5 |
| Molecular Weight (DA) | |||||||||
| Interval | 716.9 to 1269.06 | 1269.06 to 1821.21 | 1821.21 to 2373.37 | 2373.37 to 2925.52 | 2925.52 to 3477.68 | 3477.68 to 4029.83 | 4029.83 to 4581.99 | 4581.99 to 5134.14 | 5134.14 to 5686.3 |
| Data (%) | 7.6 | 11.6 | 4.8 | 12.4 | 15.2 | 22.4 | 16.8 | 5.2 | 4 |
| Isoelectric Point (pH) | |||||||||
| Interval | 2.71 to 3.73 | 3.73 to 4.75 | 4.75 to 5.76 | 5.76 to 6.78 | 6.78 to 7.8 | 7.8 to 8.82 | 8.82 to 9.83 | 9.83 to 10.85 | 10.85 to 11.87 |
| Data (%) | 8 | 12.4 | 4.4 | 6 | 11.6 | 34 | 20 | 2.4 | 1.2 |
| GRAVY | |||||||||
| Interval | −2.22 to −1.81 | −1.81 to −1.41 | −1.41 to −1 | −1 to −0.6 | −0.6 to −0.19 | −0.19 to 0.21 | 0.21 to 0.62 | 0.62 to 1.02 | 1.02 to 1.43 |
| Data (%) | 0.8 | 2 | 7.6 | 19.6 | 40.4 | 14.8 | 9.2 | 3.6 | 2 |
| Aliphatic index | |||||||||
| Interval | 0 to 17.78 | 17.78 to 35.56 | 35.56 to 53.33 | 53.33 to 71.11 | 71.11 to 88.89 | 88.89 to 106.67 | 106.67 to 124.44 | 124.44 to 142.22 | 142.22 to 160 |
| Data (%) | 18 | 21.6 | 36.4 | 13.2 | 5.2 | 2.8 | 1.2 | 1.2 | 0.4 |
| Boman index | |||||||||
| Interval | −1.83 to −1.06 | −1.06 to −0.28 | −0.28 to 0.5 | 0.5 to 1.27 | 1.27 to 2.05 | 2.05 to 2.83 | 2.83 to 3.6 | 3.6 to 4.38 | 4.38 to 5.15 |
| Data (%) | 2 | 2.8 | 8 | 20 | 31.2 | 23.6 | 8 | 3.6 | 0.8 |
References
- Cournoyer, A.; Bernier, M.-E.; Aboubacar, H.; De Toro-Martín, J.; Vohl, M.-C.; Ravallec, R.; Cudennec, B.; Bazinet, L. Machine Learning-Driven Discovery of Bioactive Peptides from Duckweed (Lemnaceae) Protein Hydrolysates: Identification and Experimental Validation of 20 Novel Antihypertensive, Antidiabetic, and/or Antioxidant Peptides. Food Chem. 2025, 482, 144029. [Google Scholar] [CrossRef]
- Correas, N.H.; Martínez, A.R.; Abellán, A.; Sánchez, H.P.; Tejada, L. Curing Strategies and Bioactive Peptide Generation in Ham: In Vitro Digestion and in Silico Evaluation. Food Chem. 2025, 484, 144360. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, S.; Cao, R.; Hu, M. Review on Bioactive Peptides from Antarctic Krill: From Preparation to Structure-Activity Relationship and Tech-Functionality. Curr. Res. Food Sci. 2025, 10, 101093. [Google Scholar] [CrossRef] [PubMed]
- Purohit, K.; Pathak, R.; Hayes, E.; Sunna, A. Novel Bioactive Peptides from Ginger Rhizome: Integrating in Silico and in Vitro Analysis with Mechanistic Insights through Molecular Docking. Food Chem. 2025, 484, 144432. [Google Scholar] [CrossRef]
- Garmidolova, A.; Desseva, I.; Terziyska, M.; Pavlov, A. Food-Derived Bioactive Peptides-Methods for Purification and Analysis. BIO Web Conf. 2022, 45, 02001. [Google Scholar] [CrossRef]
- Terziyski, Z.; Terziyska, M.; Deseva, I.; Hadzhikoleva, S.; Krastanov, A.; Mihaylova, D.; Hadzhikolev, E. PepLab Platform: Database and Software Tools for Analysis of Food-Derived Bioactive Peptides. Appl. Sci. 2023, 13, 961. [Google Scholar] [CrossRef]
- Chen, L.; Hu, Z.; Rong, Y.; Lou, B. Deep2Pep: A Deep Learning Method in Multi-Label Classification of Bioactive Peptide. Comput. Biol. Chem. 2024, 109, 108021. [Google Scholar] [CrossRef]
- Kang, Y.; Peng, Y.; Zheng, D.; Zhang, H.; Yang, X. Multi-View Framework for Multi-Label Bioactive Peptide Classification Based on Multi-Modal Representation Learning. Appl. Soft Comput. 2025, 175, 113007. [Google Scholar] [CrossRef]
