A Comparative Study Between Clinical Optical Coherence Tomography (OCT) Analysis and Artificial Intelligence-Based Quantitative Evaluation in the Diagnosis of Diabetic Macular Edema
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Data Collection
2.2. OCT Image Acquisition and Analysis
2.3. Feature Vector and Preprocessing
- Demographic and clinical data: Patient ID, age, sex (male/female), and eye laterality (right/left).
- Visual acuity: Patient’s visual acuity.
- ETDRS parameters: 18 features derived from ETDRS thickness and volume maps, covering the nine macular sectors (e.g., etdrs9_1 to etdrs9_9 for thickness and etdrs9v_1 to etdrs9v_9 for volume).
- Other OCT metrics: Fovea minima (foveamin) and total area volume (whole/total).
- Diagnosis: The phenotype verified by the physician, which served as the label for supervised learning.
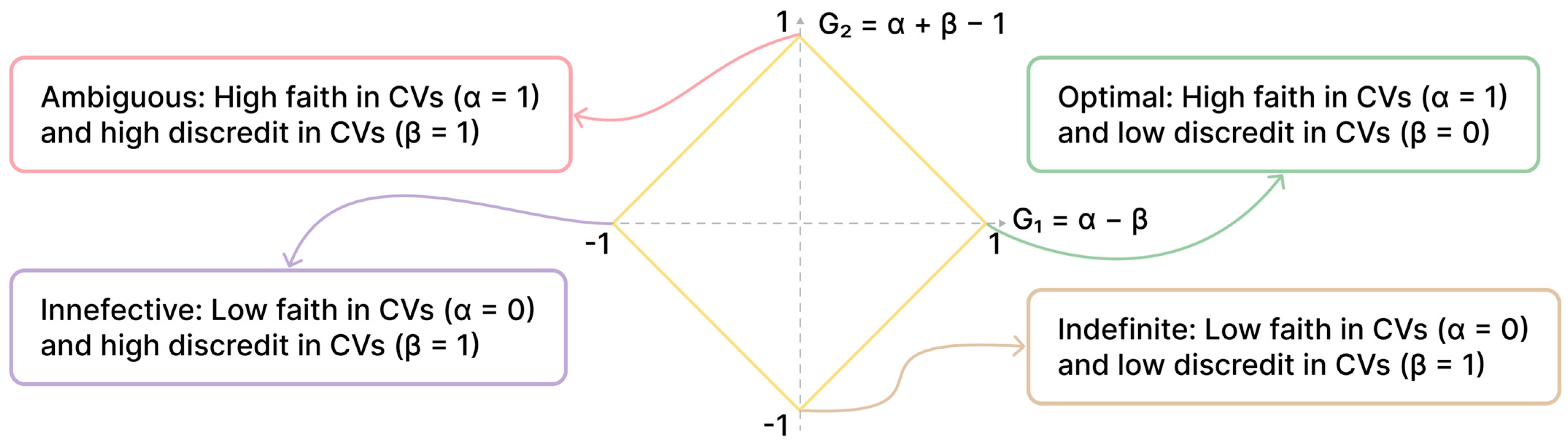
2.4. Paraconsistent Feature Engineering (PFE)
- α (Intraclass Similarity): Measures how similar the values of a feature are within the same class (e.g., all patients with DME).
- β (Interclass Dissimilarity): Measures how different the values of a feature are between different classes (e.g., between patients with and without DME).
- ‘R/L eye’: The laterality of the examined eye.
- ‘etdrs9v_7’: The volume of the external nasal ring.
- ‘sex’: The patient’s sex.
- ‘etdrs9_6’: The thickness of the superior external ring.
2.5. Artificial Intelligence Models
- Logistic Regression (LR): A linear classifier commonly used in medical diagnosis due to its interpretability and effectiveness in binary classification tasks [28].
- Support Vector Machines (SVM): A robust algorithm that finds an optimal hyperplane to separate data into classes. It is particularly effective in high-dimensional spaces and for nonlinear problems when combined with kernel functions [29].
- K-Nearest Neighbors (KNN): A nonparametric method that classifies a new sample based on the majority class of its ‘k’ nearest neighbors in the feature space. It is intuitive and useful when the relationship between variables is complex and nonlinear [29].
