Development and Characterization of Fifteen Polymorphic Microsatellite Loci for Rare and Endangered Species within Luciobarbus Heckel, 1843 Genus in the Aral Basin and Their Conservation Application
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Froese, R.; Pauly, D. (Eds.) FishBase. Worldwide Web Electronic Publication. Version (06/2023). 2023. Available online: www.fishbase.org (accessed on 11 November 2023).
- Doadrio, I. Freshwater fish fauna of North Africa and its biogeography. Ann. R. Cent. Afr. Mus. 1994, 275, 21–34. [Google Scholar]
- Kottelat, M.; Freyhof, J. Handbook of European Freshwater Fishes; Cornol: Berlin, Germany, 2007; 646p. [Google Scholar]
- Berg, L.S. Fish of Freshwater of USSR and Neighboring Counties, 3rd ed.; AS USSR: Moscow, Russia, 1949; Part 2; p. 458. [Google Scholar]
- Xenarios, S.; Schmidt-Vogt, D.; Qadir, M.; Janusz-Pawletta, B.; Abdullaev, I. The Aral Sea Basin: Water for Sustainable Development in Central Asia; Routledge: Oxfordshire, UK, 2019. [Google Scholar]
- Reimov, P.; Fayzieva, D. The Present State of the South Aral Sea Area. In The Aral Sea; Micklin, P., Aladin, N., Plotnikov, I., Eds.; Springer Earth System Sciences; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Anchita; Zhupankhan, A.; Khaibullina, Z.; Kabiyev, Y.; Persson, K.M.; Tussupova, K. Health impact of drying Aral Sea: One health and socio-economical approach. Water 2021, 13, 3196. [Google Scholar] [CrossRef]
- Esmaeili, H.R.; Mehraban, H.; Abbasi, K.; Keivany, Y.; Coad, B.W. Review and updated checklist of freshwater fishes of Iran: Taxonomy, distribution and conservation status. Iran J. Ichthyol. 2017, 4, 1–114. [Google Scholar]
- Dumont, H.J. Endemism in the Ponto—Caspian Fauna, with Special Emphasis on the Onychopoda (Crustacea). Adv. Ecol. Res. 2000, 31, 181–196. [Google Scholar]
- Nikolski, G.V. Structura vida i Zakonomernosti Izmenchivosti ryb; Pischevaya Prom.: Moskva, Russia, 1980; 183 s. [Google Scholar]
- Rass, T.S. Animal Life. T. Lancelets, Cyclostomes, Cartilaginous Fishes, Bony Fishes; Education: Moscow, Russia, 1983; 576p. [Google Scholar]
- Baimbetov, A.A.; Dukravets, G.M.; Ereshchenko, V.I.; Kulikov, E.V.; Kulikova, E.V.; Melnikov, V.A.; Mitrofanov, V.P.; Timirkhanov, S.R. The Red Data Book of the Republic of Kazakhstan, 4th ed.; revised and updated; Animals; Part 1: Vertebrates—Almaty, “DPS”; 2010; Volume I, p. 324. Available online: https://zool.kz/wp-content/uploads/2021/02/red-data-book-rk_v1_1_2010.pdf (accessed on 11 November 2023)ISBN 9965-32-738-6.
- Orlova, I.V.; Tereshchenko, A.M.; Murova, E.V.; Klimov, F.V. Features of the biology and morphology of the Aral and Turkestan barbels in the basin of the river. Syrdarya Tethys Aqua Zool. Res. III 2007, 1, 93–102. [Google Scholar]
- Maksunov, V.A. Materials for the morphometric and biological characteristics of the Turkestan barbel Barbus capito conocephalus Kessler in the upper reaches of the Syrdarya River. Issues Ichthyol. 1962, 4, 592–596. [Google Scholar]
- Turdakov, F.A. Fishes of Kyrgyzstan. In Academy of Sciences of the KirgSSR; KirgSSR: Frunze, Kyrgyzstan, 1963; p. 284. [Google Scholar]
- Pivnev, I.A. Fish from the Basins of the Chu and Talas Rivers; KirgSSR: Frunze, Kyrgyzstan, 1985; p. 190. [Google Scholar]
- Mitrofanov, V.P.; Dukravets, G.M.; Melnikov, V.A.; Baimbetov, A.A. Fishes of Kazakhstan: Vol.3; Carps (Continuation); Nauka: Alma-Ata, Kazakhstan, 1988; p. 304. [Google Scholar]
- Coad, B.W. Fresh water fishes of Iran. Acta Sci. Nat. Acad. Sci. Bohemicae 1995, 29, 1–64. [Google Scholar]
- Fricke, R.; Eschmeyer, W.N.; van der Laan, R. (Eds.) Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2023. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 25 July 2023).
