Image-Based Histological Evaluation of Scaffold-Free 3D Osteoblast Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Cell Culture
2.3. Biomechanical Loading
2.4. Van-Gieson Staining
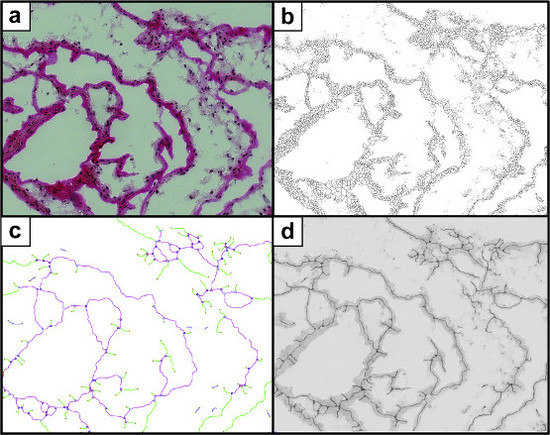
2.5. Semi-Automated Image Evaluation Process
2.5.1. Feature Definition
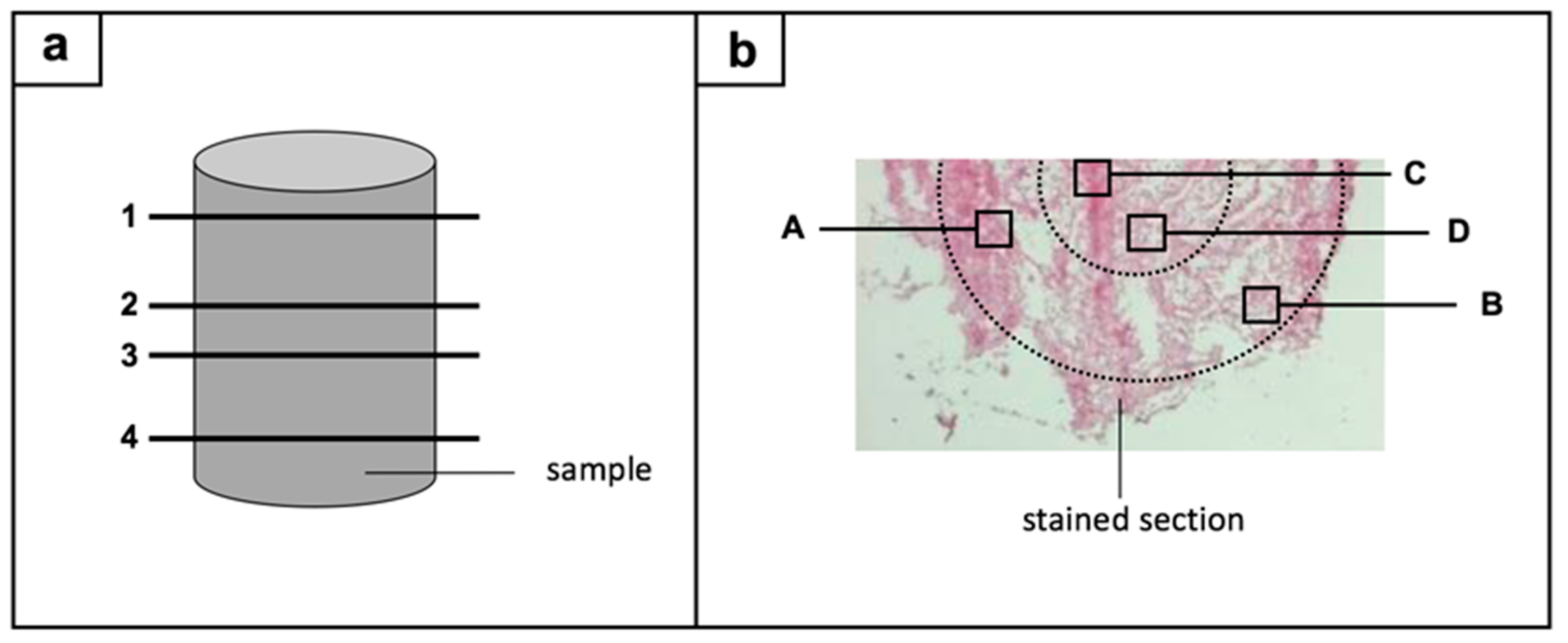
2.5.2. Definition of Regions-of-Interest
2.5.3. Image Acquisition
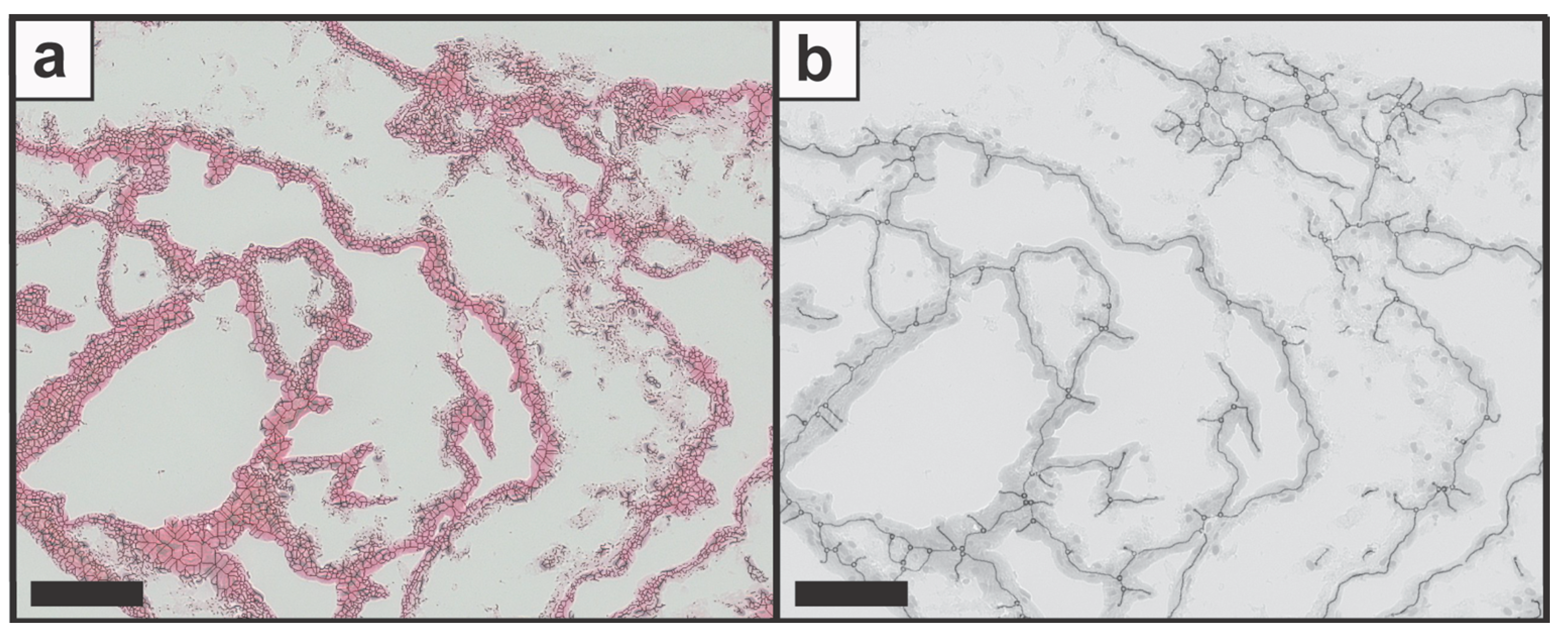
2.5.4. Image Segmentation
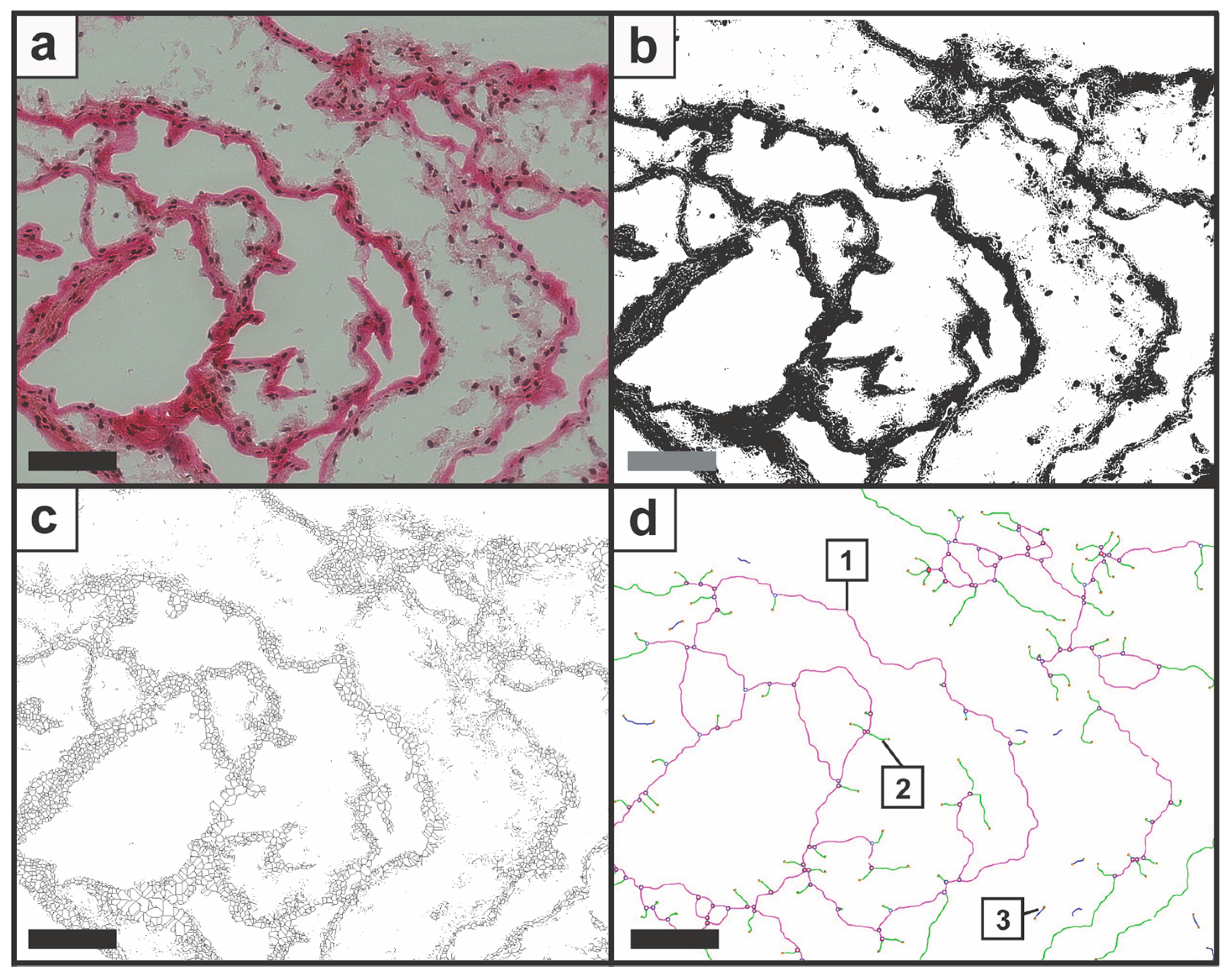
2.5.5. Feature Identification
2.5.6. Analysis
3. Results
3.1. Demonstrating the Feasibility of the Image Processing and the Feature Identification
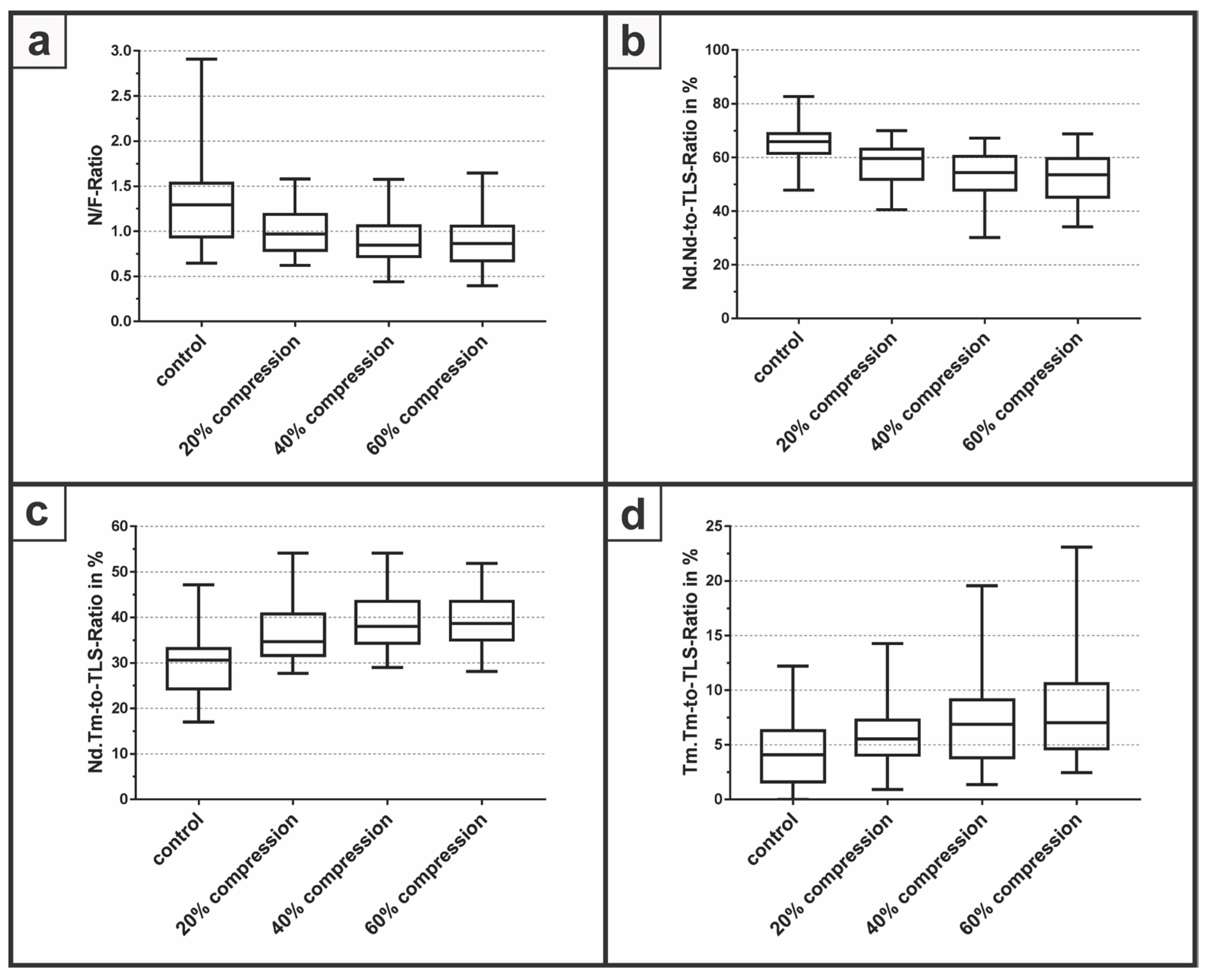
3.2. Determination of Node-to-Free-Ratio and Strut Analysis
4. Discussion
4.1. Future Work
4.2. Study Limitations
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviation
| Nd | Node |
| N/F-Ratio | Node-to-Free-Ratio |
| Nd.Nd | Node-to-Node-Strut-Length |
| Nd.Tm | Node-to-Terminus-Strut-Length |
| N.Nd | Number-of-Nodes |
| N.Tm | Number-of-Termini |
| Tm | Terminus |
| Tm.Tm | Terminus-to-Terminus-Strut-Length |
| TLS | Total-Strut-Length |
| ROI | Regions-of-Interest |
References
- McCarthy, E.F. The histology of metabolic bone disease. Diagn. Histopathol. 2016, 22, 378–383. [Google Scholar] [CrossRef]
- Coleman, R. The long-term contribution of dyes and stains to histology and histopathology. Acta Histochem. 2006, 108, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Canfield, P.J.; Hemsley, S. The roles of histology and immunohistology in the investigation of marsupial disease and normal lymphoid tissue. Dev. Comp. Immunol. 2000, 24, 455–471. [Google Scholar] [CrossRef]