- Decision Trees (DTREE): Highly interpretable models that use a hierarchical tree structure to make decisions, dividing the feature space into homogeneous subsets [30].
2.6. Experimental Scenarios and Performance Evaluation
- Scenario 1 (Binary Classification): This task classified the scans into two categories: Y (Yes), for patients with DME, and N (No), for patients without DME. This scenario included 131 positive cases (Y) and 569 negative cases (N).
- Scenario 2 (Multiclass Classification): A more complex task with six phenotypes: Y (Yes, with DME), Y-Mer (Yes, with epiretinal membrane), Y-Perifoveal (Yes, with perifoveal edema), N (No), N-Anomalies (No, but with other anomalies), and N-Mer (No, but with epiretinal membrane).
2.7. Statistical Analysis
3. Results
- Demographic Data
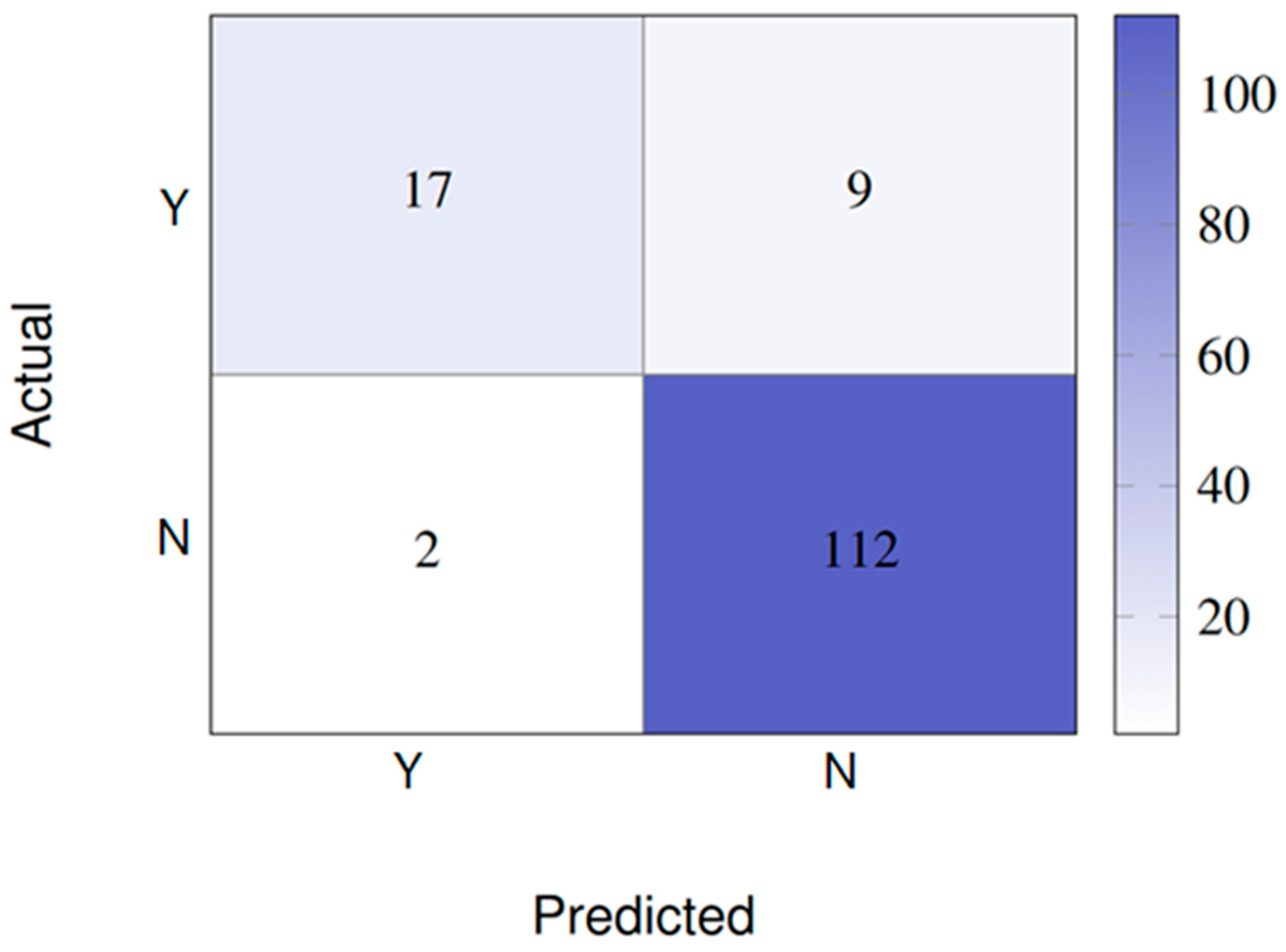
- Scenario 1: binary classification (presence vs. absence of DME)
- With 24 features:
- With four features (PFE):
- Scenario 2: Multiclass classification (six phenotypes)
- With 24 features:
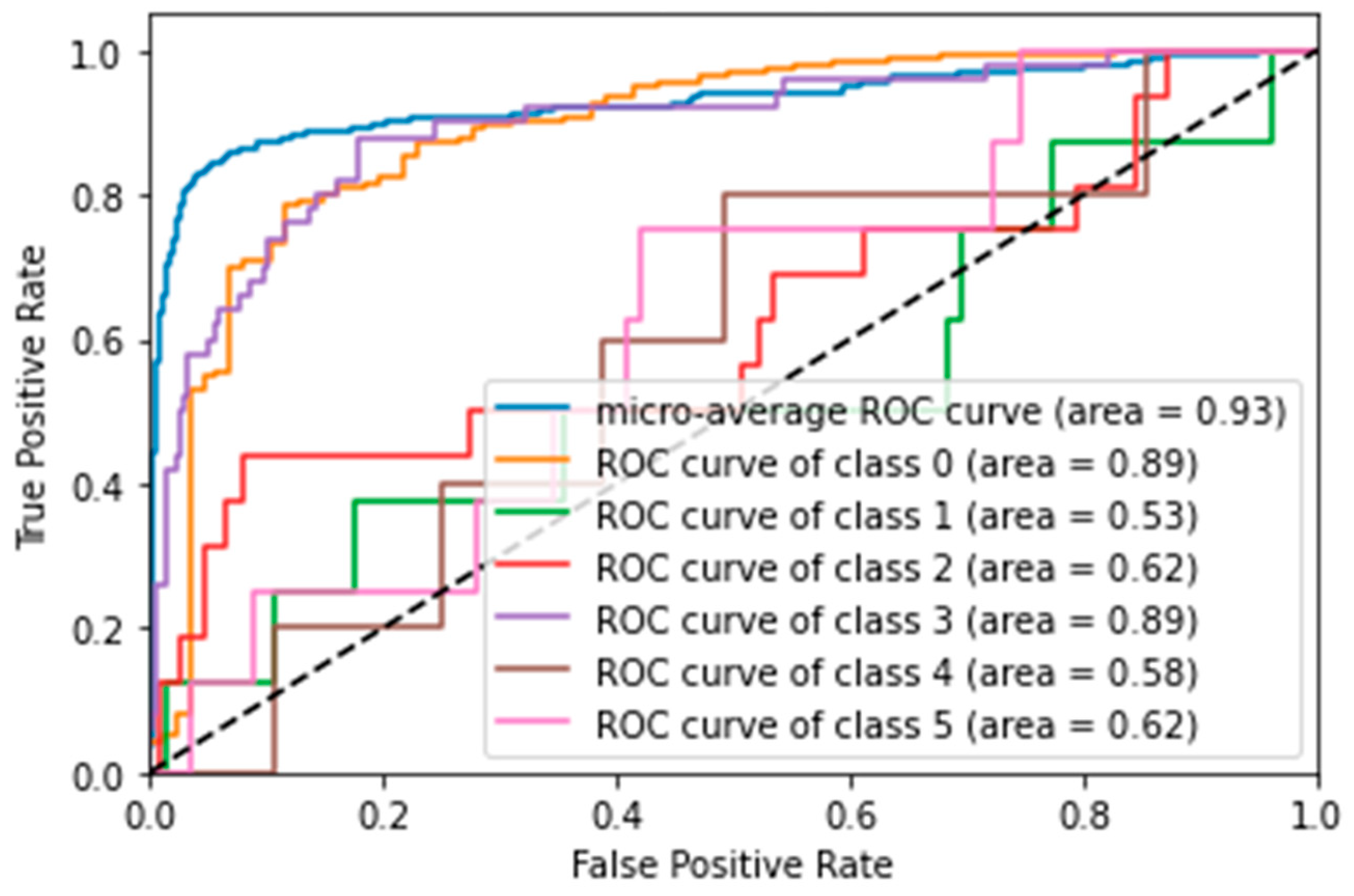
- SVM was the best-performing model, achieving an accuracy of 84.3% and an AUC score of 82.7%. ROC curve analysis (Figure 3) showed that the model was particularly good at distinguishing the ‘N’ (No: AUC = 0.89) and ‘Y’ (Yes: AUC = 0.89) classes, but struggled with less frequent classes, such as ‘N-Anomalies’ (AUC = 0.53).
