Abstract
Accurate and rapid taxonomy identification is the initial step in spider image recognition. More than 50,000 spider species are estimated to exist worldwide; however, their identification is still challenging due to the morphological similarity in their physical structures. Deep learning is a known modern technique in computer science, biomedical science, and bioinformatics. With the help of deep learning, new opportunities are available to reveal advanced taxonomic methods. In this study, we applied a deep-learning-based approach using the YOLOv7 framework to provide an efficient and user-friendly identification tool for spider species found in Taiwan called Spider Identification APP (SpiderID_APP). The YOLOv7 model is integrated as a fully connected neural network. The training of the model was performed on 24,000 images retrieved from the freely available annotated database iNaturalist. We provided 120 genus classifications for Taiwan spider species, and the results exhibited accuracy on par with iNaturalist. Furthermore, the presented SpiderID_APP is time- and cost-effective, and researchers and citizen scientists can use this APP as an initial entry point to perform spider identification in Taiwan. However, for detailed species identification at the species level, additional methods like DNA barcoding or genitalic structure dissection are still considered necessary.
1. Introduction
The populace and diversity of arachnids are adversely influenced by anthropogenic factors, such as the alteration of their natural habitat due to human activities or the active extermination of these repugnant yet beneficial indoor predators [1,2]. Scientists estimate that approximately 4300 spider genera and 50,000 spider species exist globally [3]. Accurate spider identification holds paramount importance across ecological, agricultural, and public health domains [4,5]. Though many spider species have distinct patterns and shapes that ease identification, a plethora of them require intensive morphological studies. In addressing these challenges, we conceptualize a broader AI-aided classification system. The primary layer, central to this paper, involves our application providing initial spider classification based on images. This tool is designed to offer non-specialists a methodological introduction to spider taxonomy and to alleviate the initial screening process for specialists. Although this layer is integral to our current research, it represents only the inaugural phase of a more encompassing strategy. For instances where our application’s primary identification yields low-confidence results, subsequent steps in the proposed pipeline would necessitate detailed microscopic procedures, such as microdissection and lactic acid treatment, followed by high-resolution imaging. These images could then be analyzed either by experts or potentially by another AI model adept at microscopic imagery of spider anatomy. It is imperative to emphasize that while this comprehensive pipeline is outlined, the current research is dedicated exclusively to its first step.
Historical taxonomic approaches, spanning centuries, have grappled with challenges, notably in differentiating sex dimorphism [6,7,8,9]. While the image-based recognition approach has demonstrated potential, as within a pool of three spider genera, 100% genus recognition and 81% species recognition were recorded [10], a study in a large scope indicated that spiders (Araneae) yielded only a 50% accuracy when identified using a morphospecies approach [11]. Notably, in this study, all specimens were separated into morphospecies by a non-specialist and later correctly identified by specialized taxonomists, suggesting that the low accuracy might stem not only from the inherent challenges of image-based taxonomic classification but also from potential human error. This underscores the pressing need and opportunity for specialized tools like our image-based AI to enhance spider identification. The same study reported up to 91% accuracy for other taxa like Lepidoptera, demonstrating the potential of tailored approaches in taxonomy. Additionally, the minor 3.3% overall difference between morphospecies and taxonomic species estimates, even though it was claimed as an artifact due to the balancing-out phenomenon of splitting (one species separated into more than one morphospecies) and lumping (more than one species classified as a single morphospecies), suggests that, with the right refinements, image-based tools can closely approach taxonomic accuracy. By integrating AI, and specifically YOLO-based models, into the scope of genus-level identification, our study aims to circumvent these challenges with a thorough understanding of its capabilities and limitations.
Our hypothesis in this study is that integrating AI, specifically YOLO-based models, for genus-level spider identification can significantly improve the accuracy and efficiency of identifying spiders compared to traditional methods. We expect that this approach will not only streamline the identification process but also reduce errors associated with manual classification, especially in cases where species exhibit subtle morphological differences. Our prediction is that our AI-assisted tool will demonstrate higher accuracy in genus-level identification than current non-expert methods, thus providing a valuable resource for both researchers and citizen scientists. Moreover, we anticipate that the application will serve as a steppingstone towards developing more advanced AI tools capable of achieving even greater precision in spider identification at the species level.
Modern biological research’s breadth is vast, but gathering large-scale biological data and long-term monitoring often hits resource constraints in which citizen science presents a compelling solution [12,13,14,15]. A notable example within spider projects includes the Spider in da House application, a collaborative effort between taxonomists and citizen scientists, originally designed for spider identification [16]. Another prominent platform is iNaturalist, a widely utilized online database both for contributing observations and referencing species [17,18]. Current identification methodologies of non-expert spider enthusiasts, major contributions of observations in citizen science projects, like seeking reference images online or consulting expert-authored spider atlases [19,20,21], are labor-intensive and prone to error. Having a tool for fast spider identification, integrated with AI capabilities, would potentially empower non-expert spider enthusiasts to make more precise contributions to online databases, thereby reducing the time experts must invest in making corrections. However, it is crucial to underline that while an AI-assisted tool can streamline and expedite the identification process, it cannot entirely supplant the role of experts in the field of spider taxonomy. This is because spider taxonomic identification based purely on external appearance, or habitus, is often misleading. Many spider species exhibit identical or nearly identical colorations and patterns, making it an unreliable metric for accurate identification. In numerous cases, precise spider identification necessitates the examination of “the finer microscopic characteristics” (“Es geht aus allem wol hervor, dasz die feinern microscopischen merkmale bei untersscheidung der arten wol nicht zu entbehren sind”) (“It is clear from everything that the finer microscopic features cannot be dispensed with when distinguishing between species”) [22]. These nuanced, internal distinctions are the gold standard in spider taxonomy, and while technology can assist, it cannot yet replicate the expert eye’s subtlety and depth.
Using deep learning model training on a large dataset of labeled images, which can identify the key features and provide the probability of detection based on such extracted features, is essential for this task [23,24]. The field of automatic classification is rapidly growing with the application of deep learning and artificial intelligence [25,26]. Mainly, image classification for spider identification using a deep learning approach still needs improvement compared to other fields [27,28]. It is observed that accuracy evaluation on testing datasets in such studies is high due to the low diversity in the dataset used. For example, a study conducted by Sinnott et al. in 2020 used nine classification classes belonging to nine different families within two infraorders [28], while the research by Chen et al. in 2021 used multi-angle images of 30 species within a single genus of Pseudopoda for gender classification [27].
You Only Look Once (YOLO) is the state-of-the-art deep learning model designed for real-time object detection that was first introduced by Redmon et al. [29]. YOLO gained attention for its single-stage detection architecture and fast detection speed without sacrificing accuracy [30]. Furthermore, YOLO works in the background with convolutional neural networks (CNNs) as feature maps to generate bounding box predictions rather than relying on generated features as in region-based convolutional neural networks (R-CNNs) [31]. YOLO exhibited successful results in a wide range of applications, such as mammals [32], fish [33], plants [34], and insects [35]. Several YOLO versions have been released by different research groups. The model advancements often include architectural changes and optimization techniques to enhance performance. However, it is important to note that the subsequent ones are not necessarily better than the previous ones since each version may also have hidden caveats that could impact its overall effectiveness in specific tasks. Therefore, in this study, we have included several recent YOLO models, namely YOLOv5 [36], YOLOv6 [37], YOLOv7, and YOLOv8 [38], to construct a deep-learning-based framework for spider classification using image datasets available on online open-source databases.
This study’s chief ambition is to pioneer an effective genus-level spider identification method for spider species found in Taiwan. The species-level distinction often demands intricate procedures like multi-angle microscopic imagery combined with lactic acid treatments and microdissection, especially when examining genitalia [39,40]. Our approach to genus-level classification, on the other hand, sidesteps the intricacies associated with polymorphism and sibling species [41,42,43]. A visual representation of this study’s pipeline is illustrated in Figure 1.
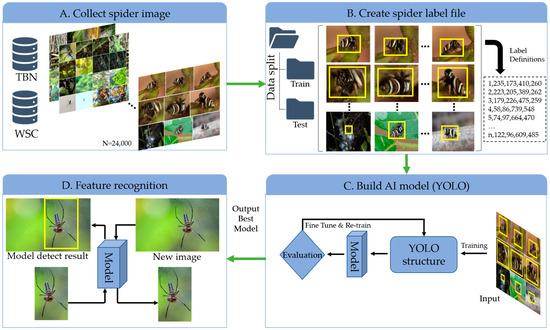
Figure 1.
Overall schema of developing a fully automated approach for spider classification (SpiderID_APP) using a YOLO-based deep learning model. (A) Collection of spider images of different genera from open-access databases. (B) Labeling of the dataset to perform training and testing using a YOLO-based model. (C) YOLO-based model architecture for given different parameters (e.g., batch size). (D) Classification results presented of spider image with respect to genus.
