Transcriptomics Analysis of Tomato Ripening Regulated by Carbon Dioxide
Abstract
1. Introduction
2. Materials and Methods
2.1. Tomato Fruit and CO2 Treatment
2.2. Characterization of Physical and Chemical Properties
2.2.1. Ethylene Production
2.2.2. Measurements of Color Change
2.2.3. Firmness Test
2.2.4. Total Soluble Carbohydrate Determination
2.2.5. RNA Extraction
2.3. RNA-Seq and Data Analysis
2.4. Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Assays
2.5. Statistical Analysis
3. Results
3.1. Changes in the Phenotypes of Tomatoes Induced by Carbon Dioxide
3.2. Transcriptome and Bioinformatics Analysis
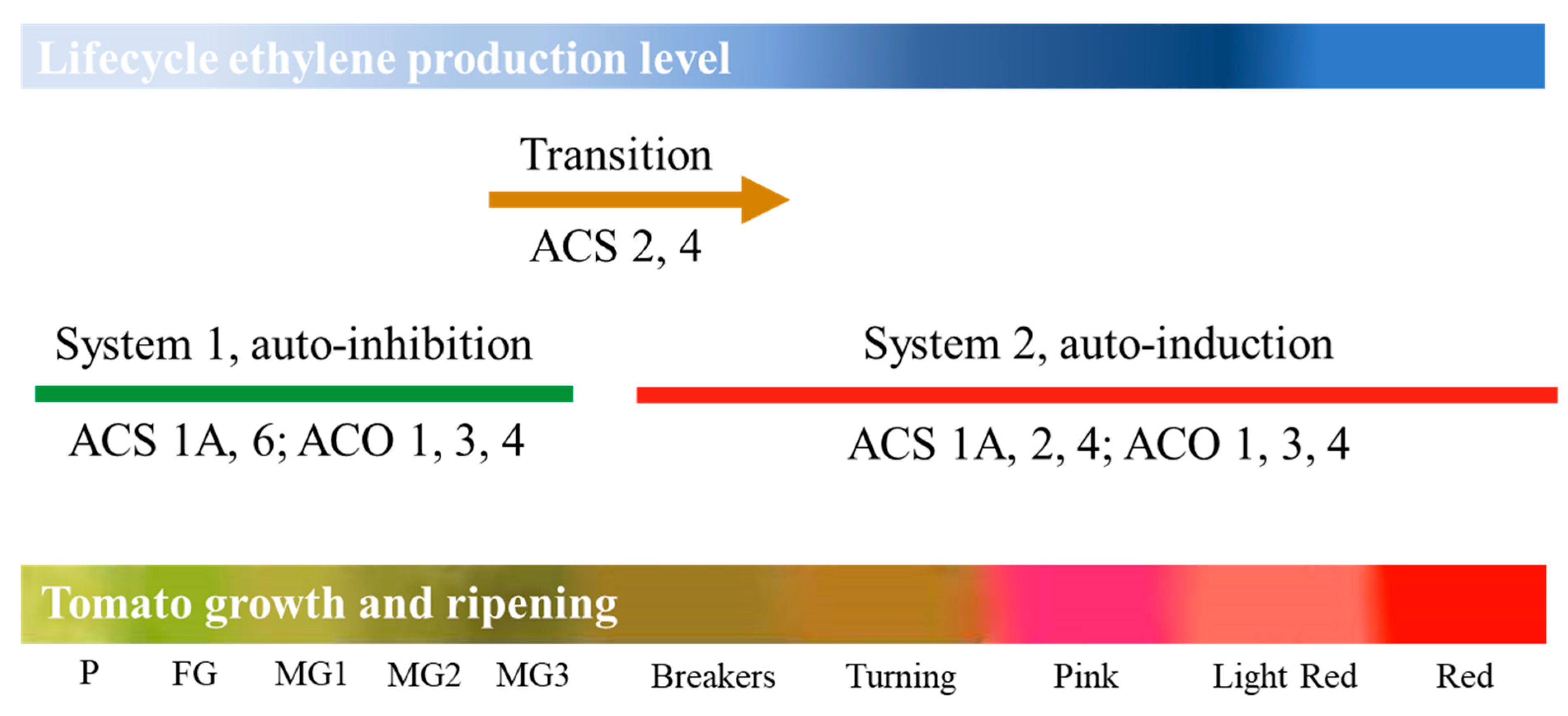
3.3. DEGs Encoding for Ethylene Synthesis and Signal Transduction
3.4. DEGs Encoding for Color Change and Cell-Wall Degradation
3.5. DEGs Encoding for Stress Resistance Induced by CO2 Treatment
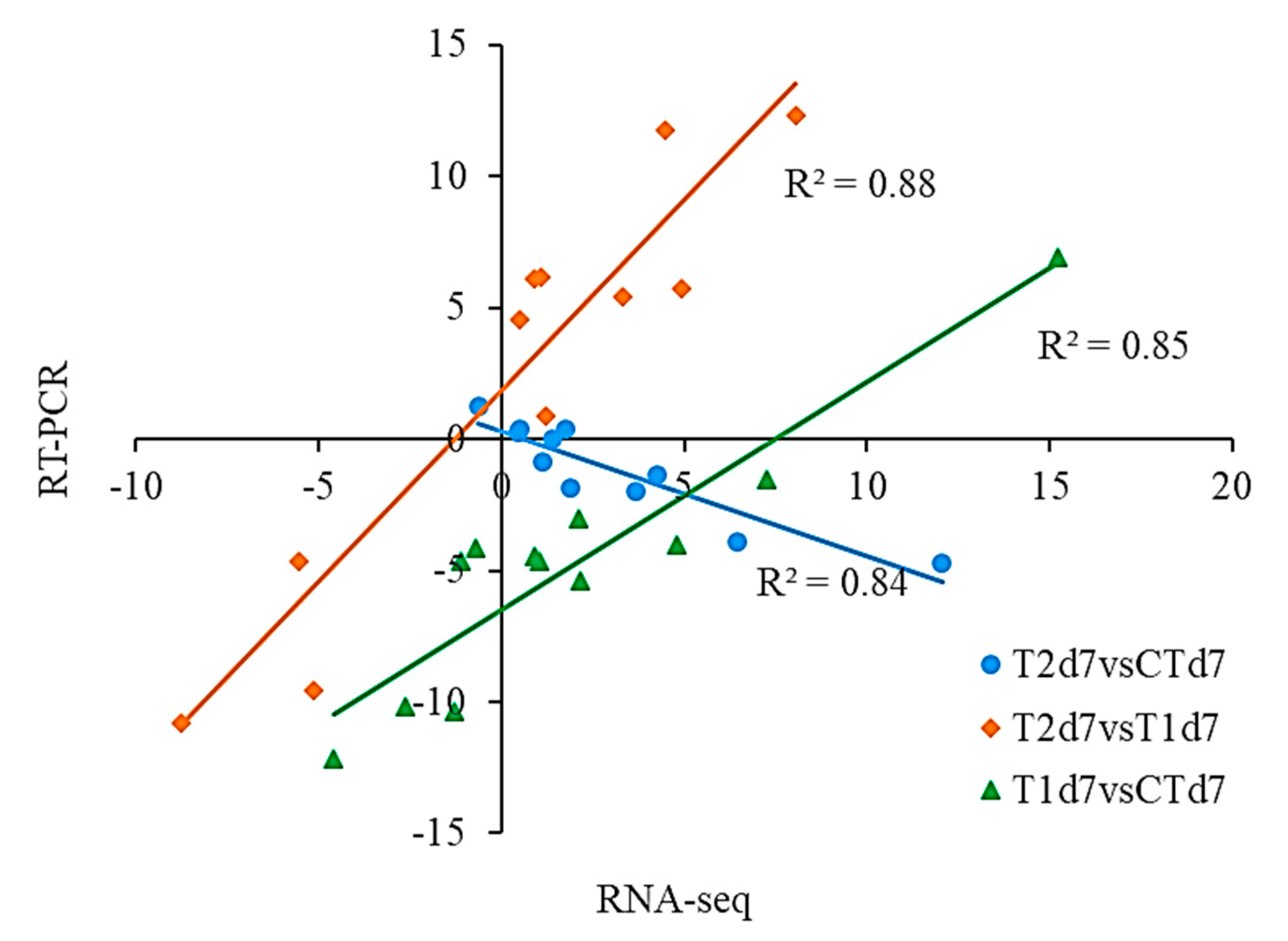
3.6. Correlation of Gene Expression Data from RNA-Seq and qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Story, E.N.; Kopec, R.E.; Schwartz, S.J.; Harris, G.K. An update on the health effects of tomato lycopene. Annu. Rev. Food Sci. Technol. 2010, 1, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Raiola, A.; Rigano, M.M.; Calafiore, R.; Frusciante, L.; Barone, A. Enhancing the health-promoting effects of tomato fruit for biofortified food. Mediat. Inflamm. 2014, 2014, 139873. [Google Scholar] [CrossRef]
- Kapotis, G.; Passam, H.C.; Akoumianakis, K.; Olympios, C.M. Storage of tomatoes in low oxygen atmospheres inhibits ehylene action and polygalacturonase activity. Russ. J. Plant. Physiol. 2004, 51, 112–115. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, S.-K.; Jung, J.; Jeong, M.-J.; Ryu, C.-M. Exploring the sound-modulated delay in tomato ripening through expression analysis of coding and non-coding RNAs. Ann. Bot. 2018, 122, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.-P.; Lutts, S. Tomato fruit development and metabolism. Front. Plant. Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Li, T.; Tan, D.; Yang, X.; Wang, A. Exploring the apple genome reveals six ACC synthase genes expressed during fruit ripening. Sci. Hortic. 2013, 157, 119–123. [Google Scholar] [CrossRef]
- Dilshad, S.; Kalair, A.R.; Khan, N. Review of carbon dioxide (CO2) based heating and cooling technologies: Past, present, and future outlook. Int. J. Energy Res. 2020, 44, 1408–1463. [Google Scholar] [CrossRef]
- Tomasko, D.L.; Li, H.; Liu, D.; Han, X.; Wingert, M.J.; Lee, L.J.; Koelling, K.W. A review of CO2 applications in the processing of polymers. Ind. Eng. Chem. Res. 2003, 42, 6431–6456. [Google Scholar] [CrossRef]