- LR also performed well, with an accuracy of 81%.
- KNN and DTREE had accuracies of 80.7% and 68.6%, respectively.
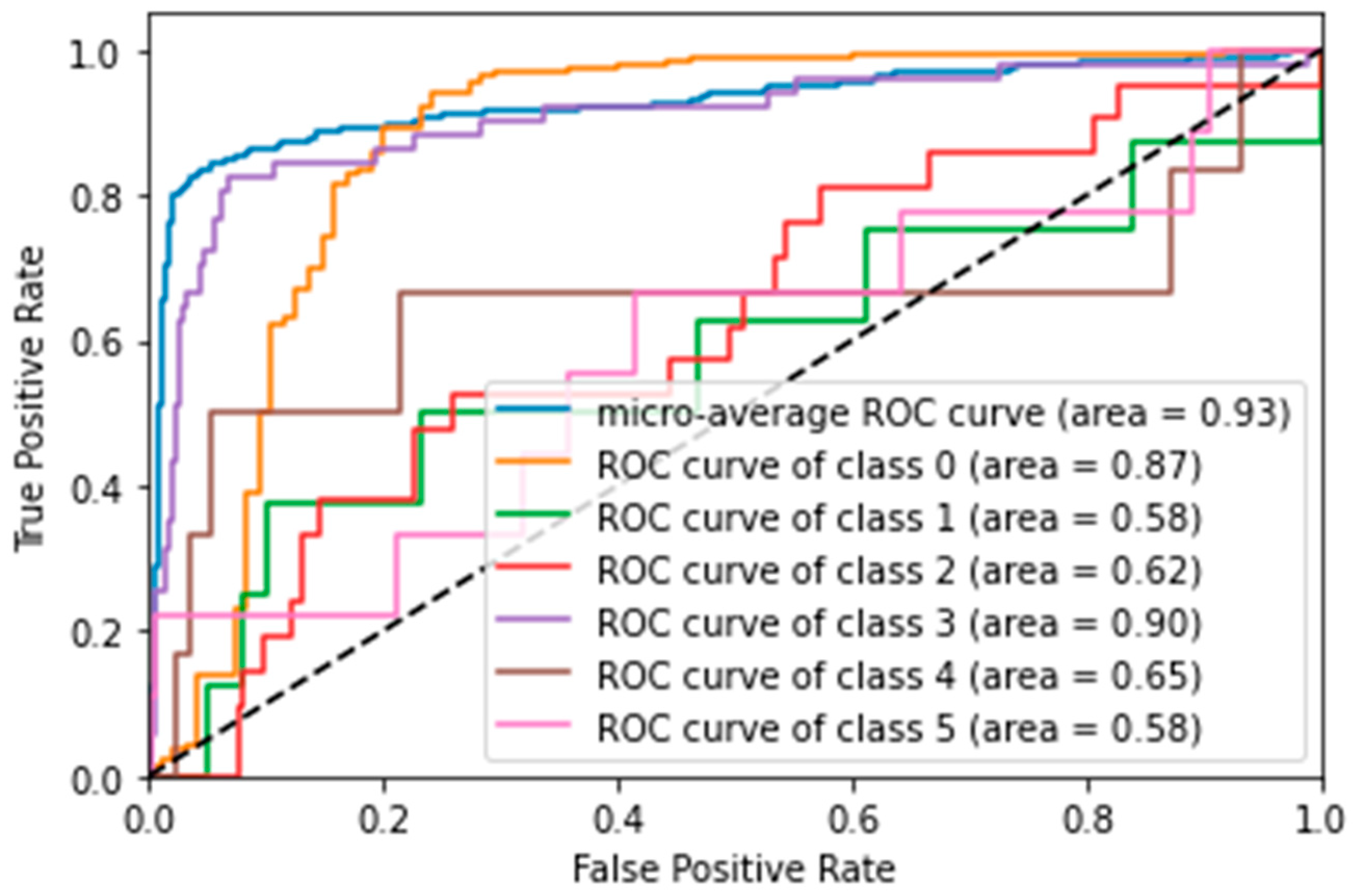
- With four features (PFE):
- Again, performance decreased with the reduced feature set. LR was the best model, with an accuracy of 78%, closely followed by SVM with 77%.