- Jouladeh-Roudbar, A.; Ghanavi, H.R.; Doadrio, I. Ichthyofauna from Iranian freshwater: Annotated checklist, diagnosis, taxonomy, distribution and conservation assessment. Zool. Stud. 2020, 59, 21. [Google Scholar] [CrossRef]
- Galaktionova, E.L. Status, Reproduction and Conservation of the Syrdarya Population of the Aral Barbel Barbus Brachycephalus Kessler in Conditions of Regulated Flow. Protection and Rational Use of the Wildlife of Kazakhstan’s Water Bodies: Materials of the International conferencei; KazSSR: Alma-Ata, Kazakhstan, 1969; pp. 86–89. [Google Scholar]
- Markun, M.I. Aral barbel, its taxonomy and biology. In Proceedings of the Aral Branch, VNIMORKH, Aralsk, USSR; 1933; pp. 5–47. [Google Scholar]
- Pavlovskaya, L.P. Morphological characteristics and some questions of the biology of the Aral barbel in the river period of life. In Fishes and the Hydrobiological Regime of the South Aral Basin; USSR: Tashkent, Uzbekistan, 1966; pp. 51–120. [Google Scholar]
- Usmanova, R.G. Sexual Dimorphism, Age and Local Variability of the Turkestan Barbel Barbus Capito Conocephalus Kessler in the Basin of the River; Questions of Ichthyology; Kashkadarya, USSR; 1971; Volume 11, pp. 203–216. Available online: https://www.iucn.org (accessed on 11 November 2023).
- Machordom, A.; Doadrio, I. Evidence of a Cenozoic Betic–Kabilian connection based on freshwater fish phylogeography (Luciobarbus, Cyprinidae). Mol. Phylogenetics Evol. 2001, 18, 252–263. [Google Scholar] [CrossRef]
- Brahimi, A.; Freyhof, J.; Henrard, A.; Libois, R. Luciobarbus chelifensis and L. mascarensis, two new species from Algeria (Teleostei: Cyprinidae). Zootaxa 2017, 4277, 32–50. [Google Scholar] [CrossRef]
- Brahimi, A.; Libois, R.; Henrard, A.; Freyhof, J. Luciobarbus lanigarensis and L. numidiensis, two new species of barbels from the Mediterranean Sea basin in North Africa (Teleostei: Cyprinidae). Zootaxa 2018, 4433, 542–560. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.A.; Freyhof, J.; Lajbner, Z.; Perea, S.; Abdoli, A.; Gaffaroğlu, M.; Doadrio, I. Phylogenetic relationships of the algae scraping cyprinid genus Capoeta (Teleostei: Cyprinidae). Mol. Phylogenetics Evol. 2012, 62, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Ouassal, K.; Perea, S.; Doadrio, I.; Casal-Lopez, M.; Yahyaoui, A. Assessment of genetic diversity in Luciobarbus rifensis Doadrio, Casal-López & Yahyaoui, 2015 (Teleostei: Cyprinidae) using cytochrome b. Aquac. Aquar. Conserv. Legis. 2021, 14, 282–290. [Google Scholar]
- Touil, A.; Casal-Lopez, M.; Bouhadad, R.; Doadrio, I. Phylogeny and phylogeography of the genus Luciobarbus (Haeckel, 1843) in Algeria inferred from mitochondrial DNA sequence variation. Mitochondrial DNA Part A 2019, 30, 332–344. [Google Scholar] [CrossRef]
- Yang, L.; Sado, T.; Hirt, M.V.; Pasco-Viel, E.; Arunachalam, M.; Li, J.; Mayden, R.L. Phylogeny and polyploidy: Resolving the classification of cyprinine fishes (Teleostei: Cypriniformes). Mol. Phylogenetics Evol. 2015, 85, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Funk, D.J.; Omland, K.E. Species level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 397–423. [Google Scholar] [CrossRef]
- Chan, K.M.A.; Levin, S.A. Leaky prezygotic isolation and porous genomes: Rapid introgression of maternally inherited DNA. Evolution 2005, 59, 720–729. [Google Scholar] [PubMed]