- Boz, Z.; Welt, B.; Brecht, J.; Pelletier, W.; McLamore, E.; Kiker, G.; Butler, J. Review of challenges and advances in modification of food package headspace gases. J. Appl. Packag. Res. 2018, 10, 62–97. [Google Scholar]
- Singh, P.A.; Wani, A.; Karim, A.A.; Langowski, H.-C. The use of carbon dioxide in the processing and packaging of milk and dairy products: A review. Int. J. Dairy Technol. 2012, 65, 161–177. [Google Scholar] [CrossRef]
- Caleb, O.J.; Mahajan, P.V.; Al-Said, F.A.; Opara, U.L. Modified atmosphere packaging technology of fresh and fresh-cut produce and the microbial consequences—A review. Food Bioprocess. Technol. 2013, 6, 303–329. [Google Scholar] [CrossRef] [PubMed]
- Terai, H.; Tsuchida, H.; Mizuno, M.; Matsui, N. Influence of short-term treatment with high CO2 and N2 on ethylene biosynthesis in tomato fruit. HortScience 1998, 33, 103–104. [Google Scholar] [CrossRef]
- Mathooko, F.M. Regulation of ethylene biosynthesis in higher plants by carbon dioxide. Postharvest Biol. Technol. 1996, 7, 1–26. [Google Scholar] [CrossRef]
- De Wild, H.P.J.; Balk, P.A.; Fernandes, A.E.C.A.; Peppelenbos, H.W. The action site of carbon dioxide in relation to inhibition of ethylene production in tomato fruit. Postharvest Biol. Technol. 2005, 36, 273–280. [Google Scholar] [CrossRef]
- May, P.; Liao, W.; Wu, Y.; Shuai, B.; McCombie, W.R.; Zhang, W.Q.; Liu, Q.A. The Effects of Carbon Dioxide and Temperature on MicroRNA Expression in Arabidopsis Development. Nat. Commun. 2013, 4, 2145. [Google Scholar] [CrossRef]
- Thompson, M.; Gamage, D.; Hirotsu, N.; Martin, A.; Seneweera, S. Effects of elevated carbon dioxide on photosynthesis and carbon partitioning: A perspective on root sugar snsing and hormonal crosstalk. Front. Phys. 2017, 8, 578. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Pirrello, J.; Chervin, C.; Roustan, J.-P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant. Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef]
- Martín-Pizarro, C.; Posé, D. Genome editing as a tool for fruit ripening manipulation. Front. Plant. Sci. 2018, 9, 1415. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar] [CrossRef]
- Pereira, L.; Domingo, M.S.; Ruggieri, V.; Argyris, J.; Phillips, M.A.; Zhao, G.; Lian, Q.; Xu, Y.; He, Y.; Huang, S.; et al. Genetic dissection of climacteric fruit ripening in a melon population segregating for ripening behavior. Hortic. Res. 2020, 7, 187. [Google Scholar] [CrossRef]