- Dapson, R.W. Dye-tissue interactions: Mechanisms, quantification and bonding parameters for dyes used in biological staining. Biotech. Histochem. 2005, 80, 49–72. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.E. The molecular biology of histochemical staining by cationic phthalocyanin dyes: The design of replacements for Alcian Blue. J. Microsc. 1980, 119, 373–381. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.A.; Moradzadeh, A.; Whitlock, E.L.; Brenner, M.J.; Myckatyn, T.M.; Wei, C.H.; Tung, T.H.H.; Mackinnon, S.E. Binary imaging analysis for comprehensive quantitative histomorphometry of peripheral nerve. J. Neurosci. Methods 2007, 166, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Dalle Carbonare, L.; Valenti, M.T.; Bertoldo, F.; Zanatta, M.; Zenari, S.; Realdi, G.; Lo Cascio, V.; Giannini, S. Bone microarchitecture evaluated by histomorphometry. Micron 2005, 36, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, T.; Laib, A.; Müller, R.; Dequeker, J.; Rüegsegger, P. Direct three-dimensional morphometric analysis of human cancellous bone: Microstructural data from spine, femur, iliac crest, and calcaneus. J. Bone Miner. Res. 1999, 14, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Kamentsky, L.; Eliceiri, K.W. A call for bioimaging software usability. Nat. Methods 2012, 9, 666–670. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.; Kirsten, R.; Rupp, N.J.; Moch, H.; Fend, F.; Wernert, N.; Kristiansen, G.; Perner, S. Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol. Histopathol. 2013, 28, 605–610. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, V.; Zen, Y.; Chan, J.K.; Yi, E.E.; Sato, Y.; Yoshino, T.; Klöppel, G.; Heathcote, J.G.; Khosroshahi, A.; Ferry, J.A.; et al. Consensus statement on the pathology of IgG4-related disease. Mod. Pathol. 2012, 25, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Meijering, E.; Carpenter, A.E.; Peng, H.; Hamprecht, F.A.; Olivo-Marin, J.-C. Imagining the future of bioimage analysis. Nat. Biotechnol. 2016, 34, 1250–1255. [Google Scholar] [CrossRef] [PubMed]
- Blasi, T.; Hennig, H.; Summers, H.D.; Theis, F.J.; Cerveira, J.; Patterson, J.O.; Davies, D.; Filby, A.; Carpenter, A.E.; Rees, P. Label-free cell cycle analysis for high-throughput imaging flow cytometry. Nat. Commun. 2016, 7, 10256. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, Q.; Voss, T.C.; Quick, K.L.; LaBarbera, D.V. High-throughput imaging: Focusing in on drug discovery in 3D. Methods 2016, 96, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Ollion, J.; Cochennec, J.; Loll, F.; Escudé, C.; Boudier, T. TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 2013, 29, 1840–1841. [Google Scholar] [CrossRef] [PubMed]
- De Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Eliceiri, K.W.; Berthold, M.R.; Goldberg, I.G.; Ibanez, L.; Manjunath, B.S.; Martone, M.E.; Murphy, R.F.; Peng, H.; Plant, A.L.; Roysam, B.; et al. Biological imaging software tools. Nat. Methods 2012, 9, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, J.C.; Cooper, S.; Heigwer, F.; Warchal, S.; Qiu, P.; Molnar, C.; Vasilevich, A.S.; Barry, J.D.; Bansal, H.S.; Kraus, O.; et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Eulenberg, P.; Köhler, N.; Blasi, T.; Filby, A.; Carpenter, A.E.; Rees, P.; Theis, F.J.; Wolf, F.A. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 2017, 8, 463. [Google Scholar] [CrossRef] [PubMed]
