Abstract
Double-strand breaks of DNA may lead to discontinuous DNA and consequent loss of genetic information, which may result in mutations or, ultimately, carcinogenesis. To avoid such potentially serious situations, cells have evolved efficient DNA damage repair systems. It is thought that DNA-repair processes involve drastic alterations of chromatin and histone structures, but detection of these altered structures in DNA-damaged cells remains rare in the literature. Recently, synchrotron radiation circular dichroism (SRCD) spectroscopy, which can provide secondary structural information of proteins in solution, has identified structural alterations of histone proteins induced by DNA damage responses. In this review, these results and experimental procedures are discussed with the aim of facilitating further studies of the chromatin remodeling and DNA damage repair pathways using SRCD spectroscopy.
1. Introduction
Nucleosomes are the basic building blocks of chromatin in eukaryotic nuclei. The nucleosome comprises a core histone, around which 146–147 base pairs of DNA are wrapped. The core histone is an octamer of histone proteins; two H2A–H2B dimers and an H3–H4 tetramer. Histone proteins have been shown to play a significant role in repairing potentially harmful DNA damage caused by various stress factors such as endogenously arising reactive oxygen species [1], ultraviolet radiation [2], and ionizing radiation [3]. These factors cause various types of DNA damage including modification of bases, formation of DNA adducts, cross-linking of the DNA strands, and single- and double-strand breaks (DSBs) [4]. DSBs are particularly harmful as they may create discontinuous DNA and a loss of genetic information, resulting in mutation or, ultimately, carcinogenesis if they are not properly removed from the genome by enzymatic systems. To attenuate these effects, cells have evolved DNA damage response (DDR) mechanisms, including the DSB repair pathway and cell-cycle regulation.
There have been a considerable number of studies and reviews devoted to DSB repair processes [3,4,5,6,7,8,9,10]. The MRN complex, which consists of Mre11, Rad50, and Nbs1 proteins, recognizes the free ends of broken DNA and recruits ATM protein kinase [11] which phosphorylates serine 139 of histone H2AX—a variant of H2A—in the vicinity of the DSB site [12]. Phosphorylated H2AX, which is well known as γ-H2AX, then binds to MDC1 protein to create a docking site for an additional MRN-ATM complex [13]. The additional ATM further phosphorylates the proximal H2AX around the DSB site [14]. This cycle leads to the formation of a megabase-sized γ-H2AX domain surrounding the DSB site, which is microscopically observed as γ-H2AX foci [15]. The γ-H2AX foci are widely used as a marker with which to observe the induction of DSBs in living cells. Furthermore, the histone acetyltransferase TIP60 is recruited and acetylates γ-H2AX, after which it associates with the E2-ubiquitin-conjugating enzyme UBC13 to regulate polyubiquitination of acetylated γ-H2AX [16]. The polyubiquitinated and acetylated γ-H2AX is then removed from the chromatin. Phosphorylation of MDC1 is carried out by ATM [17,18,19], and the phosphorylated protein recruits the RNF8-UBC13 complex to ubiquitinate H2A and H2AX [19,20,21]. The E3-ubiquitin ligase RNF168 binds to ubiquitinated H2A and H2AX to promote the formation of ubiquitin conjugates [22]. Finally, BRCA1 binds to the polyubiquitinated histone through RAP80, and locates near the DSB site [23,24,25]. Together, these events facilitate DSB repair and/or cell-cycle checkpoints. After the accumulation of repair proteins, DSB repair proceeds through two distinct mechanisms, the nonhomologous-end-joining (NHEJ) and homologous-recombination (HR) pathways [4,8,9]. The former involves minimal processing of the damage by nucleases, followed by direct re-ligation of the damaged DNA ends. It can be activated in all cell-cycle phases, but it is an error-prone repair pathway. In contrast, although HR is error-free, it requires adjacent sister chromatids for homology searching, and is therefore restricted to the S and G2 phases. Thus, sequential recruitment of DSB repair proteins and post-translational modifications of histone proteins are preferentially induced at early stages of DSB repair processes.