- ROC curve analysis for SVM with four features (Figure 4) showed that the discrimination ability for the ‘Y’ class improved slightly (AUC = 0.90), but overall, the performance remained inferior to the model with 24 features.
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapoor, R.; Walters, S.P.; Al-Aswad, L.A. The Current State of Artificial Intelligence in Ophthalmology. Surv. Ophthalmol. 2019, 64, 233–240. [Google Scholar] [CrossRef]
- Jabeen, A. Beyond Human Perception: Revolutionizing Ophthalmology with Artificial Intelligence and Deep Learning. J. Clin. Ophthalmol. Res. 2024, 12, 287–292. [Google Scholar] [CrossRef]
- Waisberg, E.; Ong, J.; Kamran, S.A.; Masalkhi, M.; Paladugu, P.; Zaman, N.; Lee, A.G.; Tavakkoli, A. Generative Artificial Intelligence in Ophthalmology. Surv. Ophthalmol. 2025, 70, 1–11. [Google Scholar] [CrossRef]
- Alam, M.; Le, D.; Lim, J.I.; Chan, R.V.P.; Yao, X. Supervised Machine Learning Based Multi-Task Artificial Intelligence Classification of Retinopathies. J. Clin. Med. 2019, 8, 872. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Orlova, E.V. Artificial Intelligence-Based System for Retinal Disease Diagnosis. Algorithms 2024, 17, 315. [Google Scholar] [CrossRef]
- Parmar, U.P.S.; Surico, P.L.; Singh, R.B.; Romano, F.; Salati, C.; Spadea, L.; Musa, M.; Gagliano, C.; Mori, T.; Zeppieri, M. Artificial Intelligence (AI) for Early Diagnosis of Retinal Diseases. Medicina 2024, 60, 527. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Selvaraj, J.; Mani, I.; Kumaragurupari, T.; Shang, X.; Mudgil, P.; Ravilla, T.; He, M. Diagnostic Accuracy of Artificial Intelligence-Based Automated Diabetic Retinopathy Screening in Real-World Settings: A Systematic Review and Meta-Analysis. Am. J. Ophthalmol. 2024, 263, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Hayati, A.; Abdol Homayuni, M.R.; Sadeghi, R.; Asadigandomani, H.; Dashtkoohi, M.; Eslami, S.; Soleimani, M. Advancing Diabetic Retinopathy Screening: A Systematic Review of Artificial Intelligence and Optical Coherence Tomography Angiography Innovations. Diagnostics 2025, 15, 737. [Google Scholar] [CrossRef]
- Alqahtani, A.S.; Alshareef, W.M.; Aljadani, H.T.; Hawsawi, W.O.; Shaheen, M.H. The Efficacy of Artificial Intelligence in Diabetic Retinopathy Screening: A Systematic Review and Meta-Analysis. Int. J. Retin. Vitr. 2025, 11, 48. [Google Scholar] [CrossRef]
- Crincoli, E.; Sacconi, R.; Querques, L.; Querques, G. Artificial Intelligence in Age-Related Macular Degeneration: State of the Art and Recent Updates. BMC Ophthalmol. 2024, 24, 121. [Google Scholar] [CrossRef] [PubMed]
- Frank-Publig, S.; Birner, K.; Riedl, S.; Reiter, G.S.; Schmidt-Erfurth, U. Artificial Intelligence in Assessing Progression of Age-Related Macular Degeneration. Eye 2025, 39, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Gandhewar, R.; Guimaraes, T.; Sen, S.; Pontikos, N.; Moghul, I.; Empeslidis, T.; Michaelides, M.; Balaskas, K. Imaging Biomarkers and Artificial Intelligence for Diagnosis, Prediction, and Therapy of Macular Fibrosis in Age-Related Macular Degeneration: Narrative Review and Future Directions. Graefes Arch. Clin. Exp. Ophthalmol. 2025, 263, 1789–1800. [Google Scholar] [CrossRef]
- Martucci, A.; Gallo Afflitto, G.; Pocobelli, G.; Aiello, F.; Mancino, R.; Nucci, C. Lights and Shadows on Artificial Intelligence in Glaucoma: Transforming Screening, Monitoring, and Prognosis. J. Clin. Med. 2025, 14, 2139. [Google Scholar] [CrossRef]