- Centurion, V.B.; Bizzotto, E.; Tonini, S.; Filannino, P.; Di Cagno, R.; Zampieri, G.; Campanaro, S. FEEDS, the Food wastE biopEptiDe claSsifier: From Microbial Genomes and Substrates to Biopeptides Function. Curr. Res. Biotechnol. 2024, 7, 100186. [Google Scholar] [CrossRef]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar] [CrossRef]
- Azodi, C.B.; Tang, J.; Shiu, S.-H. Opening the Black Box: Interpretable Machine Learning for Geneticists. Trends Genet. 2020, 36, 442–455. [Google Scholar] [CrossRef] [PubMed]
- Tonekaboni, S.; Joshi, S.; McCradden, M.D.; Goldenberg, A. What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use. arXiv 2019, arXiv:1905.05134. [Google Scholar] [CrossRef]
- Yang, P.Q.; Wilson, M.L. Explaining Personal and Public Pro-Environmental Behaviors. Sci 2023, 5, 6. [Google Scholar] [CrossRef]
- Dayan, H.; Khoury-Kassabri, M.; Pollak, Y. Sense of Coherence Is Associated with Functional Impairment in Individuals Diagnosed with ADHD. Sci 2025, 7, 60. [Google Scholar] [CrossRef]
- Yuan, Y.; Cao, K.; Gao, P.; Wang, Y.; An, W.; Dong, Y. Extracellular Vesicles and Bioactive Peptides for Regenerative Medicine in Cosmetology. Ageing Res. Rev. 2025, 107, 102712. [Google Scholar] [CrossRef]
- Terziyska, M.; Vladev, V.; Terziyski, Z.; Ilieva, I.; Bozhkov, S. Application of Peptide Nanostructures in the Food Industry. BIO Web Conf. 2025, 170, 01002. [Google Scholar] [CrossRef]
- Phyo, S.H.; Siddique, M.S.; Mushtaq, A.; Yiasmin, M.N.; Alahmad, K.; Khan, I.; Ghamry, M.; Zhao, W. Plant-Derived Peptides and Bioactive Compounds: Mechanisms of AGEs Formation, Detection, and Innovative Approaches for Prevention in Food Processing. Food Biosci. 2025, 69, 106818. [Google Scholar] [CrossRef]
- Terziyski, Z.; Terziyska, M.; Hadzhikoleva, S.; Desseva, I. A Software Tool for Data Mining of Physicochemical Properties of Peptides. BIO Web Conf. 2023, 58, 03007. [Google Scholar] [CrossRef]
- Sturges, H.A. The Choice of a Class Interval. J. Am. Stat. Assoc. 1926, 21, 65–66. [Google Scholar] [CrossRef]
- Fontela, E.; Gabus, A. The DEMATEL Observer; Battelle Geneva Research Center: Geneva, Switzerland, 1976. [Google Scholar]
- Arik, S.O.; Pfister, T. TabNet: Attentive Interpretable Tabular Learning. arXiv 2019, arXiv:1908.07442. [Google Scholar] [CrossRef]
- Manoharan, S.; Shuib, A.S.; Abdullah, N. Structural characteristics and antihypertensive effects of angiotensin-iconverting enzyme inhibitory peptides in the renin-angiotensin and kallikrein kinin systems. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 383–406. [Google Scholar] [CrossRef]
- Daskaya-Dikmen, C.; Yucetepe, A.; Karbancioglu-Guler, F.; Daskaya, H.; Ozcelik, B. Angiotensin-I-Converting Enzyme (ACE)-Inhibitory Peptides from Plants. Nutrients 2017, 9, 316. [Google Scholar] [CrossRef]
- Sitanggang, A.B.; Putri, J.E.; Palupi, N.S.; Hatzakis, E.; Syamsir, E.; Budijanto, S. Enzymatic Preparation of Bioactive Peptides Exhibiting ACE Inhibitory Activity from Soybean and Velvet Bean: A Systematic Review. Molecules 2021, 26, 3822. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Y.; Zhang, J.-T.; Miyakawa, T.; Li, G.-M.; Gu, R.-Z.; Tanokura, M. Antioxidant Properties and Inhibition of Angiotensin-Converting Enzyme by Highly Active Peptides from Wheat Gluten. Sci. Rep. 2021, 11, 5206. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Jiménez, J.; Berraquero-García, C.; Pérez-Gálvez, R.; García-Moreno, P.J.; Espejo-Carpio, F.J.; Guadix, A.; Guadix, E.M. Peptides and Protein Hydrolysates Exhibiting Anti-Inflammatory Activity: Sources, Structural Features and Modulation Mechanisms. Food Funct. 2022, 13, 12510–12540. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, X.; Zhang, X.-L.; Xie, Q.-F. Purification and Identification of Anti-Inflammatory Peptides Derived from Simulated Gastrointestinal Digests of Velvet Antler Protein (Cervus elaphus Linnaeus). J. Food Drug Anal. 2016, 24, 376–384. [Google Scholar] [CrossRef]
- Gozes, I. NAP (Davunetide) Provides Functional and Structural Neuroprotection. Curr. Pharm. Des. 2011, 17, 1040–1044. [Google Scholar] [CrossRef]
- Banasiak-Cieślar, H.; Wiener, D.; Kuszczyk, M.; Dobrzyńska, K.; Polanowski, A. Proline-Rich Polypeptides (Colostrinin®/COLOCO®) Modulate BDNF Concentration in Blood Affecting Cognitive Function in Adults: A Double-Blind Randomized Placebo-Controlled Study. Food Sci. Nutr. 2023, 11, 1477–1485. [Google Scholar] [CrossRef]
- Janusz, M.; Zabłocka, A. Colostrinin: A Proline-Rich Polypeptide Complex of Potential Therapeutic Interest. Cell. Mol. Biol. Noisy—Gd. Fr. 2013, 59, 4–11. [Google Scholar]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The Expanding Scope of Antimicrobial Peptide Structures and Their Modes of Action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.-J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P.S. CancerPPD: A Database of Anticancer Peptides and Proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From Antimicrobial to Anticancer Peptides. A Review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Host Defense Peptides as New Weapons in Cancer Treatment. CMLS Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, M.; Awad, S. Milk Bioactive Peptides: Antioxidant, Antimicrobial and Anti-Diabetic Activities. Adv. Biochem. 2019, 7, 22. [Google Scholar] [CrossRef]
- Tavano, O.L.; Berenguer-Murcia, A.; Secundo, F.; Fernandez-Lafuente, R. Biotechnological Applications of Proteases in Food Technology. Compr. Rev. Food Sci. Food Saf. 2018, 17, 412–436. [Google Scholar] [CrossRef]
- Soltaninejad, H.; Zare-Zardini, H.; Ordooei, M.; Ghelmani, Y.; Ghadiri-Anari, A.; Mojahedi, S.; Hamidieh, A.A. Antimicrobial Peptides from Amphibian Innate Immune System as Potent Antidiabetic Agents: A Literature Review and Bioinformatics Analysis. J. Diabetes Res. 2021, 2021, 2894722. [Google Scholar] [CrossRef]
- Rivero-Pino, F.; Espejo-Carpio, F.J.; Guadix, E.M. Antidiabetic Food-Derived Peptides for Functional Feeding: Production, Functionality and In Vivo Evidences. Foods 2020, 9, 983. [Google Scholar] [CrossRef]
- Elam, E.; Feng, J.; Lv, Y.-M.; Ni, Z.-J.; Sun, P.; Thakur, K.; Zhang, J.-G.; Ma, Y.-L.; Wei, Z.-J. Recent advances on bioactive food-derived anti-diabetic hydrolysates and peptides from natural resources. J. Funct. Foods 2021, 86, 104674. [Google Scholar] [CrossRef]
- Fernández De Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal Peptides as Therapeutic Agents. Front. Cell. Infect. Microbiol. 2020, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Van Der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. Properties and Mechanisms of Action of Naturally Occurring Antifungal Peptides. Cell. Mol. Life Sci. 2013, 70, 3545–3570. [Google Scholar] [CrossRef]