The access/prime-repair-restore model [26,27] is a well-known representation of chromatin remodeling which assumes that the abovementioned DSB repair processes involve drastic alterations of chromatin structure. For example, the model predicts that core histone proteins are displaced from chromatin around the DNA damage sites in order to give repair proteins access to the damage site before the repair process is initiated, and are repositioned after repair is completed. Eviction of the H2A–H2B dimer from chromatin does occur within 30 min of the induction of DSBs, but histone levels around the DSB sites return to normal within 2 h after the induction of DSBs [28]. On the other hand, chromatin remodeling processes, which are thought to be induced by post-translational modifications of the histones, have not yet been completely elucidated. Although some groups have reported that acetylation [29,30,31,32] and methylation [33,34,35] of histone peptides and proteins induce structural changes in vitro, reports of structural alterations of chromatin or histones during DDR or relationships between such structural alterations and post-translational modifications in vivo are scarce.
Recently, our group discovered that the secondary structure alterations of histones are induced by X-ray irradiation of human cancer cells [36,37,38] through circular dichroism (CD) spectroscopic analysis. In the present review, these results and experimental procedures are discussed with the aim of facilitating further studies of chromatin remodeling and DNA damage repair pathways using CD spectroscopy. In the next section, CD spectroscopy for protein structure analysis is briefly introduced. In Section 3, experimental results are discussed. Future perspectives are described in the final section.
2. CD Spectroscopy
CD spectra provide data relating to the secondary structures of proteins; namely, α-helices, β-strands, turns, unordered structures, and so on. This information is somewhat limited compared with X-ray crystallography and nuclear magnetic resonance (NMR), both of which can be used to obtain three-dimensional structures with atomic-level resolution. Nonetheless, CD spectroscopy is a powerful tool because it provides structural information, including structural dynamics, which is difficult for general X-ray crystallography, with the following advantages over the above techniques. In particular, samples can be prepared by simply dissolving the protein in a solvent, that is, neither crystallization or isotopic substitution are required. Therefore, sample losses and accidental denaturation during sample preparation are negligible in most cases. In addition, smaller sample quantities are required than for general X-ray crystallography or NMR. Most importantly, CD spectroscopy is highly sensitive to structural changes. Thus, CD can detect even subtle structural changes of histones due to DNA lesions.
2.1. CD and Its Notation
The phenomenon of CD is observed in the absorption bands of optically active molecules such as proteins, and is defined as the difference between the molar absorption coefficients for left circularly polarized light (LCPL) εL and right circularly polarized light (RCPL) εR (Equation (1)):
The CD value Δε is sometimes referred to as the “molar CD.”
For historical reasons, CD intensity is often expressed as ellipticity. Indeed, commercial CD spectrophotometers often produce CD spectra in terms of ellipticity (θ, in millidegrees). Ellipticity can be converted to the molar CD using Equation (2):
where MRW is mean residue weight, defined as the molecular weight of a protein divided by its number of amino-acid residues; c is the concentration of the sample (in mg/mL); and l is the path length of the sample cell (in cm). The mean residue ellipticity [θ] (in degrees cm2 dmol−1) is also commonly used to describe CD intensity, and is calculated using Equation (3):
2.2. Advantages of CD Spectroscopy Using Synchrotron Radiation
Although CD spectroscopy can be performed using commercial benchtop CD spectrophotometers, the author would like to recommend the use of synchrotron radiation (SR) facilities for CD analysis. The advantages of this approach are reviewed elsewhere [39]. Briefly, the photon flux of SR is higher than that of xenon lamps (used as light sources in commercial CD spectrophotometers) in the vacuum ultraviolet (VUV) region (wavelength <~200 nm). Therefore, SR can extend the CD spectra to the wavelength region below 190 nm and thus provide information which is unobtainable using commercial CD instruments. The use of SR is essential for CD measurements of chiral molecules that contain only single bonds, such as saccharides, because the CD peaks are detectable only below 190 nm [40,41].
At the time of writing, to my knowledge, synchrotron radiation circular dichroism (SRCD) spectroscopy can be performed at eight SR facilities: Hiroshima Synchrotron Radiation Center (HiSOR) in Japan [42], Institute for Storage Ring Facilities (ISA) in Denmark [43], Beijing Synchrotron Radiation Facility (BSRF) in China [44], National Synchrotron Radiation Research Center (NSRRC) in Taiwan [45], Diamond Light Source (DLS) in UK [46], Synchrotron SOLEIL in France [47], BESSY-II [48], and ANKA [49] in Germany. In general, SRCD experiments can be performed after the acceptance of proposal(s), which can be referred to the websites of each SR facility. The experimental data discussed in this review were obtained using the HiSOR and Synchrotron SOLEIL [37,38].