2. Methodology
2.1. Data Collection
Data were collected from the online open-source database iNaturalist (https://www.iNaturalist.org/, accessed on 18 July 2022) and the Taiwan Biodiversity Network (TBN, https://www.tbn.org.tw/, accessed on 20 July 2022), while ensuring that the downloaded images were research-graded with given genus names (Figure 1A). Data collection was further enhanced by using a self-developed, specifically designed Python web crawling script to hasten the data gathering. Out of more than 524,000 images of those 325 spider species identified across iNaturalist and TBN databases, more than 28,000 research-graded images of spiders with a history of being found in Taiwan were collected. The image set was quality-controlled by removing images of spiders with low visibility (such as out-of-focus images, over- or under-exposed images) to minimize the potential negative impacts on model performance.
2.2. Computer Specification
The training process was performed on a system equipped with a graphic processing unit (GPU) and CUDA kernel; details of our computer configuration are as follows: CPU of i7-12700, 2.1 GHz, memory (RAM) of 16 GB, and Nvidia GeForce GTX 3050 with 2560 CUDA cores [44], graphic memory of 8 GB and essential software such as Python 3.9.13 (https://www.python.org/downloads/release/python-3913/, accessed on 28 May 2022), Pytorch 1.13 (https://pytorch.org/get-started/previous-versions/, accessed on 27 July 2022) [45], NVIDIA CUDA toolkits version 11 (https://developer.nvidia.com/cuda-11.0-download-archive, accessed on 27 July 2022), and NVIDIA CUDA Deep Neural Network Library version 8.4.1 (cuDNN, https://developer.nvidia.com/rdp/cudnn-download, accessed on 27 July 2022) [46,47].
2.3. Dataset Labeling and Preparation Process
Training of an accurate model to identify spider genera and gender significantly depends on the input dataset with reliable bounding box specifications [48]. Images were annotated by drawing bounding boxes around spiders in each image. When the cephalothorax (head compartment) of the spider was visible, gender was determined based on the presence or absence of palps; typically, male spiders exhibit noticeable palps, while females do not [49]. Additionally, for some spider genera, such as Chikunia and Argiope, sexual dimorphism is distinctly evident, with clear and distinctive differences in habitus between males and females [50]. In some cases, gender annotation was not feasible due to camera angle, low image quality, or spider morphology that did not exhibit clear gender distinctions. For optimal analysis, images should adhere to certain minimum requirements, such as clarity, proper angle showcasing the distinctive anatomical features, and adequate resolution [51]. The labeling script used was a customized version of the OpenLabeling script developed by Joao Cartucho (https://github.com/Cartucho/OpenLabeling, accessed on 28 July 2022) with added features for faster labeling and relabeling processes. The necessary information for precise labeling was procured from the sources, examination of taxonomic literature, or consultation with experts in spider taxonomy, particularly of spider species in Taiwan. Subsequently, the labeled dataset was duplicated and processed to generate two distinct types of datasets: the NoGender (NG) dataset, which comprises 120 classes corresponding to 120 spider genera without gender involvement, and the WithGender (WG) dataset, which includes 240 classes due to the differentiation between male and female individuals within each genus, thus involving gender. Both datasets were then partitioned into datasets for training and validation (NG_TVD and WG_TVD) and testing (NG_TeD and WG_TeD) datasets, employing a stratified sampling technique to guarantee that each subset preserved a comparable distribution of genera and genders. Both TVDs underwent further division using an 80/20 holdout ratio before being input into the training process [52]. The annotation files were stored in YOLO format, with a separate text file for each labeled image. Each text file contained five arguments: an integer-type number representing the labeling class and four float-type numbers representing bounding box characteristics of relative center point coordinates and their width and height.
2.4. Data Augmentation and Dataset Balancing
Data augmentation is a common technique used in deep learning studies, with various data augmentation techniques for images being considered, where original images undergo random transformations such as flip, rotation, shear, mosaic, or color channel manipulations such as HSV augmentation. This process helps the models increase their generalizability by balancing the dataset and enhancing the robustness, enabling models to learn to recognize spiders under different lighting conditions, orientations, and other variations that may be encountered in real-world situations. Data augmentation for balancing purposes in this study was performed using a self-developed algorithm to generate a random combination of transformation based on probability across three types of augmentation techniques: random vertical flip, random rotation up to 90 degrees, and x-axis and y-axis random shear up to 45 degrees. Several transformation matrices were applied to maintain the accurate position of the bounding box on the augmented images (Figure 2). Other image augmentations were applied for robustness enhancement by adjusting the hyperparameters in the YOLO data augmentation feature [53].
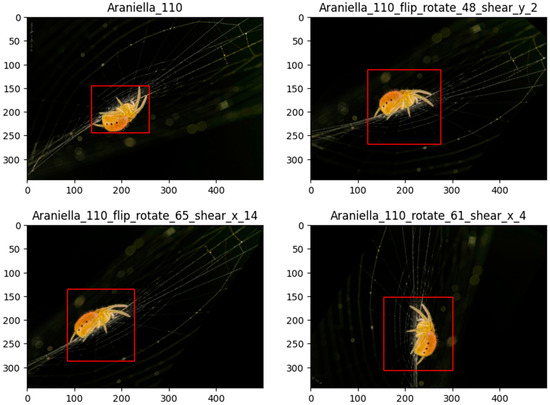
Figure 2.
Spider images undergo three distinct combinations of image augmentations, ensuring variety in the dataset. Post-augmentation, the bounding boxes are recalculated to continue encompassing the spiders, preserving accurate object localization tightly.
2.5. Model Fine-Tuning Process and Evaluation Metrics
This study employed YOLO-based models (versions 5, 6, 7, and 8) for spider identification due to their strong performance in object detection and classification tasks. These models are acclaimed for their optimal balance between accuracy, complexity, and computational efficiency. It is important to note that this study did not aim to alter the already optimized architecture of the YOLO models, as the team did not possess the mathematical expertise required for such architectural modifications. Instead, the focus was on optimizing other critical factors such as the input dataset and training parameters, which were within the scope of our capabilities. To compare the performance of each YOLO iteration and select the best model for this study, each model was initialized with pre-trained weights from large-scale datasets such as ImageNet [54] and Microsoft Common Objects in Context (COCO) [55]. Each YOLO-based model’s pre-trained weights were fine-tuned using WG_TVD and NG_TVD, resulting in two lines of models: model_WG and model_NG. Fine-tuning of the models was performed using the custom WG_TVD and NG_TVD datasets, adjusting hyperparameters to optimize spider identification. The training process was monitored using Tensorboard 2.9.0 [56], with a learning rate scheduler and early stop mechanism to ensure precise convergence and avoid overfitting. Model performance was evaluated using TeD and iteratively refined through hyperparameter tuning and cross-validation. An internally designed scoring system was developed for cross-model performance comparison, and statistical analyses were conducted to determine the significance of the difference between models’ performances (Table 1).

Table 1.
Evaluation of trained models on TeD regarding speed benchmark metrics.
We fine-tuned two types of datasets, “NG” and “WG”, using four selected YOLO models, resulting in eight distinct models. Input hyperparameters such as batch size, number of epochs, initial learning rate, and learning rate decay were considered. Input data resolution and batch size required tuning, so experiments were designed with image resolutions of 416 × 416, 512 × 512, and 640 × 640 and batch sizes of 16, 32, 64, and 128. YOLO models’ optimization steps depend on three loss values.
In the training process evaluation, loss values were monitored. All four models use localization loss (Lbox) and classification loss (Lcls) in their total loss calculation, while YOLOv5 and YOLOv7 use confidence loss (Lobj), and YOLOv6 and YOLOv8 use distribution focal loss (LDFL). Total loss was calculated with corresponding weights for each loss function (Formulas (1) and (2)).
where wi are the weights of corresponding loss functions. There has been a proposal of new loss functions such as SIoU, which greatly speeds up the training convergence process so that the prediction box first moves to the nearest axis. However, such a function greatly penalizes a large margin of error, which goes against the variety of spider species within a genus [57,58]. Monitoring classification loss values during training is crucial to avoid overfitting and enhance model generalization. The YOLO training code incorporates measures to address overfittings, such as Pytorch scheduler algorithms and best-weight-saving scripts.
Loss = w1 × Lbox + w2 × Lcls + w3 × Lobj
Loss = w1 × Lbox + w2 × Lcls + w3 × LDFL
To determine the optimal version of YOLO-based image recognition for spiders, a performance evaluation was performed between the 8 models obtained from the training process using the standard F1-score, precision, and recall, [59] along with other factors regarding the easiness of implementation into a spider detection APP such as inference time (IT) and trainable parameters. These evaluation measurements were computed as per standard true positive (TP), false positive (FP), and false negative (FN) cases [60]:
Similarly, the F1-score was calculated based on a previously reported formula [61]:
Additionally, due to the aim of this study being to identify spider genera, accuracy and area under the receiver operating characteristic curve (AUC-ROC), or area under the curve, were also considered for inclusion in the benchmark metric for the performance evaluation process of the model, which was calculated using sklearn.metrics, a machine learning library available in Python packages, the inputs of which are arrays of ground truth values and prediction values [62].