- De Lucca, A.J.; Walsh, T.J. Antifungal Peptides: Novel Therapeutic Compounds against Emerging Pathogens. Antimicrob. Agents Chemother. 1999, 43, 1–11. [Google Scholar] [CrossRef]
- Li, T.; Li, L.; Du, F.; Sun, L.; Shi, J.; Long, M.; Chen, Z. Activity and Mechanism of Action of Antifungal Peptides from Microorganisms: A Review. Molecules 2021, 26, 3438. [Google Scholar] [CrossRef]
- Freitas, C.G.; Felipe, M.S. Candida Albicans and Antifungal Peptides. Infect. Dis. Ther. 2023, 12, 2631–2648. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Zhang, L.; Gallo, R.L. Antimicrobial Peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial Peptides of Multicellular Organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Haney, E.F.; Gill, E.E. The Immunology of Host Defence Peptides: Beyond Antimicrobial Activity. Nat. Rev. Immunol. 2016, 16, 321–334. [Google Scholar] [CrossRef]
- Zou, T.-B.; He, T.-P.; Li, H.-B.; Tang, H.-W.; Xia, E.-Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Nirmal, N.; Khanashyam, A.C.; Shah, K.; Awasti, N.; Sajith Babu, K.; Ucak, İ.; Afreen, M.; Hassoun, A.; Tuanthong, A. Plant Protein-Derived Peptides: Frontiers in Sustainable Food System and Applications. Front. Sustain. Food Syst. 2024, 8, 1292297. [Google Scholar] [CrossRef]
- Sila, A.; Bougatef, A. Antioxidant Peptides from Marine By-Products: Isolation, Identification and Application in Food Systems. A Review. J. Funct. Foods 2016, 21, 10–26. [Google Scholar] [CrossRef]
- You, L.; Zhao, M.; Regenstein, J.M.; Ren, J. Purification and Identification of Antioxidative Peptides from Loach (Misgurnus anguillicaudatus) Protein Hydrolysate by Consecutive Chromatography and Electrospray Ionization-Mass Spectrometry. Food Res. Int. 2010, 43, 1167–1173. [Google Scholar] [CrossRef]
- Ma, J.; Su, K.; Chen, M.; Wang, S. Study on the Antioxidant Activity of Peptides from Soybean Meal by Fermentation Based on the Chemical Method and AAPH-Induced Oxidative Stress. Food Sci. Nutr. 2023, 11, 6634–6647. [Google Scholar] [CrossRef]
- Udenigwe, C.C.; Aluko, R.E. Food Protein-Derived Bioactive Peptides: Production, Processing, and Potential Health Benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Guillen Schlippe, Y.V.; Hartman, M.C.T.; Josephson, K.; Szostak, J.W. In Vitro Selection of Highly Modified Cyclic Peptides That Act as Tight Binding Inhibitors. J. Am. Chem. Soc. 2012, 134, 10469–10477. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, N.; Katkar, R.; Pham, P.V.; Downey, T.; Kashyap, P.; Anastasiu, D.C.; Ramasubramanian, A.K. Prospection of Peptide Inhibitors of Thrombin from Diverse Origins Using a Machine Learning Pipeline. Bioengineering 2023, 10, 1300. [Google Scholar] [CrossRef] [PubMed]
- Kretz, C.A.; Tomberg, K.; Van Esbroeck, A.; Yee, A.; Ginsburg, D. High Throughput Protease Profiling Comprehensively Defines Active Site Specificity for Thrombin and ADAMTS13. Sci. Rep. 2018, 8, 2788. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Gabrani, R. Antiviral Peptides: Identification and Validation. Int. J. Pept. Res. Ther. 2021, 27, 149–168. [Google Scholar] [CrossRef]