2.3. Experimental Procedure for CD Spectroscopy
2.3.1. Sample Preparation
Proteins are usually prepared in liquid solutions for CD analysis. The solvent must be carefully selected through samples can be prepared by simply dissolving the protein in an appropriate solvent. Chloride ions interfere with CD measurements, even when using SR, because those exhibit strong absorption bands in the VUV region. Therefore, sodium chloride—which is often included in protein solutions—should be removed. Phosphate buffers and sodium fluoride (NaF), as a salt, are preferable, but concentrations should be kept as low as possible.
To reduce absorption of solvents, the path length of the sample cell used for SRCD measurements is shorter than for conventional ones (below 100 μm for SRCD vs. 1–10 mm for conventional CD). Therefore, the sample concentration needs to be much higher for SRCD than conventional CD because the measured intensities, such as θ, depend on the concentration of the sample and the path length of the sample cell (Equation (2)). In general, the optimum protein concentration for analysis with a sample-cell path length of 10 μm is 10 mg/mL for α-helical proteins and 15–20 mg/mL for β-sheet-rich proteins [39].
2.3.2. The Sample Cell
Demountable cells are often used for SRCD spectroscopy due to their ease of cleaning because short path length is preferable. Since SRCD measurements are sometimes carried out in the wavelength region down to 160 nm [39,42], sample cells are often made of calcium fluoride (CaF2) around which transmittance is higher than quartz (SiO2) used for conventional CD spectroscopy (the cutoff wavelength is ~140 nm for CaF2 and ~160 nm for SiO2).
As an example, the sample cell used in the HiSOR [50] is comprised of two circular CaF2 glasses and a donut-shaped Teflon spacer. The thickness of the spacer corresponds with the path length of the sample cell. After placing the sample solution on one glass, the other glass is placed on top of the first glass with the spacer sandwiched between them. The glasses are then fixed into the sample-cell holder. To avoid foaming, the recommended sample volume is generally 15 and 20 μL for 10 and 50 μm cells, respectively, although convenient volumes depend on the type of solution and skill of the user. Small-capacity sample cells (2–3 μL in volume) are also used in combination with a focusing mirror, although the measurable region is narrower owing to the use of SiO2 glasses [35].
2.3.3. Measurements and Analyses of CD Spectra
The CD measurement systems used in SR facilities are similar to those of commercial CD spectrophotometers, although important details should be checked in publications [42,43,44,45,46,47,48,49] and on the websites of each facility. In general, SR is monochromated and converted LCPL or RCPL using a phase shifter (photo-elastic modulator). The intensities of transmitted LCPL and RCPL passing through the sample are detected and the CD spectra are then generated.
Prior to analysis, measured CD intensities must be converted to Δε values using Equation (2). Spectra can then be analyzed using empirical programs that are publicly available, see [51,52,53,54] for example. Because the results tend to depend on the reference data set, which is used in the programs, it is advisable to select the program that is recommended by the staff of the SRCD beamline being used. Analyses are easily performed using these programs by inputting the measured CD data, and calculations can be completed in a short time on standard personal computers or over the internet.
3. Secondary Structure Analyses of Histones Extracted from X-Irradiated Cells
3.1. Experimental Procedure
The sample preparation procedures used in the present study have been previously described in detail [36,37,38]. Briefly, cultured human cancer cells (HeLa.S-FUCCI cells) were irradiated with X-rays at a dose of 40 Gy. An X-ray generator equipped with a tungsten anode (M-150WE; SOFTEX, Kanagawa, Japan) was used. The tube voltage and current were set at 150 kV and 6 mA. The energies of the characteristic X-rays emitted from the anode were 59.318 (Kα1 line), 57.982 (Kα2 line), 67.244 (Kβ1 line), and 69.067 (Kβ2 line) keV [55]. An aluminum filter (0.2 mm thick) was used to reduce the low energy component (<10 keV) of bremsstrahlung X-ray. The dose of 40 Gy is thought to induce ~1600 DSBs per cell nucleus. It is thought that those damages are produced by the radiation-induced breakage of the chemical bonds and the reaction with free radicals produced by ionization of the surrounding water molecules [56]. Assuming such DSBs are randomly spaced in DNA, an average DNA fragment length is ~2 megabase pairs (Mbp) since the human genome is ~3000 Mbp. Since phosphorylation of H2AX spreads along ~2 Mbp from the DSB sites in the early stage of DNA repair processes [15], it is expected that most of the histones in the X-irradiated cells are involved in DDRs. After irradiation, cells were incubated for 30 min to allow for DDRs. H2A–H2B or H3–H4 proteins were extracted using a Histone Purification Kit (Active Motif, Carlsbad, CA, USA) and dissolved in 10 mM Tris-HCl buffer supplemented with 250 mM NaF. Absorption spectroscopy was carried out to determine the sample concentration and evaluate the decomposition of the histones. The spectra discussed hereafter were obtained using the V-630 Bio spectrophotometer (JASCO, Tokyo, Japan).