Unlike existing taxonomical identifier tools such as the one provided by iNaturalist, which rely on a suggestion mode to present an identification of higher taxonomic rank when the confidence of the current rank is insufficient, our final application would display the genus prediction and its confidence score. Due to such a difference in result demonstration, in this study, we present the novel Spider Identification Scoring System (SISS) specifically designed for the classification task. For identifiers with a confidence value, SISS awards higher points for high-confidence classifications (>0.75), lower points for lower-confidence classifications (0.5 to 7.5), and no points for very-low-confidence classifications (<0.5). Identifiers such as the iNaturalist tool use a scoring method for identifiers without a confidence value (Table 1).
For model complexity and time evaluation, we used the same set of hyperparameters for the initial set of models, considering training time for future updates to the APP. Training time was calculated automatically by YOLO training scripts during the training process. In contrast, the inference time was calculated using the Python time package to obtain the value of time at the beginning of the prediction process and time at the end of the prediction process, hence resulting in time taken (in milliseconds) for the prediction process. It must be noted that the model loading process is excluded from such calculations, so the actual time for processing individual images is much higher than the given values. However, if multiple images are selected for identification, the model must be loaded once at the beginning of the process, hence reducing the total time taken significantly. The trainable parameters of a CNN network like YOLO are determined by the complexity of each layer in the architecture, which includes factors such as the size of the input channel, output channel, and filter layer. The number of parameters also depends on the type of layer, such as convolutional, pooling [63], or fully connected layers [64], and the specific architecture of the model. These parameters are represented by each layer’s learnable weights and biases, which are adjusted during training to minimize the loss function. A high number of parameters can increase the model’s capacity to learn complex features and improve its performance. Still, it can also increase the risk of overfitting and require more computational resources for training and inference.
2.6. GUI Design
The implementation of a user-friendly GUI for the SpiderID application, which utilized Python’s Tkinter package [65] and YOLO detection script (https://github.com/WongKinYiu/yolov7, accessed on 19 September 2022), was essential in providing a seamless and intuitive experience for users aiming to identify spider genera. The incorporation of an aesthetically pleasing and easy-to-navigate graphical interface allows users to efficiently access the application’s features without prior knowledge or technical expertise. Furthermore, this approachable design fosters user engagement and accessibility, encouraging more individuals to explore the fascinating world of spiders.
3. Result
3.1. Data Collection and Preparation
According to data from TBN and the WSC, there are currently 325 registered spider species across 201 genera and 44 families in Taiwan. Interestingly, across 201 genera, 120 of those possess 99.69% image records, while the other genera either have no or very few images (Supplemental Figure S1A). It can be observed that online databases are susceptible to data imbalance due to the disparity in how commonly a spider genus is encountered by humans. Some genera possess tens of thousands of images, while some only have around 100 images available. Downloading and using this whole dataset would cause a serious imbalance in the dataset, which is not recommended for effective model training. Hence, in this study, we have downloaded 28,000 images across 120 spider genera. After performing quality control on the downloaded images, a total of 23,962 images were selected for further processing, and the quantity details are provided in Supplemental Table S1B.
Out of the total pool of 23,962 images, 2449 images were probed pseudorandomly to preserve the ratio between classes, for TeD (Supplemental Figure S1B,C); the rest were TVD, which would be later split into a training dataset and validation dataset using an 80/20 holdout ratio. The TVD datasets were then augmented by applying three transformations: 50% to flip vertically, a random rotation of 0 ÷ 90 degrees, and a random shear on the x-axis or y-axis with shear degree varying between 0 and 45 degrees to generate up to 600 images per labeling class in the training dataset and 150 images per labeling class in the validation and testing dataset.
3.2. Model Complexity and Training Time Evaluation
To best fit the capability of the computer specification, the training on all models runs with the default settings of each YOLO version to determine the optimizable range of batch size since a higher batch size would require much more computational power. Compared to other YOLO versions, we observed that YOLOv7 models have the highest GPU requirement (4.21–4.66 Gb) and average training time per epoch (31.2–59.8 min), showing higher model complexity compared to the other models tested (Table 2).

Table 2.
Comparison of training time on GPU with the same hyperparameter settings on different YOLO-based model versions.
3.3. Cross-Model Evaluation in the Training Process
Each YOLO version was used for training on both the WG and NG datasets, resulting in eight different models. The models were trained over 500 epochs as the validation mAP0.5:0.95 values, and recorded loss values are plotted in Figure 3. It can be observed that even though the training loss values keep decreasing in each subsequent epoch, the validation mAP values do not follow, which is a normal phenomenon in model training. YOLOv7 models perform the best among the models used, showing better mAP validation values in both the WG (Figure 3A) and NG (Figure 3B) models.
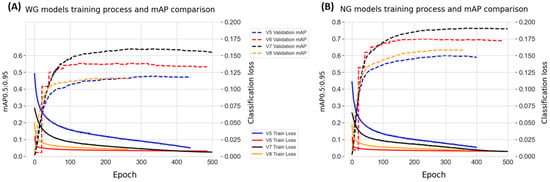
Figure 3.
Comparative analysis of classification loss for WG and NG models across various YOLO iterations. (A) The classification loss for WG models, showcasing eight distinct lines representing both training loss and validation mAP0.5:0.95 values for YOLO iterations v5, v6, v7, and v8. (B) Analogously, the classification loss for NG models is displayed, featuring eight corresponding lines for the training and validation losses of YOLO versions v5, v6, v7, and v8.
3.4. Cross-Model Evaluation on TeD
Figure 4 describes the low false positive rate of identification for all models. Regarding the tradeoff between true positive and false positive rates, YOLOv7 has an AUC score ranked among the highest in both the WG (Figure 4A) and NG (Figure 4B) model comparisons. Even though v5_WG and v6_NG perform slightly better than their respective v7 models, with the ultimate aim of building an application for identification, we found that the support scripts of YOLOv6 are not path-friendly for application deployment. The optimization steps would be conducted on YOLOv7 models; the fully optimized configurations would be applied to other YOLO models to train the final versions for performance comparison.
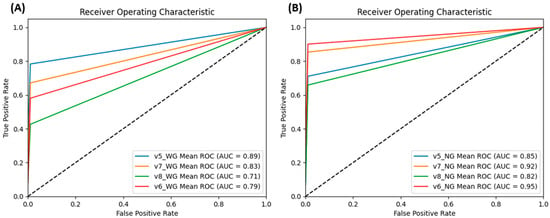
Figure 4.
Mean ROC of the classes in YOLOv7_WG model (A) and YOLOv7_NG model (B) when evaluated on TeD. The consistent positioning of YOLOv7 variants in the top two ranks among the four models under comparison underscores their superiority and robustness in performance.
3.5. Optimization
3.5.1. Data Resolution Selection
To evaluate the performance of models using different input image size values, a partial dataset was probed from the whole dataset in a pseudorandom manner to keep the image ratio per class. The image size values must satisfy the conditions of being a multiple of pixel stride, which is 32 for YOLO models, and remain close to the mean value of both the training and real-life data the model would encounter. A pool of three image size values—416 × 416, 512 × 512, and 640 × 640 pixels, among which 416 × 416 and 640 × 640 are the common image size settings in the YOLO network and 512 × 512 was determined based on a dataset analysis (Supplemental Figure S1D)—was then selected for performance analysis. The input image size of 512 × 512 was proven to be the best configuration for YOLOv7 models according to classification loss, F1-score, and SISS evaluation in both the WG and NG model comparisons (Figure 5 and Table 3).
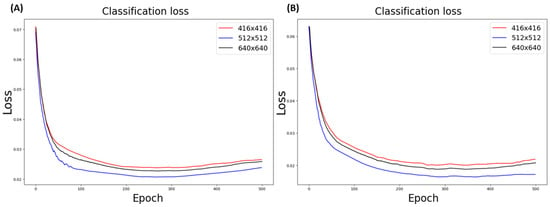
Figure 5.
YOLOv7 model loss curve with different image size configurations: (A) classification loss in YOLOv7_WG model; (B) classification loss in YOLOv7_NG model.

Table 3.
YOLOv7 models’ performance with different image size configurations, evaluated by F1-score and SISS.
3.5.2. Batch Size Optimization
Due to the large dataset, training with a batch size of eight images already allocated a considerable amount of GPU memory using the mentioned computer system (Table 2). Thus, to fine-tune the batch size parameter, nominal batch size (NBS) was used, in which the model would accumulate the loss after each batch κ times before optimization.
k = NBS ÷ BS
This method enables the possibility of simulating model training with a high batch size, even when working with a large dataset on a limited computational budget. However, NBS is a hidden parameter set at 64 by default across all versions of YOLO used; hence, source code modification has to be performed to enable fine-tuning such parameters. In this study, the conventional loss functions of YOLO were preserved by respective evaluation. An optimization based on the nominal batch size was conducted on the whole dataset, in which images were resized to 512 × 512, as the classification loss values were used as assessment metrics. NBS 64 has proven to be the best configuration for YOLOv7 models as it has a lower classification loss than other models (Figure 6A). NBS 64 performed slightly better than NBS 128 and more than 10 percent better than lower NBS settings, as shown quantitatively in Figure 6B.