- Sharma, P.; Takahashi, N.; Ninomiya, T.; Sato, M.; Miya, T.; Tsuda, S.; Nakazawa, T. A Hybrid Multi Model Artificial Intelligence Approach for Glaucoma Screening Using Fundus Images. NPJ Digit. Med. 2025, 8, 130. [Google Scholar] [CrossRef]
- Ravindranath, R.; Stein, J.D.; Hernandez-Boussard, T.; Fisher, A.C.; Wang, S.Y.; Amin, S.; Edwards, P.A.; Srikumaran, D.; Woreta, F.; Schultz, J.S.; et al. The Impact of Race, Ethnicity, and Sex on Fairness in Artificial Intelligence for Glaucoma Prediction Models. Ophthalmol. Sci. 2025, 5, 100596. [Google Scholar] [CrossRef]
- Jan, C.; He, M.; Vingrys, A.; Zhu, Z.; Stafford, R.S. Diagnosing Glaucoma in Primary Eye Care and the Role of Artificial Intelligence Applications for Reducing the Prevalence of Undetected Glaucoma in Australia. Eye 2024, 38, 2003–2013. [Google Scholar] [CrossRef]
- Fantozzi, C.B. Propostas de Algoritmos de Inteligência Artificial para Screening de Edema Macular Diabético; Doctorate in Health Sciences Program; Faculdade de Medicina de São José do Rio Preto: São José do Rio Preto, Brazil, 2024; 68p; Official Depository Library: Faculdade de Medicina de São José do Rio Preto. Available online: https://sucupira-legado.capes.gov.br/sucupira/public/consultas/coleta/trabalhoConclusao/viewTrabalhoConclusao.jsf?popup=true&id_trabalho=15195332 (accessed on 18 August 2025).
- Abràmoff, M.D.; Lavin, P.T.; Birch, M.; Shah, N.; Folk, J.C. Pivotal Trial of an Autonomous AI-Based Diagnostic System for Detection of Diabetic Retinopathy in Primary Care Offices. NPJ Digit. Med. 2018, 1, 39. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Cheung, C.Y.-L.; Lim, G.; Tan, G.S.W.; Quang, N.D.; Gan, A.; Hamzah, H.; Garcia-Franco, R.; San Yeo, I.Y.; Lee, S.Y.; et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images from Multiethnic Populations with Diabetes. JAMA 2017, 318, 2211. [Google Scholar] [CrossRef]
- Cole, E.D.; Moult, E.M.; Dang, S.; Choi, W.J.; Ploner, S.B.; Lee, B.K.; Louzada, R.; Novais, E.; Schottenhamml, J.; Husvogt, L.; et al. The Definition, Rationale, and Effects of Thresholding in OCT Angiography. Ophthalmol. Retin. 2017, 1, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Vilela, M.A.P.; Arrigo, A.; Parodi, M.B.; da Silva Mengue, C. Smartphone Eye Examination: Artificial Intelligence and Telemedicine. Telemed. e-Health 2024, 30, 341–353. [Google Scholar] [CrossRef]
- Christopher, M.; Hallaj, S.; Jiravarnsirikul, A.; Baxter, S.L.; Zangwill, L.M. Novel Technologies in Artificial Intelligence and Telemedicine for Glaucoma Screening. J. Glaucoma 2024, 33, S26–S32. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Pasquale, L.R.; Peng, L.; Campbell, J.P.; Lee, A.Y.; Raman, R.; Tan, G.S.W.; Schmetterer, L.; Keane, P.A.; Wong, T.Y. Artificial Intelligence and Deep Learning in Ophthalmology. Br. J. Ophthalmol. 2019, 103, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Abràmoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5200–5206. [Google Scholar] [CrossRef] [PubMed]
- Early Treatment Diabetic Retinopathy Study Design and Baseline Patient Characteristics: ETDRS Report Number 7. Ophthalmology 1991, 98, 741–756. [CrossRef]
- Guido, R.C. Paraconsistent Feature Engineering [Lecture Notes]. IEEE Signal Process. Mag. 2019, 36, 154–158. [Google Scholar] [CrossRef]
- Prabhakaran, S. Logistic Regression-a Complete Tutorial with Examples in R. Machine Learning Plus. 2017. Available online: https://www.machinelearningplus.com/machine-learning/logistic-regression-tutorial-examples-r/ (accessed on 1 July 2022).