CD spectroscopy of H2A–H2B and H3–H4 was carried out at the DISCO beamline of Synchrotron SOLEIL [47] and BL-12 of HiSOR [42], respectively. The CD spectra that were obtained at 25 °C are discussed hereafter.
The contents of α-helices, β-strands, turns, and unordered structures were analyzed using BeStSel [54] and SELCON3 [51,52] programs for H2A–H2B and H3–H4, respectively.
3.2. Absorption and CD Spectra of Histones
Figure 1a,b shows the absorption spectra of H2A–H2B and H3–H4 extracted from unirradiated and X-irradiated cells, respectively. In both cases, apparent spectral changes were not observed. It shows that the decomposition of the histones induced by X-ray irradiation to the cells was negligible and/or the fragments of histones were discarded during the extraction procedures.
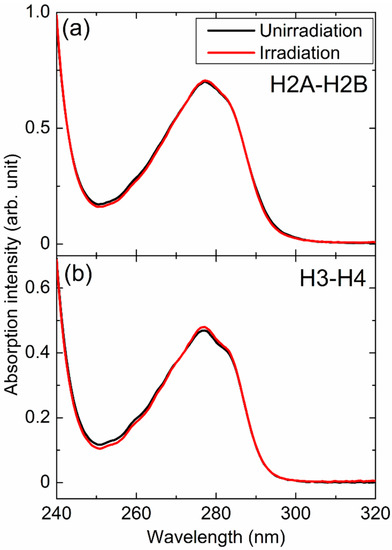
Figure 1.
Absorption spectra of (a) H2A–H2B and (b) H3–H4 extracted from unirradiated (black) and X-irradiated (red) cells.
Figure 2a,b shows the CD spectra of H2A–H2B and H3–H4 extracted from X-irradiated cells, respectively, along with spectra of equivalent samples extracted from unirradiated cells. The CD spectra of H2A–H2B [37] and H3–H4 [38] were reproduced from the previous papers. All spectra showed a positive peak around 190 nm and two negative peaks around 208 nm and 222 nm. These peaks are characteristic of the CD peaks of α-helix structures [57] and it suggests H2A–H2B and H3–H4 contain α-helix structures. The absolute values of CD peak intensities were higher for H2A–H2B extracted from irradiated cells than from unirradiated cells. In the case of H3–H4, no differences were observed between irradiated and nonirradiated samples in terms of the negative peaks, but the intensities of the positive peaks around 190 nm were lower for irradiated samples than for unirradiated samples.
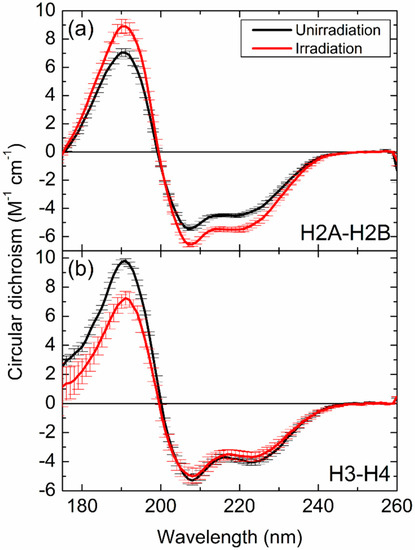
Figure 2.
Circular dichroism spectra of (a) H2A–H2B and (b) H3–H4 extracted from unirradiated (black) and X-irradiated (red) cells. Data represent the mean values; error bars represent the standard deviation. The circular dichroism (CD) spectra were reproduced from the previous papers [37,38].