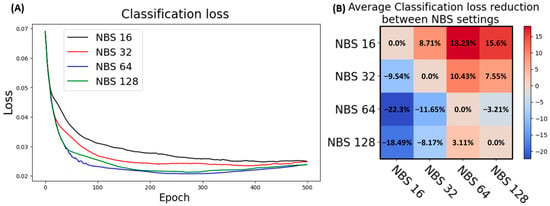
Figure 6.
Nominal batch size optimization in YOLOv7 model. (A) Classification loss in validation of YOLOv7 model at four different NBS settings. (B) Heatmap demonstrating the reduction in classification loss between NBS settings; the blue tone signifies a lower decrease, and the red tone signifies a higher decrease in loss values, which means a potential increase in performance.
3.6. Assessment of Final Models through Internal Evaluation
All eight models that were retrained based on the optimized parameters were then used to run prediction on TeD. When the models are evaluated using the F1-score, a similar trend can be observed in pre-optimized models. While comparing YOLOv7 to YOLOv6, it was observed that despite the F1-score of the NG version being 4% lower, the YOLOv7 model pair (YOLOv7_NG and YOLOv7_WG) was still better overall; the F1-score of the WG version for YOLOv7 was 19.1% higher than that of YOLOv6. In terms of detection speed, although YOLOv7 inference time is three times longer compared to other models when running on a system that uses only CPU, this difference in milliseconds (ms) is not considered a significant drawback for the project because the aim of the project does not involve real-time identification. Moreover, while running on a GPU and CUDA-supported system, YOLOv7 models have much lower inference time (IT) than YOLOv6 models (Table 4). Additionally, the YOLOv7 pair of models (WG and NG) exhibits overall better performance than model pairs from other YOLO versions when the models are evaluated on TeD using SISS. Eventually, the overall weights of the YOLOv7 model pair were selected as default weights for SpiderID_APP deployment.

Table 4.
Evaluation of trained models on TeD.
Inference time (IT) is calculated for predictions running on a GPU (IT-GPU) and on a CPU (IT-CPU); IT only considers the time (in milliseconds, ms) required to identify each image; the model loading time is excluded. The computational requirements, which are proportional to the number of parameters within the models in a nonlinear manner, are displayed in GFLOPS, equivalent to 2 billion (109) floating-point operations per second (FLOPS).
3.7. Assessment of Final Models on Flexibility and Robustness
Real-life spider records can be susceptible to various occlusions; images or video shapes can be captured in a variety of resolutions and angles. Due to the need to evaluate the models’ performance in real-life situations, where the confidence level of the identification is important, SISS was used as the evaluation metric. Firstly, we evaluated the performance of models on TeD at their original size and through the resizing process. Table 5 demonstrated a drop in performance with TeD images resized to a resolution other than 512 × 512; such decrease is small (<1%) for YOLOv7_WG models but by a considerable amount (>5%) for YOLOv7_NG. Secondly, to evaluate the robustness of the models, we utilized an augmented version of the TeD dataset (TeD_Aug) in which the images had been subjected to synthetic image perturbations. Three distinct versions of TeD_Aug were generated using different random seeds in the transformation script, an extended version of the one employed for augmenting the training and validation datasets since we had to include the augmentations performed by YOLO to account for a broad range of image occlusions. The performances of YOLOv7 models tested on TeD and TeD_Aug are displayed in Table 6. Considering SISS as an evaluation metric, the final model of YOLOv7_NG is overall better than its version that does not use the balanced dataset, with 0.3% and 4.9% increases in performance when tested on TeD and TeD_Aug, respectively. Meanwhile, the YOLOv7_WG model that was trained on the dataset augmented for balance interestingly has a slight drop in performance (~0.5%) compared to the non-augmented one when tested on TeD but a considerable boost (~4.4%) in performance when tested on TeD_Aug. Moreover, the final models have better robustness since their SISS decreases when tested on augmented images instead of original ones are only 3.06% and 1.28%, which are 54 to 75 percent lower than those of the non-balanced models, 6.76% and 5.13%, for WG and NG models, respectively. The accuracy of the v7_WG and v7_NG models across genera and families within the testing dataset can be further visualized in Figure 7.

Table 5.
Evaluation of final YOLOv7 models when receiving images at different resolutions.

Table 6.
Evaluation of final YOLOv7 models when being tested on original test dataset (TeD) and augmented test dataset (TeD_Aug). The TeD_Aug evaluations were performed in triplicate and are expressed as mean with standard deviation.
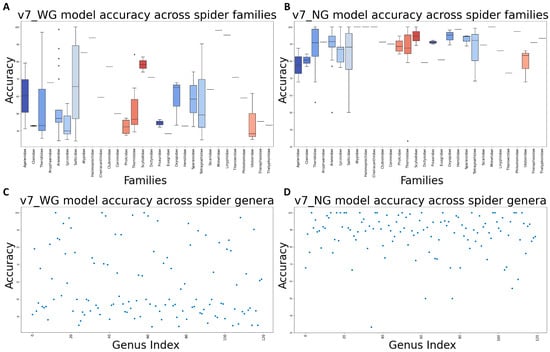
Figure 7.
Model accuracy across different spider taxa: (A) v7_WG model accuracy across spider families; (B) v7_NG model accuracy across spider families; (C) v7_WG model accuracy across spider genera; (D) v7_NG model accuracy across spider genera.
3.8. Assessment of Final Models through External Evaluation
In this study, we compared the identification performance between our SpiderID_APP and the iNaturalist classification tool. As the most expansive citizen science database, iNaturalist boasts an extensive metadata collection of myriad fauna and insects derived from user contributions and potentially other online publicly available databases. Given iNaturalist’s expansive and diverse metadata, it was necessary to create an evaluation set composed of locally stored images annotated by arachnology experts to establish reliable ground truth values. The evaluation set consisted of 51 images of spiders found in the Taiwan region, with 40 images featuring spider genera present in both tools’ databases and 11 images containing spider genera not included in our training dataset. These images were identified by the SpiderID_APP utilizing two distinct weight sets of YOLOv7 (v7_WG and v7_NG) to generate two sets of results accompanied by confidence scores. These images were subsequently graded by using the iNaturalist tool, yielding a third set of results without confidence scores, as iNaturalist needs to furnish such information. It can be observed from the iNaturalist website that their tool’s algorithm avoids results with low confidence scores, opting instead for higher taxonomic ranks (such as subfamily or family) until a specific confidence threshold is met. In contrast, our models are limited to identifying spiders at the genus level, albeit with the provision of confidence scores. The SISS method was employed to ensure a fair evaluation of the results. Upon evaluating the initial set of 40 images, SpiderID_APP and iNaturalist manifested virtually identical performance, with respective scores of 59 and 53 (Figure 8A). However, when the assessment was expanded to include an additional 11 images featuring spider genera absent from our training dataset—a circumstance guaranteed to yield a score of zero for SpiderID_APP—iNaturalist’s performance showcased only a minimal uptick (Figure 8B). Of the potential 22 additional points that could be obtained, iNaturalist’s score increased by 4 points. Even though all spider genera within the 51 images were represented in the iNaturalist database, the score of the iNaturalist tool only slightly improved when these unfamiliar genera were included.
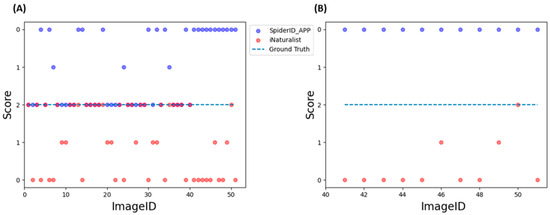
Figure 8.
Evaluating SpiderID_APP and iNaturalist using SISS on locally stored images provided by spider experts. (A) Evaluation of 40 images that contain spider genera that are presented in both tools. (B) Evaluation of 11 images that contain spider genera that are not presented in SpiderID_APP models training dataset. Blue points are SpiderID_APP scores; red points are iNaturalist scores; purple points are where both SpiderID_APP and iNaturalist were given a maximum point of 2 points, overlapping the ground truth line. Detailed evaluation can be found in Table S1A.
3.9. Easy operation of SpiderID_APP for Spider Identification at the Genus Level
To enable users to identify spiders efficiently, we developed a user-friendly application, SpiderID_APP, in which users can select one or several spider images for identification (Figure 9A). After selecting the appropriate weight, SpiderID_APP can recognize spiders at the genus level, providing gender information when using the v7_WG weight as the default setting or omitting gender information when the v7_NG weight is selected (Figure 9B). Additionally, SpiderID_APP supports video input and can halt the prediction process prematurely, saving the prediction results obtained at that point instead of waiting for the entire video to be identified, as is the case with the built-in prediction function of YOLOv7.
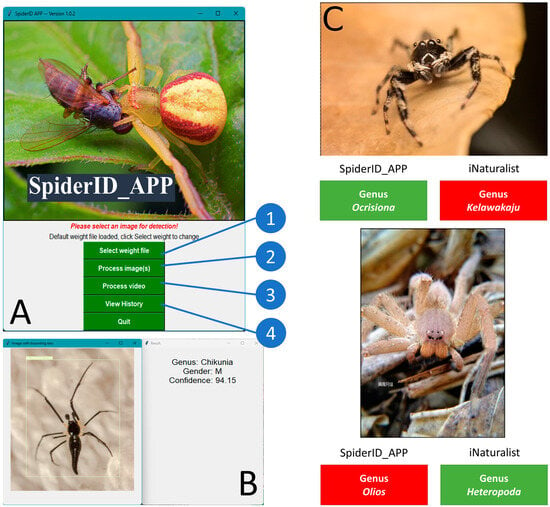
Figure 9.