- Cuesta, G.; Suarez, N.; Bessio, M.I.; Ferreira, F.; Massaldi, H. Quantitative determination of pneumococcal capsular polysaccharide serotype 14 using a modification of phenol–sulfuric acid method. J. Microbiol. Methods 2003, 52, 69–73. [Google Scholar] [CrossRef]
- Bobokalonov, J.T.; Liu, Y.; Shahrin, T.; Liu, L.S. Transcriptomic Analysis on the Regulation of Tomato Ripening by the Ethylene Inhibitor 1-Methylcyclopropene. J. Plant. Stud. 2018, 7, 49. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Murachi, S.; Okunishi, H.; Shiomi, S.; Nakano, R.; Kubo, Y.; Inaba, A. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant. Physiol. 1998, 118, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Cantwell, M. Melon Quality and Ripening. In Proceedings of the Fruit Ripening and Retail Handling Workshop Postharvest Technology Center, UC Davis, Davis, CA, USA, 18–19 April 2017; Available online: https://postharvest.ucdavis.edu/files/261288.pdf (accessed on 27 May 2023).
- Gagne, J.M.; Smalle, J.; Gingerich, D.J.; Walker, J.M.; Yoo, S.-D.; Yanagisawa, S.; Vierstra, R.D. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and pomote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 2004, 101, 6803–6808. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tao, X.; Li, L.; Mao, L.; Luo, Z.; Khan, Z.U.; Ying, T. Comprehensive RNA-seq analysis on the regulation of tomato ripening by exogenous auxin. PLoS ONE 2016, 11, e0156453. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Ju, Z.; Gao, C.; Mei, X.; Fu, D.; Zhu, H.; Luo, Y.; Zhu, B. Genome-wide identification of cytosine-5 DNA methyltransferases and demethylases in solanum lycopersicum. Gene 2014, 550, 230–237. [Google Scholar] [CrossRef]
- Zhong, S.; Fei, Z.; Chen, Y.-R.; Zheng, Y.; Huang, M.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Alelú-Paz, R.; Ashour, N.; González-Corpas, A.; Ropero, S. DNA methylation, histone modifications, and signal transduction pathways: A close relationship in malignant gliomas pathophysiology. J. Signal. Transduct. 2012, 2012, 956958. [Google Scholar] [CrossRef]
- Omidvar, V.; Fellner, M. DNA methylation and transcriptomic changes in response to different lights and stresses in 7B-1 male-sterile tomato. PLoS ONE 2015, 10, e0121864. [Google Scholar] [CrossRef]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Schwarz, N.; Armbruster, U.; Iven, T.; Brückle, L.; Melzer, M.; Feussner, I.; Jahns, P. Tissue-specific accumulation and regulation of zeaxanthin epoxidase in arabidopsis reflect the multiple functions of the enzyme in plastids. Plant. Cell. Physiol. 2015, 56, 346–357. [Google Scholar] [CrossRef]