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4. [Google Scholar]
- Quinlan, J.R. Induction of Decision Trees. Mach. Learn. 1986, 1, 81–106. [Google Scholar] [CrossRef]
- Yao, J.; Lim, J.; Lim, G.Y.S.; Ong, J.C.L.; Ke, Y.; Tan, T.F.; Tan, T.-E.; Vujosevic, S.; Ting, D.S.W. Novel Artificial Intelligence Algorithms for Diabetic Retinopathy and Diabetic Macular Edema. Eye Vis. 2024, 11, 23. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Leng, X.; Wu, Y.; Zhu, S.; Cai, X.; Lu, X. Hybrid Deep Learning Models for the Screening of Diabetic Macular Edema in Optical Coherence Tomography Volumes. Sci. Rep. 2023, 13, 18746. [Google Scholar] [CrossRef]
- Shahriari, M.H.; Sabbaghi, H.; Asadi, F.; Hosseini, A.; Khorrami, Z. Artificial Intelligence in Screening, Diagnosis, and Classification of Diabetic Macular Edema: A Systematic Review. Surv. Ophthalmol. 2023, 68, 42–53. [Google Scholar] [CrossRef]
- Sayres, R.; Hammel, N.; Liu, Y. Artificial Intelligence, Machine Learning and Deep Learning for Eye Care Specialists. Ann. Eye Sci. 2020, 5, 18. [Google Scholar] [CrossRef]
- Haykin, S. Neural Networks and Learning Machines, 3/E; Pearson Education India: Noida, India, 2012; ISBN 933258625X. [Google Scholar]
- Ting, D.S.W.; Lee, A.Y.; Wong, T.Y. An Ophthalmologist’s Guide to Deciphering Studies in Artificial Intelligence. Ophthalmology 2019, 126, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Qin, Y. Current Status, Hotspots, and Prospects of Artificial Intelligence in Ophthalmology: A Bibliometric Analysis (2003–2023). Ophthalmic Epidemiol. 2025, 32, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y. The Right Direction Needed to Develop White-Box Deep Learning in Radiology, Pathology, and Ophthalmology: A Short Review. Front. Robot. AI 2019, 6, 24. [Google Scholar] [CrossRef] [PubMed]
| Order | Abbreviation | Meaning |
|---|---|---|
| 1 | ID | Patient ID |
| 2 | R/L eye | Definition of the examined eye (right or left) |
| 3 | VisualAcuity | Patient’s visual acuity level |
| 4 | etdrs9_2 | Upper inner ring |
| 5 | etdrs9_4 | Lower inner ring |
| 6 | etdrs9_6 | Upper outer ring |
| 7 | etdrs9_8 | Lower outer ring |
| 8 | foveamin | Measurement of the fovea minima |
| 9 | etdrs9v_2 | Upper inner ring volume |
| 10 | etdrs9v_4 | Lower inner ring volume |
| 11 | etdrs9v_6 | Upper outer ring volume |
| 12 | etdrs9v_8 | Lower outer ring volume |
| 13 | whole/total | Measurement of the volume of the total area |
| 14 | Diagnosis | Phenotype verified by doctor |
| 15 | Sex | Patient sex (male or female) |
| 16 | etdrs9_1 | ETDRS ring center |
| 17 | etdrs9_3 | Internal nasal ring |
| 18 | etdrs9_5 | Internal temporal ring |
| 19 | etdrs9_7 | External nasal ring |
| 20 | etdrs9_9 | Outer temporal ring |
| 21 | etdrs9v_1 | ETDRS ring center volume |
| 22 | etdrs9v_3 | Inner nasal ring volume |
| 23 | etdrs9v_5 | Internal temporal ring volume |
| 24 | etdrs9v_7 | External nasal ring volume |
| 25 | etdrs9v_9 | Temporal outer ring volume |
| 26 | Age | Patient’s age on the day of the examination |