A simple diagram showing how to conduct spider identification using SpiderID_APP. (A,B) The SpiderID_APP main interface with three function buttons: (1) load model from weight file, (2) select one or more images for identification, (3) select a video for identification, and (4) load analyze history. (C) Side-by-side comparison of spider identification results between SpiderID_APP and iNaturalist; the green box means correct identification, while the red box means incorrect identification.
4. Discussion
In this study, our primary endeavor was to harness the YOLO-based method to devise an initial screening tool, the SpiderID_APP, for spider identification in Taiwan. While this application marks a significant stride in the realm of spider distribution, its primary role within the scope of this study is the preliminary classification of major spider genera in Taiwan and goes some way to assist citizen science in obtaining information on spider identification. Such a tool, besides its identification capability, can also contribute to diminishing the unwarranted fear many harbor toward spiders [66]. An interesting observation from our data collection is that the 120 genera classified represent 99.69% of the recorded spider data in Taiwan (Supplemental Figure S1A). This highlights that while there are more genera in the Taiwanese habitat, their lower frequency of encounters and scant record numbers limit their inclusion and representativeness in our model training. This dominance of certain genera implies that the SpiderID_APP identification capabilities are predominantly inclined towards spiders from peridomestic areas and those residing in hiking trails and national parks. Recognizing this, we want users to be aware of the APP’s inherent bias towards certain environments, especially since the data suggest a higher representation of genera frequently encountered by humans. Considering the dominant genera in our dataset, the SpiderID_APP, developed in a Python environment [67], is tailored to classify spiders that are frequently documented in Taiwan. However, the underlying design and framework of the APP suggest the potential to facilitate the identification of other less documented or even unfamiliar genera based on shared morphological and pattern characteristics present in the data. Taxa not included in this study, like Callitrichia formosana and Loxobates daitoensis, are either non-indigenous or exceedingly rare, with minimal sightings reported, such as those in Taiwan based on the TBN records (accessed on 20 July 2022).
The proliferation of deep learning applications in image-processing models across sectors like biomedical research, plant sciences, and biodiversity has been remarkable [68,69,70,71,72]. Yet, the task of discerning spider images is daunting given the variability in imaging conditions. In our pursuit, we employed YOLO-based models for spider genus classification, utilizing an extensive image dataset. Post-collection, images were standardized to 512 × 512 pixels, ensuring uniformity for the YOLO model [73]. The data augmentation strategies employed included random flip, rotation, and dual-axis shear. Our evaluations pinpointed the YOLOv7 model as the most adept for our classification aims, mirroring its success in other domains [33,74]. However, while our tool is pioneering, it is pivotal to underscore that it does not supplant the intricate processes of taxonomic analysis [75]. As a precursor, it simplifies the process of initial screening, but the depth of taxonomy lies beyond its scope. This limitation is recognized as one of the inherent challenges in the application of machine learning for species identification, especially when considering the rich diversity of spider species within Taiwan and globally.
During the model training process, we incorporated various hyperparameter optimization steps, such as input image size and training batch size. Input image size impacts computational requirements and detail detection, with smaller sizes reducing memory consumption and training time but potentially losing critical information. In comparison, larger sizes allow for finer detail detection but increase resource usage [76,77,78]. Furthermore, using an image size configuration higher than the training dataset image size can also cause potential loss of mid-level and high-level features [76], negatively affecting the model learning process and leading to suboptimal detection performance. Regarding batch size, fine-tuning the batch size hyperparameter is significant for balancing the tradeoff between accuracy and training speed [79]. While larger batch sizes accelerate the training process by allowing the optimization algorithm to process more training samples per iteration, they may result in less noisy gradient estimates and reduced stochasticity, potentially leading to overfitting or poor generalization on validation or test data [80]. However, despite causing slower training speed, smaller batch sizes do not guarantee superior model performance. Smaller batch sizes introduce more noise in the gradient estimates since fewer samples are used to compute the gradients. This increased noise might help the model to bypass the local minima or saddle points. However, it also can lead to convergence instability, causing the model’s training loss to fluctuate and making it more challenging for the model to reach an optimal solution. This can prevent the model from escaping suboptimal regions in the loss landscape. Another issue worth considering is that a smaller batch size may impede the training process, primarily on hardware optimized for large batch sizes, and a large batch size will cause out-of-memory errors on systems that have low GPU memory [81,82]. By carefully considering these parameters, we developed an efficient and accurate model for spider identification.
It can be observed throughout the result demonstration that WG models generally do not perform as well as NG models. The assessment using Spearman rank correlation of all the training classes in WG models shows that the male percentage of the training dataset has a significant (p < 0.0001) correlation with every WG model’s SISS evaluation score on male spiders in each genus (Supplemental Figure S1E). It has been suggested in several deep learning studies that an imbalance in the dataset can lead to a decrease in the overall performance of the model, primarily due to the nature of machine learning, which relies on patterns learned from data to make predictions and may tend to favor the majority class in pursuit of minimizing loss and maximizing accuracy [83,84]. A similar phenomenon can be observed based on the assessment of individual training classes in this study. However, such imbalance is inevitable in the current study because even though dataset balancing was employed, the ratio discrepancy between male and female images is vast and can be compensated by such up-sampling techniques. Data-balancing strategies such as over-sampling and under-sampling [85] using the mentioned image augmentation techniques can only be applied to the NG dataset but not the WG dataset. This was observed to be due to the lack of male individuals in some spider genera, which consequently generated several training classes of male spiders with zero or close-to-zero image counts (Table S1B). There are several contributing factors to this observed pattern. Firstly, in certain genera, gender identification from a single-angle image proved challenging, as the dimorphic features were not clearly discernible. Secondly, for some genera, the copulatory organs are relatively diminutive. Thus, the differentiation between male and female specimens is primarily predicated on relative size disparities. For instance, genera like Pholcus, Smeringopus, and Spermophora, which are often referred to as “cellar spiders”, present challenges in gender differentiation without a comparative reference. The third reason is that the natural gender distribution in such genera is already imbalanced due to underlying bias in offspring production [86]. While removing the training classes with close-to-zero image counts could potentially increase the overall model performance, targeting a constantly updated model was valued over the temporary performance of the current study’s model. Indeed, keeping the classes intact to facilitate a smooth transfer learning process [83] and gathering sufficient training data to reach a certain male percentage threshold for each YOLO model would certainly improve the models’ performance in future studies (Supplemental Figure S1F).
In our internal evaluation of eight models, we observed varying rankings when using two different evaluation methods: F1-score and SISS. Specifically, in the final assessment, model v5_WG achieved a higher F1-score but a lower SISS score compared to v7_WG. This discrepancy stems from the inherent nature of these metrics. While F1-score assesses the balance between precision and recall without factoring in the confidence of predictions, SISS awards points based on both the correctness and confidence level of predictions. Consequently, this revealed that although v5_WG was adept at correctly identifying the spider genus, it did so with less confidence than v7_WG, resulting in overall less reliable outcomes. In this context, where an accurate and confident identification of a spider genus is paramount to prevent misclassification, SISS emerges as the more appropriate metric for final evaluation. This emphasis on high-confidence predictions underscores the reliability and value of models in real-world applications. This understanding of model performance and reliability led us to further assess the flexibility and robustness of our final models. Notably, a significant decline in performance was observed when the input images were not resized, highlighting a challenge in adapting to varying image sizes. To mitigate this, we introduced a resize variable in the prediction script, effectively addressing this limitation. Additionally, when tested on the augmented TeD dataset, our final models exhibited greater resilience compared to those trained on non-balanced data. This resilience was further evidenced by a markedly lower performance decrease—ranging from 54 to 75 percent less than the primitive models—when various synthetic image perturbations were introduced. Such findings highlight the critical role of data augmentation in enhancing the performance and robustness of our models, ensuring their efficacy in diverse and challenging identification scenarios.