- Sommer, C.; Gerlich, D.W. Machine learning in cell biology—Teaching computers to recognize phenotypes. J. Cell Sci. 2013, 126, 5529–5539. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.-A.; Singh, S.; Han, H.; Davis, C.T.; Borgeson, B.; Hartland, C.; Kost-Alimova, M.; Gustafsdottir, S.M.; Gibson, C.C.; Carpenter, A.E. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat. Protoc. 2016, 11, 49817. [Google Scholar] [CrossRef] [PubMed]
- Morin, K.T.; Carlson, P.D.; Tranquillo, R.T. Automated image analysis programs for the quantification of microvascular network characteristics. Methods 2015, 84, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Kontulainen, S.; Liu, D.; Manske, S.; Jamieson, M.; Sievänen, H.; McKay, H. Analyzing cortical bone cross-sectional geometry by peripheral QCT: Comparison with bone histomorphometry. J. Clin. Densitom. 2007, 10, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.C.; Mcgregor, N.E.; Poulton, I.J.; Pompolo, S.; Allan, E.H.; Quinn, J.M.; Gillespie, M.T.; Martin, T.J.; Sims, N.A. Cardiotrophin-1 is an osteoclast-derived stimulus of bone formation required for normal bone remodeling. J. Bone Miner. Res. 2008, 23, 2025–2032. [Google Scholar] [CrossRef] [PubMed]
- Egan, K.P.; Brennan, T.A.; Pignolo, R.J. Bone histomorphometry using free and commonly available software. Histopathology 2012, 61, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Van’t Hof, R.J.; Rose, L.; Bassonga, E.; Daroszewska, A. Open source software for