- Kumar, N.; Larkin, J.C. Why do plants need so many cyclin-dependent kinase inhibitors? Plant. Signal. Behav. 2017, 12, e1282021. [Google Scholar] [CrossRef] [PubMed]
- Carrari, F.; Baxter, C.; Usadel, B.; Urbanczyk-Wochniak, E.; Zanor, M.-I.; Nunes-Nesi, A.; Nikiforova, V.; Centero, D.; Ratzka, A.; Pauly, M.; et al. Integrated analysis of metabolite and tanscript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of mtabolic network behavior. Plant. Physiol. 2006, 142, 1380–1396. [Google Scholar] [CrossRef] [PubMed]
- Matas, A.J.; Gapper, N.E.; Chung, M.-Y.; Giovannoni, J.J.; Rose, J. Biology and genetic engineering of fruit maturation for enhanced quality and shelf-life. Curr. Opin. Biotechnol. 2009, 20, 197–203. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling feshy fruit ripening. New. Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Paliyath, G. Microarray analysis of ripening-regulated gene expression and its modulation by 1-MCP and hexanal. Plant. Physiol. Biochem. 2011, 49, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; Maagd, R.A. Transcriptionalcontrol of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef]
- Li, T.; Tan, D.; Liu, Z.; Jiang, Z.; Wei, Y.; Zhang, L.; Li, X.; Yuan, H.; Wang, A. Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor. Plant Cell Physiol. 2015, 56, 1909–1917. [Google Scholar] [CrossRef]
- Liu, L.; Shao, Z.; Zhang, M.; Wang, Q. Regulation of carotenoid metabolism in tomato. Mol. Plant 2015, 8, 28–39. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hort. Res. 2015, 2, 15036. [Google Scholar] [CrossRef]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant. Sci. 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Enfissi, E.; Nogueira, M.; Bramley, P.; Fraser, P. The regulation of carotenoid formation in tomato fruit. Plant. J. 2017, 89, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Ernesto, B.; Lira, R.B.; Monteiro, S.S.; Demarco, D.; Purgatto, E.; Rothan, C.; Rossi, M.; Freschi, L. Fruit-localized pytochromes regulate plastid biogenesis, starch synthesis, and carotenoid metabolism in tomato. J. Exp. Bot. 2018, 69, 3573–3586. [Google Scholar] [CrossRef]
- Schouten, R.E.; Farneti, B.; Tijskens, L.M.M.; Alarcón, A.A.; Woltering, E.J. Quantifying lycopene synthesis and chlorophyll breakdown in tomato fuit using remittance VIS spectroscopy. Postharvest Biol. Technol. 2014, 96, 53–63. [Google Scholar] [CrossRef]
- Cruz, A.B.; Bianchetti, R.E.; Alves, F.R.; Purgatto, E.; Peres, L.E.; Rossi, M.; Freschi, L. Light, ethylene and auxin signaling interaction regulates carotenoid biosynthesis during tomato fruit ripening. Front. Plant. Sci. 2018, 9, 1370. [Google Scholar] [CrossRef]