- Vilas Boas, L.C.P.; Campos, M.L.; Berlanda, R.L.A.; De Carvalho Neves, N.; Franco, O.L. Antiviral Peptides as Promising Therapeutic Drugs. Cell. Mol. Life Sci. 2019, 76, 3525–3542. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, Y.; Sun, X.; Ma, T.; Lao, X.; Zheng, H. DRAVP: A Comprehensive Database of Antiviral Peptides and Proteins. Viruses 2023, 15, 820. [Google Scholar] [CrossRef]
- Chia, L.Y.; Kumar, P.V.; Maki, M.A.A.; Ravichandran, G.; Thilagar, S. A Review: The Antiviral Activity of Cyclic Peptides. Int. J. Pept. Res. Ther. 2022, 29, 7. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Teng, X.; Shang, J.; Wang, D.; Liu, N. Identification of Novel DPP–IV Inhibitory Peptides from Atlantic Salmon (Salmo salar) Skin. Food Res. Int. 2020, 133, 109161. [Google Scholar] [CrossRef]
- Liu, R.; Cheng, J.; Wu, H. Discovery of Food-Derived Dipeptidyl Peptidase IV Inhibitory Peptides: A Review. Int. J. Mol. Sci. 2019, 20, 463. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; Mooney, C.; Shields, D.C.; FitzGerald, R.J. In Silico Approaches to Predict the Potential of Milk Protein-Derived Peptides as Dipeptidyl Peptidase IV (DPP-IV) Inhibitors. Peptides 2014, 57, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Wang, R.; Cheng, C.; Ma, Y.; Zhang, Y.; Lu, W. Preparation, Structural Properties, and in Vitro and in Vivo Activities of Peptides against Dipeptidyl Peptidase IV (DPP-IV) and α-Glucosidase: A General Review. Crit. Rev. Food Sci. Nutr. 2024, 64, 9844–9858. [Google Scholar] [CrossRef]
- Charoenkwan, P.; Nantasenamat, C.; Hasan, M.M.; Moni, M.A.; Lio’, P.; Manavalan, B.; Shoombuatong, W. StackDPPIV: A Novel Computational Approach for Accurate Prediction of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides. Methods 2022, 204, 189–198. [Google Scholar] [CrossRef]
- Hökfelt, T.; Broberger, C.; Xu, Z.-Q.D.; Sergeyev, V.; Ubink, R.; Diez, M. Neuropeptides—An Overview. Neuropharmacology 2000, 39, 1337–1356. [Google Scholar] [CrossRef]
- Li, C. Neuropeptides. WormBook 2008, 2008, 1–36. [Google Scholar] [CrossRef]
- DeLaney, K.; Buchberger, A.R.; Atkinson, L.; Gründer, S.; Mousley, A.; Li, L. New Techniques, Applications and Perspectives in Neuropeptide Research. J. Exp. Biol. 2018, 221, jeb151167. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Yin, S.; Jang, R.; Wang, J.; Xue, Z.; Xu, T. NeuroPep: A Comprehensive Resource of Neuropeptides. Database 2015, 2015, bav038. [Google Scholar] [CrossRef]
- Girven, K.S.; Mangieri, L.; Bruchas, M.R. Emerging Approaches for Decoding Neuropeptide Transmission. Trends Neurosci. 2022, 45, 899–912. [Google Scholar] [CrossRef]
- Brownstein, M.J. A Brief History of Opiates, Opioid Peptides, and Opioid Receptors. Proc. Natl. Acad. Sci. USA 1993, 90, 5391–5393. [Google Scholar] [CrossRef]
- Fricker, L.D.; Margolis, E.B.; Gomes, I.; Devi, L.A. Five Decades of Research on Opioid Peptides: Current Knowledge and Unanswered Questions. Mol. Pharmacol. 2020, 98, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Kumar, V.; Sharma, K.; Kaur, S.; Gat, Y.; Goyal, A.; Tanwar, B. Opioid Peptides: An Overview of Functional Significance. Int. J. Pept. Res. Ther. 2020, 26, 33–41. [Google Scholar] [CrossRef]
- Zioudrou, C.; Streaty, R.A.; Klee, W.A. Opioid Peptides Derived from Food Proteins. The Exorphins. J. Biol. Chem. 1979, 254, 2446–2449. [Google Scholar] [CrossRef]
- Possani, L.D.; Merino, E.; Corona, M.; Bolivar, F.; Becerril, B. Peptides and Genes Coding for Scorpion Toxins That Affect Ion-Channels. Biochimie 2000, 82, 861–868. [Google Scholar] [CrossRef]