In the external evaluation, iNaturalist was selected as a comparative tool due to its prominent standing as an optimal spider identification instrument in the research community [18]. Our SpiderID_APP exhibited marginally superior results compared to iNaturalist in our limited testing dataset, yielding scores of 59 and 57, respectively. When omitting the 11 data points relating to genera not encompassed in our training dataset, this score becomes 59 to 53. It is critical to underline that the design intent behind SpiderID_APP is not to overshadow iNaturalist but to act as a supplementary offline tool that is specially designed for researchers with a keen interest in natural taxonomy. Notably, the performance of SpiderID_APP varied when juxtaposed with iNaturalist, surpassing it in certain identification scenarios and lagging in others (as illustrated in Figure 9C). Multiple underlying factors dictate this differential performance. Primarily, the nature of their respective training datasets is distinct. The specialized dataset of SpiderID_APP, being spider-focused, offers it a potentially enhanced acumen in spider identification. On the other hand, iNaturalist’s dataset, by virtue of encompassing myriad animal species, offers a broader range but perhaps at the expense of specialized precision. The constrained size of our dataset, relative to iNaturalist’s expansive one, might curtail its proficiency in discerning spider genera that exhibit high morphological similarities. The focus on a subset of around 50 prevalent spider species is deliberate, considering the pronounced influence of urbanization on biodiversity. The spiders that frequently manifest in urban settings, and thus are predominantly recorded on platforms like iNaturalist, are typically from this select group. Correctly identifying this subset is of paramount importance for a range of applications, from public health to ecological studies. By refining our model to cater to this subset, our objective is to address the immediate identification challenges faced by the majority of users, while recognizing that capturing the extensive diversity of spiders is a far more intricate endeavor. Lastly, the foundational models and image classification methodologies differ between the tools. SpiderID_APP employs YOLOv7 for object detection, leveraging bounding box theory, whereas iNaturalist, over its evolution, has incorporated a variety of models, such as Inception-V3 [18], Resnet-50, Regnet-8GF [87], MAE [40], and MetaFormer, all of which are based on whole image inputs.
In the realm of academic research, two other studies have focused on spider identification, considering taxonomy and gender [27,28]. Unfortunately, comparisons with these studies are not feasible. One study did not release its model or application [27], while the other claimed to have developed an iOS application [28]. However, their associated GitHub repository is outdated, making a comparative test impossible. In the broader context of spider identification methodologies outside academic circles, it is observed that platforms like community-driven forums, while free, rely on user submissions and collective expertise, which might not always yield the most accurate or timely identifications. Conversely, many mobile applications, despite their claims of sophisticated AI and machine learning capabilities, often restrict access through payment models or advertisements, potentially limiting their use.
In contrast, SpiderID_APP stands out with its user-centric approach, providing immediate, automated identification without such barriers. This accessibility, combined with advanced technological capabilities, sets SpiderID_APP apart from other methodologies. The application includes a setup file that streamlines the installation process, making it especially user-friendly for those not well versed in Python or coding languages. Our setup file is a batch file (setup.bat) compatible with Windows and MacOS. It automatically downloads and installs the Python environment and the necessary dependencies for SpiderID_APP. Upon completion, a run.bat file is generated, which enables users to launch the application directly, bypassing the need to navigate through several installation steps. By prioritizing ease of use, SpiderID_APP offers a unique blend of technological sophistication and accessibility, catering to both researchers and enthusiasts seeking reliable and straightforward spider identifications.
5. Potential Limitations and Future Work
Our research, pioneering in its approach, is focused on spider identification within Taiwan, utilizing a dataset that encompasses 120 genera. This specific focus, while effective for the Taiwanese context, highlights a significant limitation—the challenge of generalizing this model to diverse spider populations in different geographical regions. The 120 genera covered represent a substantial portion of Taiwan’s spider diversity, but they may not encapsulate the broader spectrum of species found globally. Addressing this constraint is pivotal for the future development of SpiderID_APP. As we plan for SpiderID_APP 2.0, aiming to extend our reach beyond Taiwan, we anticipate encountering a more complex array of spider genera. This complexity is not just in numbers but also in the nuanced morphological differences that are critical for accurate identification. Some of these genera may require advanced identification techniques, such as genitalia dissection, which are beyond the current capabilities of image-based AI models. This expansion will not only test the model’s adaptability to a wider range of species but also its ability to maintain accuracy amidst greater diversity. In tackling these challenges, SpiderID_APP 2.0 will explore the integration of a microscopic-adept AI model. This model would assist in instances where the primary AI tool yields low-confidence identifications, potentially utilizing high-resolution microscopic images for a deeper level of analysis. However, this approach brings forth its own set of challenges. Ensuring the accuracy and reliability of the model across diverse species, each with unique identifying features, will be a significant undertaking. It underscores the critical role of human expertise in the taxonomic process, not only in model validation but also in guiding the AI’s learning process.
Moreover, a comprehensive analysis of the challenges faced by SpiderID_APP reveals several key areas of concern. Firstly, the current model’s reliance on image quality and specific imaging conditions can lead to limitations in real-world scenarios where such ideal conditions may not always be met. Secondly, the potential for bias in the training dataset, given the prevalence of certain spider species over others, might skew the model’s performance. This bias could manifest in less accurate identifications for less commonly encountered species. Lastly, the necessity for continual updates to the model’s training dataset cannot be overstated, especially in the rapidly evolving field of taxonomy where new species are discovered and existing classifications are frequently updated. These factors collectively emphasize the need for ongoing refinement and enhancement of the SpiderID_APP to ensure its robustness and reliability as a tool for spider identification.
6. Conclusions
In this study, we have successfully developed SpiderID_APP, an AI-driven application that leverages the YOLOv7 object detection model for the preliminary identification of major spider genera in Taiwan. This tool represents a significant step in the application of advanced AI methodologies to the field of arachnology, specifically targeting the needs of spider identification within a defined geographical region. The core achievement of our research lies in the creation of a practical, user-friendly tool that assists in the initial classification of spiders. By streamlining the identification process, SpiderID_APP facilitates the work of both professional arachnologists and citizen scientists. Its utility, however, is primarily as a preliminary screening tool, laying a foundation for further detailed taxonomic analysis by experts. Our approach demonstrates the potential of AI in enhancing traditional biological studies, particularly in biodiversity research. While SpiderID_APP is tailored for a specific ecological and geographical context, the methodologies and insights gained could serve as a valuable reference for similar projects in other areas of biological research. It is important to note that while SpiderID_APP marks a step forward in integrating technology with biodiversity studies, it does not aim to replace the nuanced and detailed work involved in spider taxonomy. Instead, it seeks to complement and support these efforts by providing a reliable and efficient means for initial species identification.
In conclusion, our study contributes to the field of arachnology by introducing an innovative tool that combines technological advancement with biological research. SpiderID_APP serves as an example of how AI can be effectively applied to specific challenges in biodiversity studies, offering a model that can be adapted and extended to other research areas within the biological sciences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/inventions8060153/s1, Figure S1: The graphical representation of spider data distribution at the genus level (% on genus and image count) which was obtained based on data from TBN (Taiwan Biodiversity Network, https://www.tbn.org.tw/taxa, accessed on 3 December 2023) and the WSC (World Spider Catalog, https://wsc.nmbe.ch/, accessed on 27 July 2022); Table S1A: Details of SpiderID_APP and iNaturalist evaluations using SISS on locally stored images provided by spider experts. Table S1B: Dataset metadata and prediction accuracies of trained models against test dataset.
Author Contributions
Conceptualization, C.-D.H. and C.T.L.; methodology, A.F.; software, C.T.L.; validation, C.-D.H., C.-H.H. and Y.-K.L.; formal analysis, C.T.L.; investigation, C.T.L.; resources, M.-D.L.; data curation, A.F.; writing—original draft preparation, C.T.L.; writing—review and editing, M.J.M.R. and R.D.V.; visualization, S.-Y.H.; supervision, Y.-K.L.; project administration, C.-H.H.; funding acquisition, C.-D.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
SpiderID_APP and its source code and supporting modules are available on GitHub at https://github.com/ThangLC304/SpiderID_APP (accessed on 2 December 2023). In addition, the system requirements and installation instructions can be found in the Readme.md file.
Acknowledgments
The authors would like to thank Gilbert Audira for the support in the editing process of the manuscript.
Conflicts of Interest
The authors declare that they have no known competing financial interest or personal relationships that could have appeared to influence the work reported in this paper. The authors declare no conflict of interest.
References
- Coyle, F.A. Effects of clearcutting on the spider community of a southern Appalachian forest. J. Arachnol. 1981, 9, 285–298. [Google Scholar]
- Nyffeler, M.; Sunderland, K.D. Composition, abundance and pest control potential of spider communities in agroecosystems: A comparison of European and US studies. Agric. Ecosyst. Environ. 2003, 95, 579–612. [Google Scholar] [CrossRef]
- Natural History Museum Bern. World Spider Catalog. Version 22.0. 2021. Available online: https://wsc.nmbe.ch/resources/archive/catalog_22.0/index.html (accessed on 17 July 2022).