- Liu, M.; Diretto, G.; Pirrello, J.; Roustan, J.-P.; Li, Z.; Giuliano, G.; Regad, F.; Bouzayen, M. The chimeric repressor vrsion of an ethylene response factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New. Phytol. 2014, 203, 206–218. [Google Scholar] [CrossRef]
- Marsic, N.K.; Vodnik, D.; Mikulic-Petkovsek, M.; Veberic, R.; Sircelj, H. Photosynthetic traits of plants and the biochemical profile of tomato fruits are influenced by grafting, salinity Stress, and growing season. J. Agric. Food Chem. 2018, 66, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Raffo, A.; Baiamonte, I.; Nardo, N.; Nicoli, S.; Moneta, E.; Peparaio, M.; Sinesio, F.; Paoletti, F. Impact of early harvesting and two cold storage technologies on eating quality of red ripe tomatoes. Eur. Food Res. Technol. 2018, 244, 805–818. [Google Scholar] [CrossRef]
- Wei, Z.; Du, T.; Li, X.; Fang, L.; Liu, F. Interactive effects of elevated CO2 and N2 fertilization on yield and quality of tomato grown under reduced irrigation regimes. Front. Plant. Sci. 2018, 9, 328. [Google Scholar] [CrossRef]
- Zhang, W.-F.; Gong, Z.-H.; Wu, M.-B.; Chan, H.; Yuan, Y.-J.; Tang, N.; Zhang, Q.; Miao, M.-J.; Chang, W.; Li, Z.; et al. Integrative comparative analyses of metabolite and transcript profiles uncovers complex regulatory network in tomato (Solanum Lycopersicum L.) fruit undergoing chilling injury. Sci. Rep. 2019, 9, 4470. [Google Scholar] [CrossRef]

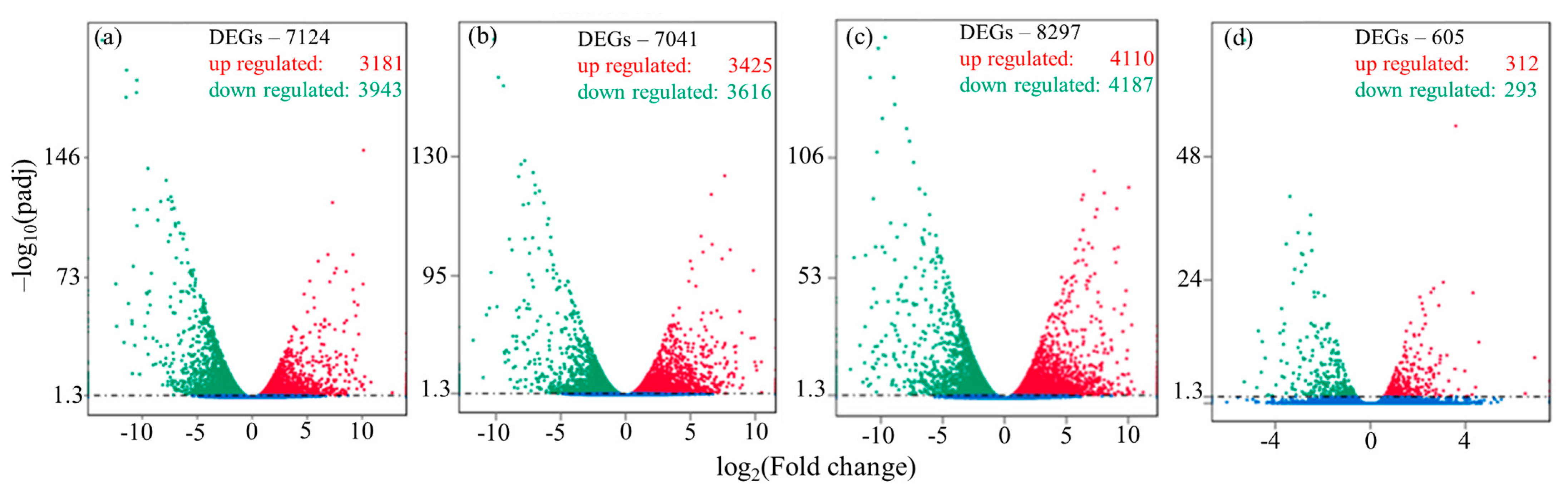
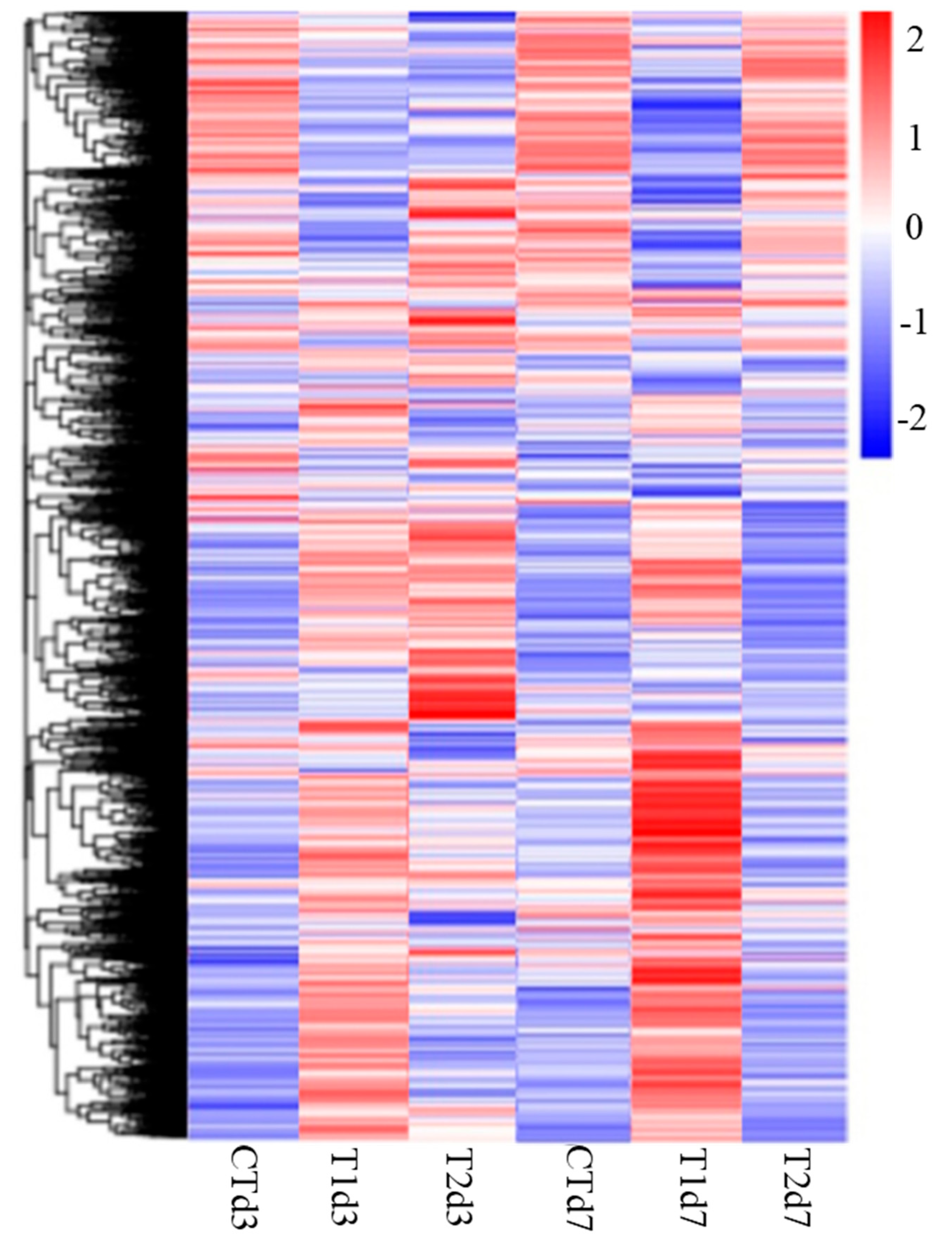

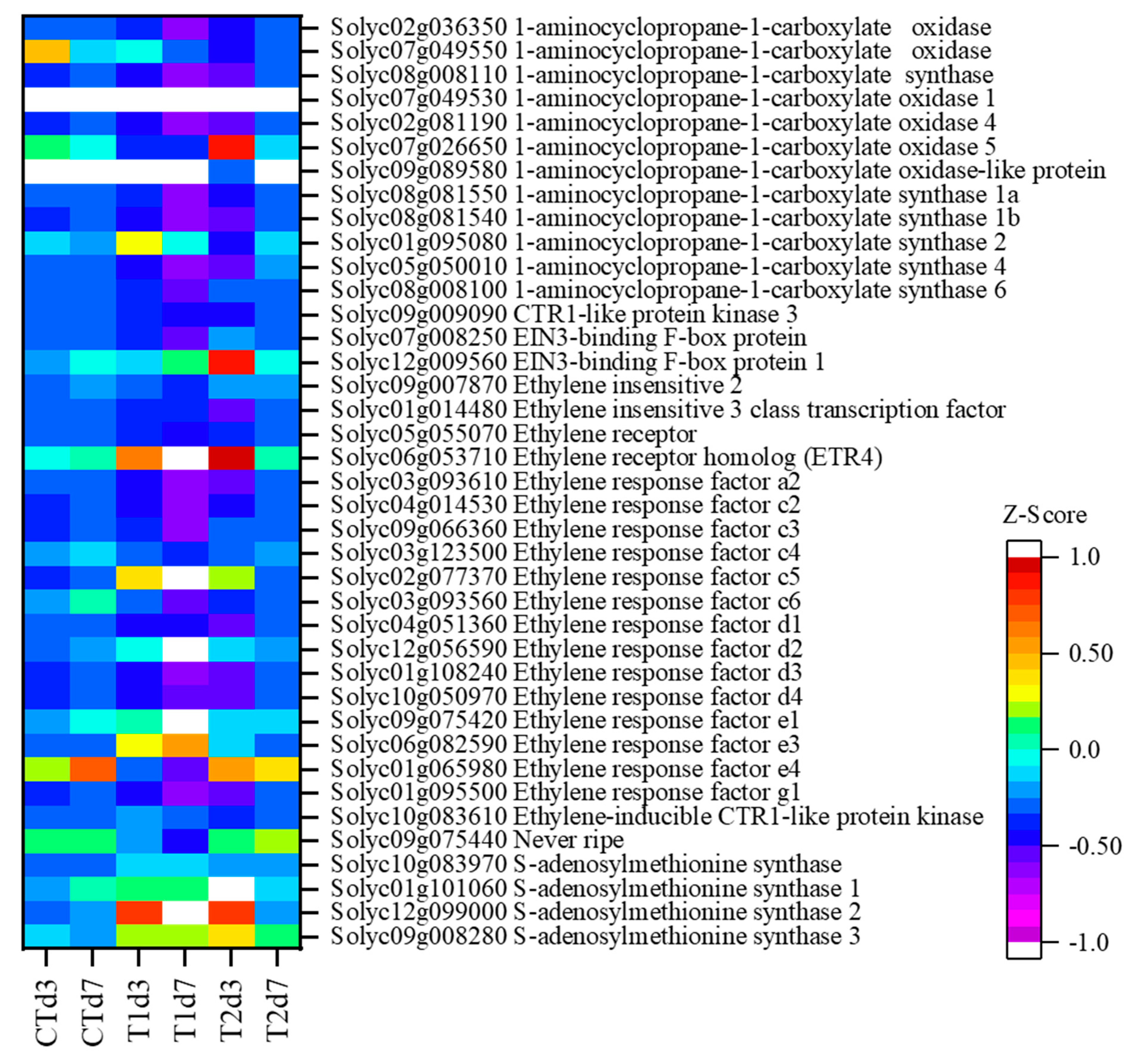
| Sample Name * | Raw Reads | Clean Reads | Q20 ** (%) | Total Mapped | Multiple-Mapped | Uniquely Mapped |
|---|---|---|---|---|---|---|
| CTd3 1 | 65,773,470 | 63,817,890 | 97.28 | 51,738,379 (81.07%) | 454,994 (0.71%) | 51,283,385 (80.36%) |
| CTd3 2 | 73,676,038 | 71,179,598 | 97.14 | 57,513,775 (80.8%) | 581,015 (0.82%) | 56,932,760 (79.98%) |
| CTd7 1 | 64,366,640 | 61,793,974 | 96.93 | 35,012,094 (56.66%) | 256,136 (0.41%) | 34,755,958 (56.24%) |
| CTd7 2 | 62,739,824 | 60,506,740 | 97.14 | 32,240,221 (53.28%) | 244,883 | 31,995,338 (52.88%) |
| −0.40% | ||||||
| T1d3 1 | 82,567,552 | 79,299,482 | 97.06 | 58,963,542 (74.36%) | 461,800 (0.58%) | 58,501,742 (73.77%) |
| T1d3 2 | 64,498,224 | 62,077,256 | 96.97 | 51,142,314 (82.38%) | 375,820 (0.61%) | 50,766,494 (81.78%) |
| T1d7 1 | 90,148,126 | 86,721,258 | 97.16 | 64,317,498 (74.17%) | 518,547 | 63,798,951 (73.57%) |
| −0.60% | ||||||
| T1d7 2 | 92,821,490 | 89,213,582 | 97.04 | 57,915,789 (64.92%) | 458,066 (0.51%) | 57,457,723 (64.4%) |