The shape of the CD spectrum reflects the secondary structures, hence, the change in spectra seen in Figure 2 indicates that the structures of H2A–H2B and H3–H4 differ between irradiated and unirradiated cells. Therefore, X-ray irradiation induces structural alterations in H2A–H2B and H3–H4 proteins. Furthermore, the spectra recorded for H2A–H2B showed different changes to that obtained for H3–H4, indicating that different structural changes occur in the proteins in response to X-irradiation. When protein decomposition or peptide-bond breaks are induced by radiation or other causes, the absolute values of CD peak intensities relating to α-helical structures tend to become smaller [58]. Indeed, direct irradiation of purified H2A–H2B in solution caused a similar peak shift compared with the spectra of H2A–H2B extracted from unirradiated or X-irradiated cells [36]. Such peak shift was not observed in the spectra shown in Figure 2; therefore, the structural alterations of H2A–H2B and H3–H4 can be concluded to be unrelated to X-ray damage to the histones themselves, instead of being caused by other cellular functions such as DDRs.
3.3. Secondary Structure Alterations of Histones
The contents of the secondary structures of the histone samples are listed in Table 1 [38]. In the case of H2A–H2B, the content of α-helices increased and that of β-strands decreased after exposing the cells to X-rays. The opposite was observed for H3–H4; the contents of β-strands and α-helices increased and decreased, respectively. This supports the above conclusion that X-irradiation of cells induces different structural alterations in H2A–H2B and H3–H4, demonstrating that various structural alterations of histones are induced by DDRs.

Table 1.
Contents of secondary structures as obtained analyzing the CD spectra [37,38]. (U) and (I) indicate the samples extracted from unirradiated and X-irradiated cells, respectively.
The underlying mechanisms of histone structural alterations have not yet been identified. However, the author assumes that post-translational modifications of histones arise during DDRs [3], because such modifications would alter steric barriers and/or electrostatic interactions between modified amino-acid residues and other residues, possibly changing the stability of structures. Indeed, trimethylation of lysine-4 or -9 of histone H3, which is a post-translational modification induced during DDRs, induces global structural changes [33,34]. The different structural alterations observed for H2A–H2B and H3–H4 might, therefore, be due to different post-translational modifications. The mechanisms underlying structural alterations may be gradually revealed by examining histone structures in DNA-damaged cells while inhibiting the function of enzymes such as ATM kinase.
Presently, it is also unclear how structural alterations of histones contribute to DDRs, such as DNA repair processes. However, the author hypothesizes the altered histones play the following roles in DDRs (Figure 3). As mentioned in Section 1 briefly, various DNA-repair enzymes and proteins bind to modified histones during DNA repair pathways [3,4,5,6,7,8,9,10]. Histones altered by post-translational modifications might be landmarks for DNA-repair enzymes and proteins recruited later to distinguish sites of DNA damage from undamaged ones in the early stage of DDRs, and therefore the repair factors might be able to work specifically around DNA damage sites. Structural alterations of histones may also contribute to chromatin relaxation because the electrostatic interactions between DNA and altered histones would be different from those that exist in normal conditions. Indeed, it is known that TIP60, which acetylates H4, is required to create open chromatin structures [5] and acetylation of H4 induces the structural alteration [29,31] and destabilizes the binding of the H4 tail at the acidic patch of the nucleosome [32]. The relaxation might help histone chaperones to evict histones at the break sites [28,59].
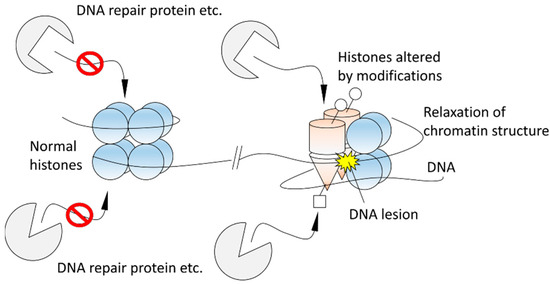
Figure 3.
A hypothesis of the role of histone structural alterations in DNA damage responses (DDRs).
If histone structural changes contribute to the DSB repair pathways, the changes might be cell-cycle dependent because the NHEJ and HR pathways predominate in the G1 and in the S and G2 phases, respectively. It would be interesting, in future work, to compare the structural alterations that occur in each phase.