semi-automated histomorphometry of bone resorption and formation parameters. Bone 2017, 99, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Doube, M.; Klosowski, M.M.; Arganda-Carreras, I.; Cordelières, F.P.; Dougherty, R.P.; Jackson, J.S.; Schmid, B.; Hutchinson, J.R.; Shefelbine, S.J. BoneJ: Free and extensible bone image analysis in ImageJ. Bone 2010, 47, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Aldinucci, D.; Quinn, J.; Degan, M.; Juzbasic, S.; De Luliis, A.; Improta, S.; Pinto, A.; Gattei, V. In vitro cellular systems for studying OC function and differentiation: Primary OC cultures and the FLG 29.1. In Human Cell Culture Protocols. Methods in Molecular Medicine; Jones, G., Ed.; Humana Press: New York, NY, USA, 1996; pp. 277–306. ISBN 978-1-59259-586-0. [Google Scholar]
- Saldamli, B.; Herzen, J.; Beckmann, F.; Tübel, J.; Schauwecker, J.; Burgkart, R.; Jürgens, P.; Zeilhofer, H.-F.; Sader, R.; Müller, B. Internal structures of scaffold-free 3D cell cultures visualized by synchrotron radiation-based micro-computed tomography. In Proceedings Volume 7078, Developments in X-Ray Tomography VI; Optical Engineering and Applications: San Diego, CA, USA, 2008; Volume 70781X. [Google Scholar] [CrossRef]
- Foehr, P.; Hautmann, V.; Prodinger, P.; Pohlig, F.; Kaddick, C.; Burgkart, R. Hochdynamisches prüfsystem zur biomechanischen charakterisierung von knorpel und seinen regeneraten. Orthopade 2012, 41, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, A.M.; Drezner, M.K.; Glorieux, F.H.; Kanis, J.A.; Malluche, H.; Meunier, P.J.; Ott, S.M.; Recker, R.R. Bone histomorphometry: Standardization of nomenclature, symbols, and units: Report of the asbmr histomorphometry nomenclature committee. J. Bone Miner. Res. 1987, 2, 595–610. [Google Scholar] [CrossRef] [PubMed]