| Model | Features | Classification Score | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Y: F1 Score | N: F1 Score | AUC Score (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| SVM | Normal (24) | 129 | 92 | 89 | 93 | 65 | 98 | 76 | 95 | 81.8 |
| SVM | Paraconsistent (4) | 117 | 84 | 64 | 85 | 27 | 96 | 38 | 91 | 61.7 |
| DTREE | Normal (24) | 120 | 86 | 64 | 90 | 54 | 93 | 58 | 91 | 73.4 |
| DTREE | Paraconsistent (4) | 106 | 76 | 33 | 84 | 31 | 86 | 32 | 85 | 58.3 |
| KNN | Normal (24) | 129 | 92 | 89 | 93 | 65 | 98 | 76 | 95 | 82 |
| KNN | Paraconsistent (4) | 108 | 77 | 35 | 84 | 27 | 89 | 30 | 86 | 57.7 |
| LR | Normal (24) | 127 | 91 | 93 | 90 | 54 | 99 | 68 | 95 | 76 |
| LR | Paraconsistent (4) | 117 | 84 | 67 | 85 | 23 | 97 | 34 | 91 | 60 |
| Model | Features | Classification Score | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Y: F1 Score | N: F1 Score | AUC Score (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| SVM | Normal (24) | 118 | 84.3 | 68 | 83 | 81 | 99 | 74 | 93 | 82.7 |
| SVM | Paraconsistent (4) | 108 | 77 | 88 | 77 | 33 | 99 | 48 | 86 | 64.6 |
| DTREE | Normal (24) | 96 | 68.6 | 43 | 82 | 29 | 88 | 34 | 85 | 53.8 |
| DTREE | Paraconsistent (4) | 85 | 61 | 38 | 75 | 29 | 77 | 32 | 76 | 48.9 |
| KNN | Normal (24) | 113 | 80.7 | 76 | 84 | 76 | 95 | 76 | 89 | 58.5 |
| KNN | Paraconsistent (4) | 108 | 69 | 60 | 74 | 29 | 89 | 39 | 81 | 47.3 |
| LR | Normal (24) | 114 | 81 | 86 | 81 | 57 | 100 | 69 | 89 | 72.8 |
| LR | Paraconsistent (4) | 117 | 78 | 80 | 78 | 38 | 99 | 52 | 87 | 56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brandão Fantozzi, C.; Peres, L.M.; Neto, J.S.; Brandão, C.C.; Guido, R.C.; Siqueira, R.C. A Comparative Study Between Clinical Optical Coherence Tomography (OCT) Analysis and Artificial Intelligence-Based Quantitative Evaluation in the Diagnosis of Diabetic Macular Edema. Vision 2025, 9, 75. https://doi.org/10.3390/vision9030075
Brandão Fantozzi C, Peres LM, Neto JS, Brandão CC, Guido RC, Siqueira RC. A Comparative Study Between Clinical Optical Coherence Tomography (OCT) Analysis and Artificial Intelligence-Based Quantitative Evaluation in the Diagnosis of Diabetic Macular Edema. Vision. 2025; 9(3):75. https://doi.org/10.3390/vision9030075
Chicago/Turabian StyleBrandão Fantozzi, Camila, Letícia Margaria Peres, Jogi Suda Neto, Cinara Cássia Brandão, Rodrigo Capobianco Guido, and Rubens Camargo Siqueira. 2025. "A Comparative Study Between Clinical Optical Coherence Tomography (OCT) Analysis and Artificial Intelligence-Based Quantitative Evaluation in the Diagnosis of Diabetic Macular Edema" Vision 9, no. 3: 75. https://doi.org/10.3390/vision9030075
APA StyleBrandão Fantozzi, C., Peres, L. M., Neto, J. S., Brandão, C. C., Guido, R. C., & Siqueira, R. C. (2025). A Comparative Study Between Clinical Optical Coherence Tomography (OCT) Analysis and Artificial Intelligence-Based Quantitative Evaluation in the Diagnosis of Diabetic Macular Edema. Vision, 9(3), 75. https://doi.org/10.3390/vision9030075