4. Future Perspectives
Previous studies have revealed the CD spectra of H2A–H2B extracted from X-irradiated cells to have no significant dependence on the incubation time after X-irradiation at least within 24 h [36]. This indicates that altered H2A–H2B persists for at least 24 h in X-irradiated cells. Since most DNA lesions are repaired within this time, even after exposure to doses greater than 50 Gy [60,61], alteration and restoration of histones may not directly accompany DNA repair. Traces of DNA damage might remain in the chromatin even after completion of the repair, which might affect epigenetic regulation and cause late-onset injury to daughter or granddaughter cells, or cells of later generations. It would be of interest to examine the structures of histones extracted from descendants of X-irradiated cells. The structural changes would be triggered by DNA lesions. Therefore, different alterations may be observed depending on the number of DNA lesions, namely the irradiation dose. It is also an interesting future work to confirm the dose-dependence.
Structural analysis of DNA-repair-related proteins is important to improve our understanding of DDRs, but research into protein structures that exist in DNA-damaged cells is in its infancy. The author hopes that this review will facilitate further structural studies of DNA-repair-related proteins in DNA-damaged cells using CD spectroscopy and other experimental methods.
Funding
The research described in this review was supported financially by JSPS KAKENHI Grant Numbers JP15K16130 and JP17K12825 and the Reimei Research Program of the Japan Atomic Energy Agency.
Acknowledgments
CD spectroscopy shown in this review was carried out with the approval of Synchrotron SOLEIL (proposal numbers 20130831 and 20140518) and Hiroshima Synchrotron Radiation Center of Hiroshima University (proposal numbers: 13B-18, 14-A-16, and 15-A-48).
Conflicts of Interest
The author declares no conflict of interest.
References
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef]
- Cadet, J.; Sage, E.; Douki, T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 2005, 571, 3–17. [Google Scholar] [CrossRef]
- Hunt, C.R.; Ramnarain, D.; Horikoshi, N.; Iyengar, P.; Pandita, R.K.; Shay, J.W.; Pandita, T.K. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013, 179, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Kanaar, R.; Hoeijmakers, J.H.J.; van Gent, D.C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998, 8, 483–489. [Google Scholar] [CrossRef]
- Price, B.D.; D’Andrea, A.D. Chromatin remodeling at DNA double-strand breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- van Attikum, H.; Gasser, S.M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009, 19, 207–217. [Google Scholar] [CrossRef]
- Pandita, T.K.; Richardson, C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009, 37, 1363–1377. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Gong, F.; Miller, K.M. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 2013, 750, 23–30. [Google Scholar] [CrossRef]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Wang, B.; Bignell, C.R.; Taylor, A.M.R.; Elledge, S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003, 421, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Moll. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Ikura, T.; Tashiro, S.; Kakino, A.; Shima, H.; Jacob, N.; Amunugama, R.; Yoder, K.; Izumi, S.; Kuraoka, I.; Tanaka, K.; et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 2007, 27, 7028–7040. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.Y.; Grant, R.; Manke, I.; Minn, K.; Yu, X.; Yaffe, M.B.; Chen, J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007, 131, 901–914. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald III, E.R.; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef]
- Kolas, N.K.; Chapman, J.R.; Nakada, S.; Ylanko, J.; Chahwan, R.; Sweeney, F.D.; Panier, S.; Mendez, M.; Wildenhain, J.; Thomson, T.M.; et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007, 318, 1637–1640. [Google Scholar] [CrossRef]
- Plans, V.; Scheper, J.; Soler, M.; Loukili, N.; Okano, Y.; Thomson, T.M. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell. Biochem. 2006, 97, 572–582. [Google Scholar] [CrossRef]