- DeHoff, R.T.; Aigeltinger, E.H.; Craig, K.R. Experimental determination of the topological properties of three-dimensional microstructures. J. Microsc. 1972, 95, 69–91. [Google Scholar] [CrossRef]
- Ridler, T.W.; Calvard, S. Picture thresholding using an iterative slection method. IEEE Trans. Syst. Man Cybern. 1978, 8, 630–632. [Google Scholar] [CrossRef]
- Zhang, T.; Suen, C. A modified fast parallel algorithm for thinning digital patterns. Commun. ACM 1984, 27, 236–239. [Google Scholar] [CrossRef]
- Carpentier, G.; Martinelli, M.; Courty, J.; Cascone, I. Angiogenesis Analyzer for ImageJ. In Proceedings of the 4th ImageJ User & Developer Conference, Mondorf-les-Bains, Luxembourg, 24–26 October 2012; pp. 198–201. [Google Scholar]
- Gholobova, D.; Decroix, L.; Van Muylder, V.; Desender, L.; Gerard, M.; Carpentier, G.; Vandenburgh, H.; Thorrez, L. Endothelial network formation within human tissue-engineered skeletal muscle. Tissue Eng. Part A 2015, 21, 2548–2558. [Google Scholar] [CrossRef] [PubMed]
- Fortenberry, Y.M.; Brandal, S.M.; Carpentier, G.; Hemani, M.; Pathak, A.P. Intracellular expression of PAI-1 specific aptamers alters breast cancer cell migration, invasion and angiogenesis. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Garrahan, N.J.; Mellish, R.W.; Compston, J.E. A new method for the two-dimensional analysis of bone structure in human iliac crest biopsies. J. Microsc. 1986, 142, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Compston, J.E.; Yamaguchi, K.; Croucher, P.I.; Garrahan, N.J.; Lindsay, P.C.; Shaw, R.W. The effects of gonadotrophin-releasing hormone agonists on iliac crest cancellous bone structure in women with endometriosis. Bone 1995, 16, 261–267. [Google Scholar] [CrossRef]