| T2d3 1 | 74,724,698 | 72,157,326 | 97.23 | 51,476,059 (71.34%) | 325,579 (0.45%) | 51,150,480 (70.89%) |
| T2d3 2 | 76,499,736 | 74,025,794 | 97.24 | 45,151,183 (60.99%) | 283,328 (0.38%) | 44,867,855 (60.61%) |
| T2d7 1 | 73,727,656 | 70,981,232 | 97.09 | 38,103,699 (53.68%) | 266,548 (0.38%) | 37,837,151 (53.31%) |
| T2d7 2 | 70,278,582 | 67,611,036 | 96.99 | 49,525,905 (73.25%) | 354,644 (0.52%) | 49,171,261 (72.73%) |
| Gene ID | Annotation | T1d3:CTd3 | T2d3:CTd3 | T1d7:CTd7 | T2d7:CTd7 |
|---|---|---|---|---|---|
| Solyc10g083970 | S-adenosylmethionine synthase | - | - | - | - |
| Solyc09g008280 | S-adenosylmethionine synthase 3 | - | - | - | 4.02 |
| Solyc12g099000 | S-adenosylmethionine synthase 2 | 3.83 | 3.73 | 2.11 | - |
| Solyc01g101060 | S-adenosylmethionine synthase 1 | - | 3.21 | 0.32 | 0.65 |
| Solyc08g008110 | 1-aminocyclopropane-1-carboxylate synthase | 0.13 | - | - | - |
| Solyc08g008100 | 1-aminocyclopropane-1-carboxylate synthase 6 | 0.34 | 5.2 | - | - |
| Solyc05g050010 | 1-aminocyclopropane-1-carboxylate synthase 4 | 0.01 | 0.01 | 0 | - |
| Solyc01g095080 | 1-aminocyclopropane-1-carboxylate synthase 2 | - | 0.09 | - | 2.79 |
| Solyc08g081550 | 1-aminocyclopropane-1-carboxylate synthase 1a | - | - | 0.29 | - |
| Solyc08g081540 | 1-aminocyclopropane-1-carboxylate synthase 1b | 0.2 | - | - | - |
| Solyc07g049530 | 1-aminocyclopropane-1-carboxylate oxidase 1 | 0.38 | 0.31 | 0.49 | - |
| Solyc09g089580 | 1-aminocyclopropane-1-carboxylate oxidase-like protein | 0.07 | 0.01 | 0.07 | - |
| Solyc02g081190 | 1-aminocyclopropane-1-carboxylate oxidase 4 | 21.22 | 73.99 | 8.96 | - |
| Solyc07g049550 | 1-aminocyclopropane-1-carboxylate oxidase | 0.12 | 0.03 | 0.31 | 0.29 |
| Solyc07g026650 | 1-aminocyclopropane-1-carboxylate oxidase 5 | 0.05 | - | 0.17 | - |
| Solyc02g036350 | 1-aminocyclopropane-1-carboxylate oxidase | 0.14 | - | 0.07 | - |
| Solyc09g089580 | 1-aminocyclopropane-1-carboxylate oxidase-like protein | 0.07 | 0.01 | 0.07 | - |
| Gene ID | Annotation | T1d3:CTd3 | T2d3:CTd3 | T1d7:CTd7 | T2d7:CTd7 |
|---|---|---|---|---|---|
| Solyc05g055070 | Ethylene receptor | 0.4 | - | 0.52 | - |
| Solyc06g053710 | Ethylene receptor homolog (ETR4) | - | - | - | - |
| Solyc09g075440 | Never ripe | 0.11 | 0.33 | 0.06 | - |
| Solyc09g009090 | CTR1-like protein kinase 3 | - | - | - | - |
| Solyc10g083610 | Ethylene-inducible CTR1-like protein kinase | - | - | - | - |
| Solyc09g007870 | Ethylene insensitive 2 | - | 1.58 | 0.52 | - |
| Solyc01g014480 | Ethylene insensitive 3 class transcription factor | - | - | 1.91 | - |
| Solyc07g008250 | EIN3-binding F-box protein | 0.37 | - | 0.24 | - |