- Mailand, N.; Bekker-Jensen, S.; Faustrup, H.; Melander, F.; Bartek, J.; Lukas, C.; Lukas, J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007, 131, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Panier, S.; Townsend, K.; Al-Hakim, A.K.; Kolas, N.K.; Miller, E.S.; Nakada, S.; Ylanko, J.; Olivarius, S.; Mendez, M.; et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009, 136, 420–434. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Matsuoka, S.; Ballif, B.A.; Zhang, D.; Smogorzewska, A.; Gygi, S.P.; Elledge, S.J. Abraxas and RAP80 from a BRCA1 protein complex required for the DNA damage response. Science 2007, 316, 1194–1198. [Google Scholar] [CrossRef] [PubMed]
- Sobhian, B.; Shao, G.; Lilli, D.R.; Culhane, A.C.; Moreau, L.A.; Xia, B.; Livingston, D.M.; Greenberg, R.A. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007, 316, 1198–1202. [Google Scholar] [CrossRef]
- Kim, H.; Chen, J.; Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007, 316, 1202–1205. [Google Scholar] [CrossRef]
- Smedon, M.J. DNA repair and the role of chromatin structure. Curr. Opin. Cell Biol. 1991, 3, 422–428. [Google Scholar] [CrossRef]
- Soria, G.; Polo, S.E.; Almouzni, G. Prime, repair, restore: The active role of chromatin in the DNA damage response. Moll. Cell. 2012, 46, 722–734. [Google Scholar] [CrossRef]
- Goldstein, M.; Derheimer, F.A.; Tait-Mulder, J.; Kastan, M.B. Nucleolin mediates nucleosome disruption critical DNA double-strand repair. Proc. Natl. Acad. Sci. USA 2013, 110, 16874–16879. [Google Scholar] [CrossRef]
- Adler, A.J.; Fasman, G.D.; Wangh, L.J.; Allfrey, V.G. Altered conformational effects of naturally acetylated histone f2al (IV) in f2al-deoxyribonucleic acid complexes. Circular dichroism studies. J. Biol. Chem. 1974, 249, 2911–2914. [Google Scholar]
- Prevelige, P.E., Jr.; Fasman, G.D. Structural studies of acetylated and control inner core histones. Biochem. 1987, 26, 2944–2955. [Google Scholar] [CrossRef]
- Wang, X.; Moore, S.C.; Laszckzak, M.; Ausió, J. Acetylation increases the α-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000, 275, 35013–35020. [Google Scholar] [CrossRef]
- Yang, D.; Arya, G. Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys. Chem. Chem. Phys. 2011, 13, 2911–2921. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Matsuo, K.; Fujii, K.; Yokoya, A.; Taniguchi, M.; Namatamae, H. Circular dichroism spectroscopic study on structural alterations of histones induced by post-translational modifications in DNA damage responses: Lysine-9 methylation of H3. J. Radiat. Res. 2018, 59, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Matsuo, K.; Namatame, H. Structural analysis of lysine-4 methylated histone H3 proteins using synchrotron radiation circular dichroism spectroscopy. Chirality 2018, 30, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Matsuo, K. Sample volume reduction using the Schwarzschild objective for a circular dichroism spectrophotometer and an application to the structural analysis of lysine-36 trimethylated histone H3 protein. Molecules 2018, 23, 2865. [Google Scholar] [CrossRef]
- Izumi, Y.; Yamamoto, S.; Fujii, K.; Yokoya, A. Secondary structure alterations of histones H2A and H2B in X-irradiated human cancer cells: Altered histones persist in cells for at least 24 hours. Radiat. Res. 2015, 184, 554–558. [Google Scholar] [CrossRef]
- Izumi, Y.; Fujii, K.; Wien, F.; Houée-Lévin, C.; Lacombe, S.; Salado-Leza, D.; Porcel, E.; Masoud, R.; Yamamoto, S.; Réfrégiers, M.; et al. Structure change from β-strand and turn to α-helix in histone H2A–H2B induced by DNA damage response. Biophys. J. 2016, 111, 69–78. [Google Scholar] [CrossRef]
- Izumi, Y.; Fujii, K.; Yamamoto, S.; Matsuo, K.; Namatame, H.; Taniguchi, M.; Yokoya, A. DNA damage response induces structural alterations in histone H3–H4. J. Radiat. Res. 2017, 58, 59–65. [Google Scholar] [CrossRef]
- Miles, A.J.; Wallace, B.A. Synchrotron radiation circular dichroism spectroscopy of proteins and applications in structural and functional genomics. Chem. Soc. Rev. 2006, 35, 39–51. [Google Scholar] [CrossRef]
- Matsuo, K.; Gekko, K. Vacuum-ultraviolet circular dichroism study of saccharides by synchrotron radiation spectrometry. Carbohydr. Res. 2004, 339, 591–597. [Google Scholar] [CrossRef]