- Riechert, S.E. Thoughts on the ecological significance of spiders. BioScience 1974, 24, 352–356. [Google Scholar] [CrossRef]
- Wise, D.H. Spiders in Ecological Webs; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- McLean, C.J.; Garwood, R.J.; Brassey, C.A. Sexual dimorphism in the Arachnid orders. PeerJ 2018, 6, e5751. [Google Scholar] [CrossRef]
- Vollrath, F.; Parker, G.A. Sexual dimorphism and distorted sex ratios in spiders. Nature 1992, 360, 156–159. [Google Scholar] [CrossRef]
- Saturnino, R.; Bonaldo, A.B. Taxonomic review of the New World spider genus Elaver O. Pickard-Cambridge, 1898 (Araneae, Clubionidae). Zootaxa 2015, 4045, 1–119. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A. Review of erigonine spider genera in the Neotropics (Araneae: Linyphiidae, Erigoninae). Zool. J. Linn. Soc. 2007, 149, 1–263. [Google Scholar] [CrossRef]
- Do, M.; Harp, J.; Norris, K. A test of a pattern recognition system for identification of spiders. Bull. Entomol. Res. 1999, 89, 217–224. [Google Scholar] [CrossRef]
- Derraik, J.G.; Closs, G.P.; Dickinson, K.J.; Sirvid, P.; Barratt, B.I.; Patrick, B.H. Arthropod morphospecies versus taxonomic species: A case study with Araneae, Coleoptera, and Lepidoptera. Conserv. Biol. 2002, 16, 1015–1023. [Google Scholar] [CrossRef]
- Bonney, R.; Cooper, C.B.; Dickinson, J.; Kelling, S.; Phillips, T.; Rosenberg, K.V.; Shirk, J. Citizen science: A developing tool for expanding science knowledge and scientific literacy. BioScience 2009, 59, 977–984. [Google Scholar] [CrossRef]
- Pocock, M.J.O.; Roy, H.E.; Preston, C.D.; Roy, D.B. The Biological Records Centre: A pioneer of citizen science. Biol. J. Linn. Soc. 2015, 115, 475–493. [Google Scholar] [CrossRef]
- Di Febbraro, M.; Bosso, L.; Fasola, M.; Santicchia, F.; Aloise, G.; Lioy, S.; Tricarico, E.; Ruggieri, L.; Bovero, S.; Mori, E. Different facets of the same niche: Integrating citizen science and scientific survey data to predict biological invasion risk under multiple global change drivers. Glob. Chang. Biol. 2023, 29, 5509–5523. [Google Scholar] [CrossRef]
- Nanglu, K.; de Carle, D.; Cullen, T.M.; Anderson, E.B.; Arif, S.; Castañeda, R.A.; Chang, L.M.; Iwama, R.E.; Fellin, E.; Manglicmot, R.C. The nature of science: The fundamental role of natural history in ecology, evolution, conservation, and education. Ecol. Evol. 2023, 13, e10621. [Google Scholar] [CrossRef] [PubMed]
- Hart, A.G.; Nesbit, R.; Goodenough, A.E. Spatiotemporal variation in house spider phenology at a national scale using citizen science. Arachnology 2018, 17, 331–334. [Google Scholar] [CrossRef]
- Nugent, J. INaturalist. Sci. Scope 2018, 41, 12–13. [Google Scholar] [CrossRef]
- Van Horn, G.; Mac Aodha, O.; Song, Y.; Cui, Y.; Sun, C.; Shepard, A.; Adam, H.; Perona, P.; Belongie, S. The inaturalist species classification and detection dataset. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8769–8778. [Google Scholar]
- Miller, J.A.; Griswold, C.E.; Scharff, N.; Řezáč, M.; Szűts, T.; Marhabaie, M. The velvet spiders: An atlas of the Eresidae (Arachnida, Araneae). ZooKeys 2012, 1–144. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Padilla, F.; Hormiga, G. Morphological and phylogenetic atlas of the orb-weaving spider family Tetragnathidae (Araneae: Araneoidea). Zool. J. Linn. Soc. 2011, 162, 713–879. [Google Scholar] [CrossRef]
- Harvey, P.R.; Nellist, D.R.; Telfer, M.G. Provisional Atlas of BRITISH Spiders (Arachnida, Araneae), Volume 1; Biological Records Centre, Centre for Ecology and Hydrology: Huntingdon, England, 2002. [Google Scholar]
- Menge, A. Preussische Spinnen; Schriften Der Naturforschenden Gesellschaft in Danzig: Frankfurt, Germany, 1873; Volume 6. [Google Scholar]
- Caci, G.; Biscaccianti, A.B.; Cistrone, L.; Bosso, L.; Garonna, A.P.; Russo, D. Spotting the right spot: Computer-aided individual identification of the threatened cerambycid beetle Rosalia alpina. J. Insect Conserv. 2013, 17, 787–795. [Google Scholar] [CrossRef]
- Willi, M.; Pitman, R.T.; Cardoso, A.W.; Locke, C.; Swanson, A.; Boyer, A.; Veldthuis, M.; Fortson, L. Identifying animal species in camera trap images using deep learning and citizen science. Methods Ecol. Evol. 2019, 10, 80–91. [Google Scholar] [CrossRef]
- Nanni, L.; Maguolo, G.; Pancino, F. Insect pest image detection and recognition based on bio-inspired methods. Ecol. Inform. 2020, 57, 101089. [Google Scholar] [CrossRef]
- Alves, A.N.; Souza, W.S.R.; Borges, D.L. Cotton pests classification in field-based images using deep residual networks. Comput. Electron. Agric. 2020, 174, 105488. [Google Scholar] [CrossRef]
- Chen, Q.; Ding, Y.; Liu, C.; Liu, J.; He, T. Research on spider sex recognition from images based on deep learning. IEEE Access 2021, 9, 120985–120995. [Google Scholar] [CrossRef]
- Sinnott, R.O.; Yang, D.; Ding, X.; Ye, Z. Poisonous spider recognition through deep learning. In Proceedings of the Australasian Computer Science Week Multiconference, Melbourne, VIC, Australia, 4–6 February 2020; pp. 1–7. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Du, Y.; Pan, N.; Xu, Z.; Deng, F.; Shen, Y.; Kang, H. Pavement distress detection and classification based on YOLO network. Int. J. Pavement Eng. 2021, 22, 1659–1672. [Google Scholar] [CrossRef]
- Radovic, M.; Adarkwa, O.; Wang, Q. Object recognition in aerial images using convolutional neural networks. J. Imaging 2017, 3, 21. [Google Scholar] [CrossRef]
- Sathvik, M.; Saranya, G.; Karpagaselvi, S. An Intelligent Convolutional Neural Network based Potholes Detection using Yolo-V7. In Proceedings of the 2022 International Conference on Automation, Computing and Renewable Systems (ICACRS), Pudukkottai, India, 13–15 December 2022; pp. 813–819. [Google Scholar]
- Priyankan, K.; Fernando, T. Mobile Application to Identify Fish Species Using YOLO and Convolutional Neural Networks. In Proceedings of the International Conference on Sustainable Expert Systems: ICSES 2020, Singapore, 31 March 2021; pp. 303–317. [Google Scholar]
- Divya, A.; Sungeetha, D.; Ramesh, S. Horticulture image based weed detection in feature extraction with dimensionality reduction using deep learning architecture. In Proceedings of the 2023 3rd International Conference on Artificial Intelligence and Signal Processing (AISP), Vijayawada, India, 18–20 March 2023; pp. 1–8. [Google Scholar]
- Liang, B.; Wu, S.; Xu, K.; Hao, J. Butterfly detection and classification based on integrated YOLO algorithm. In Proceedings of the Thirteenth International Conference on Genetic and Evolutionary Computing, Qingdao, China, 1–3 November 2019; pp. 500–512. [Google Scholar]
- Wu, W.; Liu, H.; Li, L.; Long, Y.; Wang, X.; Wang, Z.; Li, J.; Chang, Y. Application of local fully Convolutional Neural Network combined with YOLO v5 algorithm in small target detection of remote sensing image. PLoS ONE 2021, 16, e0259283. [Google Scholar] [CrossRef] [PubMed]
- Yung, N.D.T.; Wong, W.; Juwono, F.H.; Sim, Z.A. Safety helmet detection using deep learning: Implementation and comparative study using YOLOv5, YOLOv6, and YOLOv7. In Proceedings of the 2022 International Conference on Green Energy, Computing and Sustainable Technology (GECOST), Virtual, 26–28 October 2022; pp. 164–170. [Google Scholar]
- Cao, L.; Zheng, X.; Fang, L. The Semantic Segmentation of Standing Tree Images Based on the Yolo V7 Deep Learning Algorithm. Electronics 2023, 12, 929. [Google Scholar] [CrossRef]
- Nan, Y.; Zhang, H.; Zeng, Y.; Zheng, J.; Ge, Y. Faster and accurate green pepper detection using NSGA-II-based pruned YOLOv5l in the field environment. Comput. Electron. Agric. 2023, 205, 107563. [Google Scholar] [CrossRef]
- Alvarez-Padilla, F.; Galán-Sánchez, M.A.; Salgueiro-Sepúlveda, F.J. A protocol for online documentation of spider biodiversity inventories applied to a Mexican tropical wet forest (Araneae, Araneomorphae). Zootaxa 2020, 4722, 241–269. [Google Scholar] [CrossRef]
- Levi, H.W. Techniques for the study of spider genitalia. Psyche A J. Entomol. 1965, 72, 152–158. [Google Scholar] [CrossRef]
- Nelson, X.J. Polymorphism in an ant mimicking jumping spider. J. Arachnol. 2010, 38, 139–141. [Google Scholar] [CrossRef]
- Puzin, C.; Leroy, B.; Pétillon, J. Intra-and inter-specific variation in size and habitus of two sibling spider species (Araneae: Lycosidae): Taxonomic and biogeographic insights from sampling across Europe. Biol. J. Linn. Soc. 2014, 113, 85–96. [Google Scholar] [CrossRef]