- Sunagar, K.; Undheim, E.A.B.; Chan, A.H.C.; Koludarov, I.; Muñoz-Gómez, S.A.; Antunes, A.; Fry, B.G. Evolution Stings: The Origin and Diversification of Scorpion Toxin Peptide Scaffolds. Toxins 2013, 5, 2456–2487. [Google Scholar] [CrossRef] [PubMed]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin Is a Fungal Peptide Toxin Critical for Mucosal Infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Undheim, E.A.B.; Mobli, M.; King, G.F. Toxin Structures as Evolutionary Tools: Using Conserved 3D Folds to Study the Evolution of Rapidly Evolving Peptides. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.S. Peptide Toxin Structure and Function by NMR. In Modern Magnetic Resonance; Webb, G.A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1–18. ISBN 978-3-319-28275-6. [Google Scholar]
- Fernández, A.; García, S.; Herrera, F.; Chawla, N.V. SMOTE for Learning from Imbalanced Data: Progress and Challenges, Marking the 15-Year Anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- He, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Gronning, A.G.B.; Kacprowski, T.; Scheele, C. MultiPep: A hierarchical deep learning approach for multi-label classification of peptide bioactivities. Biol. Methods Protoc. 2021, 6, bpab021. [Google Scholar] [CrossRef]
- LI, Y.; Li, X.; Liu, Y.; Yao, Y.; Huang, G. MPMABP: A CNN and Bi-LSTM-Based method for predicting multi-activities of bioactive peptides. Pharmaceuticals 2022, 15, 707. [Google Scholar] [CrossRef]
- Fan, H.; Yan, W.; Wang, L.; Liu, J.; Bin, Y.; Xia, J. Deep learning-based multi-functional therapeutic peptides prediction with a multi-label focal dice loss function. Bioinformatics 2023, 39, btad334. [Google Scholar] [CrossRef]


| Activity | Number of Peptides |
|---|---|
| ACE inhibitor | 524 |
| Antimicrobial | 490 |
| Antibacterial | 351 |
| Anticancer | 279 |
| Antioxidative | 261 |
| Toxin | 251 |
| Antidiabetic | 238 |
| Opioid | 79 |
| Neuropeptide | 71 |
| DPP-IV inhibitor | 55 |
| Antiamnestic | 47 |
| Antifungal | 39 |
| Antithrombotic | 35 |
| Anti-inflammatory | 14 |
| Antiviral | 14 |
| len | MW | pI | gravy | alipha | boman | |
|---|---|---|---|---|---|---|
| len | 0.239726 | 0.095303 | −5.61042 | 0.094433 | −1.59476 | −0.76024 |
| MW | 0.095303 | −0.01738 | −4.95706 | 0.093243 | −1.35694 | −0.68756 |
| pI | −5.61042 | −4.95706 | 24.09702 | −1.20775 | 6.640577 | 3.26559 |
| gravy | 0.094433 | 0.093243 | −1.20775 | −0.52175 | −1.04366 | 0.408626 |
| alipha | −1.59476 | −1.35694 | 6.640577 | −1.04366 | 1.790399 | 1.718069 |
| boman | −0.76024 | −0.68756 | 3.26559 | 0.408626 | 1.718069 | 0.131324 |
| Causality (D − R) | Centrality (D + R) | |
|---|---|---|
| pI | 0 | 44.45590436 |
| alipha | 1.77636−15 | 12.30738337 |
| boman | 0 | 8.151630598 |
| gravy | −1.33227−15 | −4.353722692 |
| MW | 1.77636−15 | −13.6607655 |
| len | 8.88178−16 | −15.07190701 |
| Indices | Values |
|---|---|
| Chi-square | 854.171 |
| Degrees of freedom | 7 |
| CFI | 0.957 |
| TLI | 0.876 |
| RMSEA | 0.028 (90% CI: 0.020–0.037) |
| SRMR | 0.041 |
| AIC | 11,259.23 |
| BIC | 11,269.69 |
| SABIC | 11,264.86 |
| Endogenous Variable | Exogenous Variable | Estimate | Z-Value | p-Value |
|---|---|---|---|---|
| MW | len | 109.222 | 382.213 | <0.001 |
| pI | MW | 0.023 | 8.999 | <0.001 |