- Dempster, D.W.; Compston, J.E.; Drezner, M.K.; Glorieux, F.H.; Kanis, J.A.; Malluche, H.; Meunier, P.J.; Ott, S.M.; Recker, R.R.; Parfitt, A.M. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2013, 28, 2–17. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eggert, S.; Tuebel, J.; Foehr, P.; Kuntz, L.; Obermeier, A.; Marthen, C.; Grosse, C.U.; Burgkart, R. Image-Based Histological Evaluation of Scaffold-Free 3D Osteoblast Cultures. J. Funct. Morphol. Kinesiol. 2017, 2, 42. https://doi.org/10.3390/jfmk2040042
Eggert S, Tuebel J, Foehr P, Kuntz L, Obermeier A, Marthen C, Grosse CU, Burgkart R. Image-Based Histological Evaluation of Scaffold-Free 3D Osteoblast Cultures. Journal of Functional Morphology and Kinesiology. 2017; 2(4):42. https://doi.org/10.3390/jfmk2040042
Chicago/Turabian StyleEggert, Sebastian, Jutta Tuebel, Peter Foehr, Lara Kuntz, Andreas Obermeier, Carmen Marthen, Christian U. Grosse, and Rainer Burgkart. 2017. "Image-Based Histological Evaluation of Scaffold-Free 3D Osteoblast Cultures" Journal of Functional Morphology and Kinesiology 2, no. 4: 42. https://doi.org/10.3390/jfmk2040042
APA StyleEggert, S., Tuebel, J., Foehr, P., Kuntz, L., Obermeier, A., Marthen, C., Grosse, C. U., & Burgkart, R. (2017). Image-Based Histological Evaluation of Scaffold-Free 3D Osteoblast Cultures. Journal of Functional Morphology and Kinesiology, 2(4), 42. https://doi.org/10.3390/jfmk2040042