- Michalko, R.; Košulič, O.; Hula, V.; Surovcová, K. Niche differentiation of two sibling wolf spider species, Pardosa lugubris and Pardosa alacris, along a canopy openness gradient. J. Arachnol. 2016, 44, 46–51. [Google Scholar] [CrossRef]
- Luebke, D. CUDA: Scalable parallel programming for high-performance scientific computing. In Proceedings of the 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Paris, France, 14–17 May 2008; pp. 836–838. [Google Scholar]
- Imambi, S.; Prakash, K.B.; Kanagachidambaresan, G. PyTorch. Program. TensorFlow Solut. Edge Comput. Appl. 2021, 87–104. [Google Scholar] [CrossRef]
- Kanagachalam, S.; Tulkinbekov, K.; Kim, D.-H. Blosm: Blockchain-based service migration for connected cars in embedded edge environment. Electronics 2022, 11, 341. [Google Scholar] [CrossRef]
- Lin, C.-H.; Kan, C.-D.; Chen, W.-L.; Huang, P.-T. Application of two-dimensional fractional-order convolution and bounding box pixel analysis for rapid screening of pleural effusion. J. X-ray Sci. Technol. 2019, 27, 517–535. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, W.G.; Huber, B.A. The Evolution of Primary Sexual Characters in Animals; Oxford University Press: New York, NY, USA, 2010; pp. 249–284. [Google Scholar]
- Cordellier, M.; Schneider, J.M.; Uhl, G.; Posnien, N. Sex differences in spiders: From phenotype to genomics. Dev. Genes Evol. 2020, 230, 155–172. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Raschka, S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv 2018, arXiv:1811.12808. [Google Scholar]
- Xia, Y.; Luo, W.; Zhang, P.; Liu, Y.; Bei, J. Detection of insulator defects based on improved YOLOv7 model. In Proceedings of the International Symposium on Artificial Intelligence and Robotics 2022, Shanghai, China, 21–23 October 2022; pp. 157–165. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Horak, K.; Sablatnig, R. Deep learning concepts and datasets for image recognition: Overview 2019. In Proceedings of the Eleventh international conference on digital image processing (ICDIP 2019), Guangzhou, China, 10–13 May 2019; pp. 484–491. [Google Scholar]
- Martinez, M.T. An Overview of Google’s Open Source Machine Intelligence Software TensorFlow. In Proceedings of the Technical Seminar, Las Cruces, NM, USA, November 2016. [Google Scholar]
- Zheng, J.; Wu, H.; Zhang, H.; Wang, Z.; Xu, W. Insulator-defect detection algorithm based on improved YOLOv7. Sensors 2022, 22, 8801. [Google Scholar] [CrossRef]
- Gevorgyan, Z. SIoU loss: More powerful learning for bounding box regression. arXiv 2022, arXiv:2205.12740. [Google Scholar]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Mane, S.; Srivastava, J.; Hwang, S.-Y.; Vayghan, J. Estimation of false negatives in classification. In Proceedings of the Fourth IEEE International Conference on Data Mining (ICDM’04), Brighton, UK, 1–4 November 2004; pp. 475–478. [Google Scholar]
- Jabed, M.R.; Shamsuzzaman, M. YOLObin: Non-decomposable garbage identification and classification based on YOLOv7. J. Comput. Commun. 2022, 10, 104–121. [Google Scholar] [CrossRef]
- Kramer, O.; Kramer, O. Scikit-learn. Mach. Learn. Evol. Strateg. 2016, 20, 45–53. [Google Scholar]
- Sun, M.; Song, Z.; Jiang, X.; Pan, J.; Pang, Y. Learning pooling for convolutional neural network. Neurocomputing 2017, 224, 96–104. [Google Scholar] [CrossRef]
- Zhou, D.-X. Theory of deep convolutional neural networks: Downsampling. Neural Netw. 2020, 124, 319–327. [Google Scholar] [CrossRef]
- Lundh, F. An Introduction to Tkinter. 1999. Available online: http://jgaltier.free.fr/Terminale_S/ISN/TclTk_Introduction_To_Tkinter.pdf (accessed on 3 December 2023).
- Asma, S.T. Monsters on the brain: An evolutionary epistemology of horror. Soc. Res. 2014, 81, 941–968. [Google Scholar] [CrossRef]
- McKinney, W. Data structures for statistical computing in python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 51–56. [Google Scholar]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Eulenberg, P.; Köhler, N.; Blasi, T.; Filby, A.; Carpenter, A.E.; Rees, P.; Theis, F.J.; Wolf, F.A. Reconstructing cell cycle and disease progression using deep learning. Nat. Commun. 2017, 8, 463. [Google Scholar] [CrossRef]
- Lippeveld, M.; Knill, C.; Ladlow, E.; Fuller, A.; Michaelis, L.J.; Saeys, Y.; Filby, A.; Peralta, D. Classification of human white blood cells using machine learning for stain-free imaging flow cytometry. Cytom. Part A 2020, 97, 308–319. [Google Scholar] [CrossRef]
- Shoaib, M.; Shah, B.; Ei-Sappagh, S.; Ali, A.; Ullah, A.; Alenezi, F.; Gechev, T.; Hussain, T.; Ali, F. An advanced deep learning models-based plant disease detection: A review of recent research. Front. Plant Sci. 2023, 14, 1158933. [Google Scholar] [CrossRef]
- Panigrahi, S.; Maski, P.; Thondiyath, A. Deep learning based real-time biodiversity analysis using aerial vehicles. In Proceedings of the International Conference on Robot Intelligence Technology and Applications, Daejeon, Republic of Korea, 16–17 December 2021; pp. 401–412. [Google Scholar]
- Bisogni, C.; Castiglione, A.; Hossain, S.; Narducci, F.; Umer, S. Impact of deep learning approaches on facial expression recognition in healthcare industries. IEEE Trans. Ind. Inform. 2022, 18, 5619–5627. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Xin, Z. Efficient Detection Model of Steel Strip Surface Defects Based on YOLO-V7. IEEE Access 2022, 10, 133936–133944. [Google Scholar] [CrossRef]
- Simpson, G.G. Principles of Animal Taxonomy; Columbia University Press: Columbia, SC, USA, 1961. [Google Scholar]
- Radiuk, P.M. Impact of Training Set Batch Size on the Performance of Convolutional Neural Networks for Diverse Datasets. Inf. Technol. Manag. 2017, 20, 20–24. [Google Scholar] [CrossRef]
- Aldin, N.B.; Aldin, S.S.A.B. Accuracy comparison of different batch size for a supervised machine learning task with image classification. In Proceedings of the 2022 9th International Conference on Electrical and Electronics Engineering (ICEEE), Alanya, Turkey, 29–31 March 2022; pp. 316–319. [Google Scholar]
- Smistad, E.; Elster, A.C.; Lindseth, F. Real-time gradient vector flow on GPUs using OpenCL. J. Real-Time Image Process. 2015, 10, 67–74. [Google Scholar] [CrossRef]
- Keskar, N.S.; Mudigere, D.; Nocedal, J.; Smelyanskiy, M.; Tang, P.T.P. On large-batch training for deep learning: Generalization gap and sharp minima. arXiv 2016, arXiv:1609.04836. [Google Scholar]
- Smith, S.; Elsen, E.; De, S. On the generalization benefit of noise in stochastic gradient descent. In Proceedings of the International Conference on Machine Learning, Virtual Event, 13–18 July 2020; pp. 9058–9067. [Google Scholar]
- Le, T.D.; Imai, H.; Negishi, Y.; Kawachiya, K. Automatic gpu memory management for large neural models in tensorflow. In Proceedings of the 2019 ACM SIGPLAN International Symposium on Memory Management, Phoenix, AZ, USA, 23 June 2019; pp. 1–13. [Google Scholar]
- Lim, H.; Kim, T.H.; Kang, S. Prediction-based error correction for gpu reliability with low overhead. Electronics 2020, 9, 1849. [Google Scholar] [CrossRef]
- Kaur, H.; Pannu, H.S.; Malhi, A.K. A systematic review on imbalanced data challenges in machine learning: Applications and solutions. ACM Comput. Surv. (CSUR) 2019, 52, 1–36. [Google Scholar] [CrossRef]
- Japkowicz, N.; Stephen, S. The class imbalance problem: A systematic study. Intell. Data Anal. 2002, 6, 429–449. [Google Scholar] [CrossRef]
- Mohammed, R.; Rawashdeh, J.; Abdullah, M. Machine learning with oversampling and undersampling techniques: Overview study and experimental results. In Proceedings of the 2020 11th International Conference on Information and Communication Systems (ICICS), Irbid, Jordan, 7–9 April 2020; pp. 243–248. [Google Scholar]
- Vanthournout, B.; Busck, M.M.; Bechsgaard, J.; Hendrickx, F.; Schramm, A.; Bilde, T. Male spiders control offspring sex ratio through greater production of female-determining sperm. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172887. [Google Scholar] [CrossRef]
- Touvron, H.; Sablayrolles, A.; Douze, M.; Cord, M.; Jégou, H. Grafit: Learning fine-grained image representations with coarse labels. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 874–884. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).