| boman | gravy | −1.643 | −56.127 | <0.001 |
| alipha | boman | 0.883 | 39.348 | <0.001 |
| alipha | gravy | 0.167 | 7.333 | <0.001 |
| IC50 | alipha | −9.647 | −25.744 | <0.001 |
| Original Data | 4655 Entries | 5255 Entries | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | F1_score | Precision | Recall | Accuracy | F1_score | Precision | Recall | Accuracy | F1_score | Precision | Recall | |
| Random Forest | 0.5386 ± 0.014 | 0.5233 ± 0.04 | 0.5100 ± 0.076 | 0.5200 ± 0.03 | 0.6237 ± 0.016 | 0.6203 ± 0.0033 | 0.6300 ± 0.06 | 0.6200 ± 0.05 | 0.6700 ± 0.02 | 0.6400 ± 0.003 | 0.6800 ± 0.08 | 0.6300 ± 0.024 |
| XGBoost | 0.5027 ± 0.02 | 0.4928 ± 0.05 | 0.4800 ± 0.07 | 0.4900 ± 0.039 | 0.6199 ± 0.021 | 0.6154 ± 0.0042 | 0.6200 ± 0.07 | 0.6100 ± 0.09 | 0.6800 ± 0.023 | 0.6500 ± 0.002 | 0.6900 ± 0.061 | 0.6400 ± 0.008 |
| LightGBM | 0.5189 ± 0.019 | 0.5021 ± 0.037 | 0.4500 ± 0.058 | 0.5200 ± 0.03 | 0.6208 ± 0.09 | 0.6218 ± 0.018 | 0.6200 ± 0.043 | 0.6200 ± 0.05 | 0.6600 ± 0.006 | 0.6623 ± 0.007 | 0.6600 ± 0.0057 | 0.6600 ± 0.0063 |
| MLP | 0.4829 ± 0.0078 | 0.4552 ± 0.01 | 0.4200 ± 0.018 | 0.5200 ± 0.013 | 0.5707 ± 0.032 | 0.5625 ± 0.027 | 0.5800 ± 0.026 | 0.5700 ± 0.017 | 0.6609 ± 0.044 | 0.6608 ± 0.05 | 0.6600 ± 0.047 | 0.6600 ± 0.053 |
| TabNet | 0.4367 ± 0.0051 | 0.4088 ± 0.01 | 0.3900 ± 0.005 | 0.4400 ± 0.016 | 0.5443 ± 0.006 | 0.5310 ± 0.031 | 0.5500 ± 0.023 | 0.5400 ± 0.02 | 0.5869 ± 0.008 | 0.5720 ± 0.06 | 0.5900 ± 0.0063 | 0.5700 ± 0.0047 |
| Database | Scope of Peptides | Key Features | Limitations |
|---|---|---|---|
| APD3 | Antimicrobial peptides | Manually curated; sequence data | Limited to antimicrobial activity |
| CAMP | Antimicrobial peptides | Multiple prediction tools | Restricted scope; less focus on physicochemical descriptors |
| DBAASP | Antimicrobial peptides | Structural data; activity assays | Focused mainly on antimicrobial function |
| BIOPEP-UWM | Broad spectrum of bioactive peptides and proteins | Functional activities; enzymatic release | Narrow application in food/functional peptides |
| PepLab | Broad spectrum of bioactive peptides | Integrated database + physicochemical descriptors + statistical tool | Currently limited dataset size; ongoing manual curation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terziyska, M.; Terziyski, Z.; Ilieva, I.; Bozhkov, S.; Vladev, V. Knowledge Discovery from Bioactive Peptide Data in the PepLab Database Through Quantitative Analysis and Machine Learning. Sci 2025, 7, 122. https://doi.org/10.3390/sci7030122
Terziyska M, Terziyski Z, Ilieva I, Bozhkov S, Vladev V. Knowledge Discovery from Bioactive Peptide Data in the PepLab Database Through Quantitative Analysis and Machine Learning. Sci. 2025; 7(3):122. https://doi.org/10.3390/sci7030122
Chicago/Turabian StyleTerziyska, Margarita, Zhelyazko Terziyski, Iliana Ilieva, Stefan Bozhkov, and Veselin Vladev. 2025. "Knowledge Discovery from Bioactive Peptide Data in the PepLab Database Through Quantitative Analysis and Machine Learning" Sci 7, no. 3: 122. https://doi.org/10.3390/sci7030122
APA StyleTerziyska, M., Terziyski, Z., Ilieva, I., Bozhkov, S., & Vladev, V. (2025). Knowledge Discovery from Bioactive Peptide Data in the PepLab Database Through Quantitative Analysis and Machine Learning. Sci, 7(3), 122. https://doi.org/10.3390/sci7030122