| Solyc12g009560 | EIN3-binding F-box protein 1 | 0.62 | 2.31 | 0.42 | - |
| Solyc01g095500 | Ethylene response factor g1 | - | - | - | |
| Solyc01g065980 | Ethylene response factor e4 | 0.05 | 0.43 | 0.02 | - |
| Solyc06g082590 | Ethylene response factor e3 | 7.82 | 4.23 | 13.31 | 0.39 |
| Solyc09g075420 | Ethylene response factor e1 | - | 0.54 | - | - |
| Solyc10g050970 | Ethylene response factor d4 | 0.14 | - | 145.91 | - |
| Solyc01g108240 | Ethylene response factor d3 | - | - | - | - |
| Solyc12g056590 | Ethylene response factor d2 | - | 1.66 | 5.67 | - |
| Solyc04g051360 | Ethylene response factor d1 | 0.27 | - | 17.94 | - |
| Solyc03g093560 | Ethylene response factor c6 | 0.19 | 0.38 | 0.04 | 0.1 |
| Solyc02g077370 | Ethylene response factor c5 | 43.87 | 37.77 | 128.16 | - |
| Solyc03g123500 | Ethylene response factor c4 | 0.36 | 0.53 | 0.26 | - |
| Solyc09g066360 | Ethylene response factor c3 | 7.58 | 87.66 | - | - |
| Solyc04g014530 | Ethylene response factor c2 | - | - | ||
| Solyc03g093610 | Ethylene response factor a2 | 0.13 | - | 0.26 | 0.24 |
| Gene ID | Annotation | T1d3:CTd3 | T2d3:CTd3 | T1d7:CTd7 | T2d7:CTd7 |
|---|---|---|---|---|---|
| Solyc11g030600 | DNA (Cytosine-5)-methyltransferase | 1.68 | 1.69 | - | 0.61 |
| Solyc12g100330 | Cytosine-specific methyltransferase | 1.93 | 0.69 | 1.87 | - |
| Solyc08g005400 | Cytosine-specific methyltransferase | - | - | 3.19 | - |
| Solyc02g062740 | DNA (Cytosine-5-)-methyltransferase 3 | 0.59 | - | 0.63 | - |
| Solyc10g078190 | DNA (Cytosine-5-)-methyltransferase 3 | - | - | 2.15 | - |
| Solyc04g005250 | DNA (Cytosine-5-)-methyltransferase 3 | - | - | - | - |
| Solyc09g009080 | Repressor of silencing 1 | - | 2.27 | 3.1 | - |
| Solyc10g083630 | Repressor of silencing 2b | 0.06 | 0.25 | 0.05 | - |
| Solyc11g007580 | HhH-GPD family protein | 0.24 | 0.74 | 0.31 | - |
| Gene ID | Annotation | T1d3:CTd3 | T2d3:CTd3 | T1d7:CTd7 | T2d7:CTd7 |
|---|---|---|---|---|---|
| Solyc02g090890 | Zeaxanthin epoxidase | 6.63 | 16.54 | 0.46 | - |
| Solyc01g097810 | Zeta-carotene desaturase | 0.21 | 0.26 | 0.15 | - |
| Solyc02g081330 | Phytoene synthase 2 | 0.29 | - | 0.19 | - |
| Solyc03g031860 | Phytoene synthase 1 | 0.05 | 0.05 | 0.01 | - |
| Solyc10g079480 | Beta-lycopene cyclase | - | 16.09 | 61.63 | - |
| Solyc04g040190 | Lycopene beta-cyclase | 0.21 | - | 0.12 | - |
| Solyc06g074240 | Lycopene beta cyclase | 0.14 | 0.05 | - | - |
| Solyc10g083790 | Cytochrome P450 | - | - | - | - |
| Solyc10g081650 | Carotenoid isomerase | 0.1 | 0.28 | 0.08 | - |
| Solyc03g007960 | Beta-carotene hydroxylase-2 | 0.07 | 0.07 | 0.01 | - |