- Matsuo, K.; Namatame, H.; Taniguchi, M.; Gekko, K. Vacuum-ultraviolet electronic circular dichroism study of methyl α-d-glucopyranoside in aqueous solution by time-dependent density functional theory. J. Phys. Chem. A 2012, 116, 9996–10003. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, K.; Gekko, K. Construction of a synchrotron-radiation vacuum-ultraviolet circular-dichroism spectrophotometer and its application to the structural analysis of biomolecules. Bull. Chem. Soc. Jpn. 2013, 86, 675–689. [Google Scholar] [CrossRef]
- Miles, A.J.; Hoffmann, S.V.; Tao, Y.; Janes, R.W.; Wallace, B.A. Synchrotron radiation circular dichroism (SRCD) spectroscopy: New beamlines and new applications in biology. Spectroscopy 2007, 21, 245–255. [Google Scholar] [CrossRef]
- Tao, Y.; Huang, Y.; Gao, Z.; Zhuang, H.; Zhou, A.; Tan, Y.; Li, D.; Sun, S. Developing VUV spectroscopy for protein folding and material luminescence on beamline 4B8 at the Beijing Synchrotron Radiation Facility. J. Synchrotron Rad. 2009, 16, 857–863. [Google Scholar] [CrossRef]
- Liu, S.-H.; Lin, Y.-H.; Huang, L.-J.; Luo, S.-W.; Tsai, W.-L.; Chiang, S.-Y.; Fung, H.-S. Design and construction of a compact end-station at NSRRC for circular-dichroism spectra in the vacuum-ultraviolet region. J. Synchrotron Rad. 2010, 17, 761–768. [Google Scholar] [CrossRef]
- Hussain, R.; Jávorfi, T.; Siligardi, G. Circular dichroism beamline B23 at the Diamond Light Source. J. Synchrotron Rad. 2012, 19, 132–135. [Google Scholar] [CrossRef]
- Réfrégiers, M.; Wien, F.; Ta, H.-P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.-L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron Rad. 2012, 19, 831–835. [Google Scholar] [CrossRef]
- Reichardt, G.; Bahrdt, J.; Schmidt, J.-S.; Gudat, W.; Ehresmann, A.; Müller-Albrecht, R.; Molter, H.; Schmoranzer, H.; Martins, M.; Schwentner, N.; et al. A 10 m-normal incidence monochromator at the quasi-periodic undulator U125-2 at BESSY II. Nucl. Instr. Methods Phys. Res. A 2001, 467–468, 462–465. [Google Scholar] [CrossRef]
- Bürck, J.; Roth, S.; Windisch, D.; Wadhwani, P.; Moss, D.; Ulrich, A.S. UV-CD12: Synchrotron radiation circular dichroism beamline at ANKA. J. Synchrotron Rad. 2015, 22, 844–852. [Google Scholar] [CrossRef]
- Matsuo, K.; Sakai, K.; Matsushima, S.; Fukuyama, T.; Gekko, K. Optical cell with a temperature-control unit for a vacuum-ultraviolet circular dichroism spectrophotometer. Anal. Sci. 2003, 19, 129–132. [Google Scholar] [CrossRef]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of the number of α-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999, 8, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2004, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Thomson, A.C.; Attwood, D.T.; Gullikson, E.M.; Howells, M.R.; Kortright, J.B.; Robinson, A.L.; Underwood, J.H. X-ray Data Booklet, 2nd ed.; Thomson, A.C., Vaughan, D., Eds.; Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 2001. [Google Scholar]
- von Sontag, C. Free-Radical-Induced DNA Damage and Its Repair; Springer-Verlag: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Greenfield, N.; Fasman, G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry 1969, 8, 4108–4116. [Google Scholar] [CrossRef]
- Wien, F.; Miles, A.J.; Lees, J.G.; Hoffmann, V.; Wallace, B.A. VUV irradiation effects on proteins in high-flux synchrotron radiation circular dichroism spectroscopy. J. Synchrotron Rad. 2005, 12, 517–523. [Google Scholar] [CrossRef]
- Kobayashi, J.; Fujimoto, H.; Sato, J.; Hayashi, I.; Burma, S.; Matsuura, S.; Chen, D.J.; Komatsu, K. Nucleolin participates in DNA double-strand break-induced damage response through MDC1-dependent pathway. PLoS ONE 2012, 7, e49245. [Google Scholar] [CrossRef]
- Banáth, J.P.; MacPhail, S.H.; Olive, P.L. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004, 64, 7144–7149. [Google Scholar] [CrossRef]
- Kühne, M.; Riballo, E.; Rief, N.; Rothkamm, K.; Jeggo, P.A.; Löbrich, M. A double-strand break repair defect in ATM-deficient cells contributes to radiosensitivity. Cancer Res. 2004, 64, 500–508. [Google Scholar] [CrossRef]
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).