| Solyc01g009230 | Xanthine dehydrogenase/oxidase | 2.76 | 2.37 | - | 0.48 |
| Solyc01g108210 | Cytochrome P450 | 6.52 | 5.71 | - | - |
| Solyc03g123760 | Phytoene desaturase | 0.11 | 0.3 | 0.08 | - |
| Solyc04g050930 | Violaxanthin de-epoxidase | 0.06 | 0.22 | 0.06 | - |
| Solyc04g051190 | Cytochrome P450 | - | - | - | - |
| Solyc04g071940 | Xanthoxin dehydrogenase | 0.5 | 0.48 | 0.29 | - |
| Gene ID | Annotation | T1d3:CTd3 | T2d3:CTd3 | T1d7:CTd7 | T2d7:CTd7 |
|---|---|---|---|---|---|
| Solyc10g080210 | Polygalacturonase-2 precursor | 0 | 0 | 0 | 2.22 |
| Solyc06g060170 | Probable polygalacturonase-like | - | - | - | - |
| Solyc12g098340 | Probable pectinesterase 29-like | - | - | - | - |
| Solyc03g083360 | Probable pectinesterase | 0.13 | 0.52 | - | - |
| Solyc03g078090 | Probable pectinesterase | - | - | - | - |
| Solyc07g017600 | Pectinesterase | 0.03 | - | - | - |
| Solyc09g010210 | Endo-1,4-beta-glucanase precursor | 0.01 | - | 0 | - |
| Solyc02g091680 | Probable beta-D-xylosidase 6-like | 0.3 | 0.47 | 0.32 | - |
| Solyc01g104950 | Beta-xylosidase | 0.27 | - | - | - |
| Solyc10g047030 | Beta-D-xylosidase 1 precursor | 0 | - | 0.01 | - |
| Solyc09g005850 | Probable pectate lyase 4-like | - | - | - | 2.09 |
| Solyc03g111690 | Probable pectate lyase 18-like | 0 | 0.02 | 0 | - |
| Solyc09g091430 | Probable pectate lyase 15-like | 0.01 | 0.33 | 0.01 | 5.85 |
| Solyc03g031840 | Expansin precursor | - | 11.81 | - | - |
| Solyc06g051800 | Expansin 1 | 0.09 | 0.13 | 0 | - |
| Solyc10g086520 | Expansin precursor 6 | 0.22 | 0.18 | - | - |
| Solyc02g088100 | Expansin precursor 5 | - | 14.93 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bobokalonov, J.; Liu, Y.; Mahalak, K.K.; Firrman, J.A.; Sheen, S.; Zhou, S.; Liu, L. Transcriptomics Analysis of Tomato Ripening Regulated by Carbon Dioxide. Sci 2023, 5, 26. https://doi.org/10.3390/sci5030026
Bobokalonov J, Liu Y, Mahalak KK, Firrman JA, Sheen S, Zhou S, Liu L. Transcriptomics Analysis of Tomato Ripening Regulated by Carbon Dioxide. Sci. 2023; 5(3):26. https://doi.org/10.3390/sci5030026
Chicago/Turabian StyleBobokalonov, Jamshed, Yanhong Liu, Karley K. Mahalak, Jenni A. Firrman, Shiowshuh Sheen, Siyuan Zhou, and LinShu Liu. 2023. "Transcriptomics Analysis of Tomato Ripening Regulated by Carbon Dioxide" Sci 5, no. 3: 26. https://doi.org/10.3390/sci5030026
APA StyleBobokalonov, J., Liu, Y., Mahalak, K. K., Firrman, J. A., Sheen, S., Zhou, S., & Liu, L. (2023). Transcriptomics Analysis of Tomato Ripening Regulated by Carbon Dioxide. Sci, 5(3), 26. https://doi.org/10.3390/sci5030026