- Zink, R.M.; Barrowclough, G. Mitochondrial DNA under siege in avian phylogeography. Mol. Ecol. 2008, 17, 2107–2121. [Google Scholar] [CrossRef]
- Toews, D.P.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Slettan, A.; Olsaker, I.; Lie, Ø. Polymorphic Atlantic salmon, Salmo salar L., microsatellites at the SSOSL438, SSOSL439, and SSOSL444 loci. Anim. Genet. 1996, 27, 57–58. [Google Scholar] [CrossRef]
- Estoup, A.; Presa, P.; Krieg, F.; Vaiman, D.; Guyomard, R. (CT)n and (GT)n microsatellites: A new class of genetic markers for Salmo trutta L. (brown trout). Heredity 1993, 71, 488–496. [Google Scholar] [CrossRef]
- Garcia de Leon, F.J.; Dallas, J.F.; Chatain, B.; Canonne, F.; Versini, J.J.; Bonhomme, F. Development and use of microsatellite markers in sea bass, Dicentrarchus labrax (Linnaeus, 1758) (Perciformes: Serrandidae). Mol. Mar. Biol. Biotechnol. 1995, 4, 62–68. [Google Scholar]
- Tan, G.; Karsi, A.; Li, P.; Kim, S.; Zheng, X.; Kucuktas, H.; Argue, B.J.; Dunham, R.A.; Liu, Z.J. Polymorphic microsatellite markers in Ictalurus punctatus and related catfish species. Mol. Ecol. 1999, 59, 190–194. [Google Scholar] [CrossRef]
- Landínez-García, R.M.; Márquez, E.J. Development and characterization of 24 polymorphic microsatellite loci for the freshwater fish Ichthyoelephas longirostris (Characiformes: Prochilodontidae). PeerJ 2016, 4, e2419. [Google Scholar] [CrossRef]
- Mariette, S.; Le Corre, V.; Austerlitz, F.; Kremer, A. Sampling within the genome for measuring within-population diversity: Tradeoffs between markers. Mol. Ecol. 2002, 11, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G. Genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 2020, 20, 1126–1131. [Google Scholar] [CrossRef]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Bostein, D.; White, R.L.; Sckolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 317–331. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- De Santis, V.; Quadroni, S.; Britton, R.J.; Carosi, A.; Gutmann Roberts, C.; Lorenzoni, M.; Crosa, G.; Zaccara, S. Biological and trophic consequences of genetic introgression between endemic and invasive Barbus fishes. Biol. Invasions 2021, 23, 3351–3368. [Google Scholar] [CrossRef]
- Jia, Z.Y.; Zhang, Y.Y.; Shi, L.Y.; Bai, Q.L.; Jin, S.B.; Mou, Z.B. Amplification of rainbow trout microsatellites in Brachymystax lenok. Mol. Ecol. Resour. 2008, 8, 1520–1521. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Nie, Z.; Zhan, F.; Wei, J.; Wang, W.; Gao, Z. Rapid development of microsatellite markers for the endangered fish Schizothorax biddulphi (Günther) using next generation sequencing and cross-species amplification. Int. J. Mol. Sci. 2012, 13, 14946–14955. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jian, S.; Luo, W.; Wei, Q.; Wang, F.; Guo, W.; Ye, H.; Chu, Z.; Wu, J.; Zhang, S. Assignment of parentage by microsatellite analysis in the endangered Brachymystax lenok tsinlingensis (Salmonidae). Aquat. Biol. 2017, 26, 69–73. [Google Scholar]
- Hu, Y.; Liu, X.; Yang, J.; Xiao, K.; Wang, B.; Du, H. Development of Twenty-Two Novel Cross-Species Microsatellite Markers for Amur sturgeon (Acipenser schrenckii) from Chinese Sturgeon (Acipenser sinensis) via next-Generation Sequencing. Turk. J. Fish. Aquat. Sci. 2019, 19, 175–178. [Google Scholar] [CrossRef]
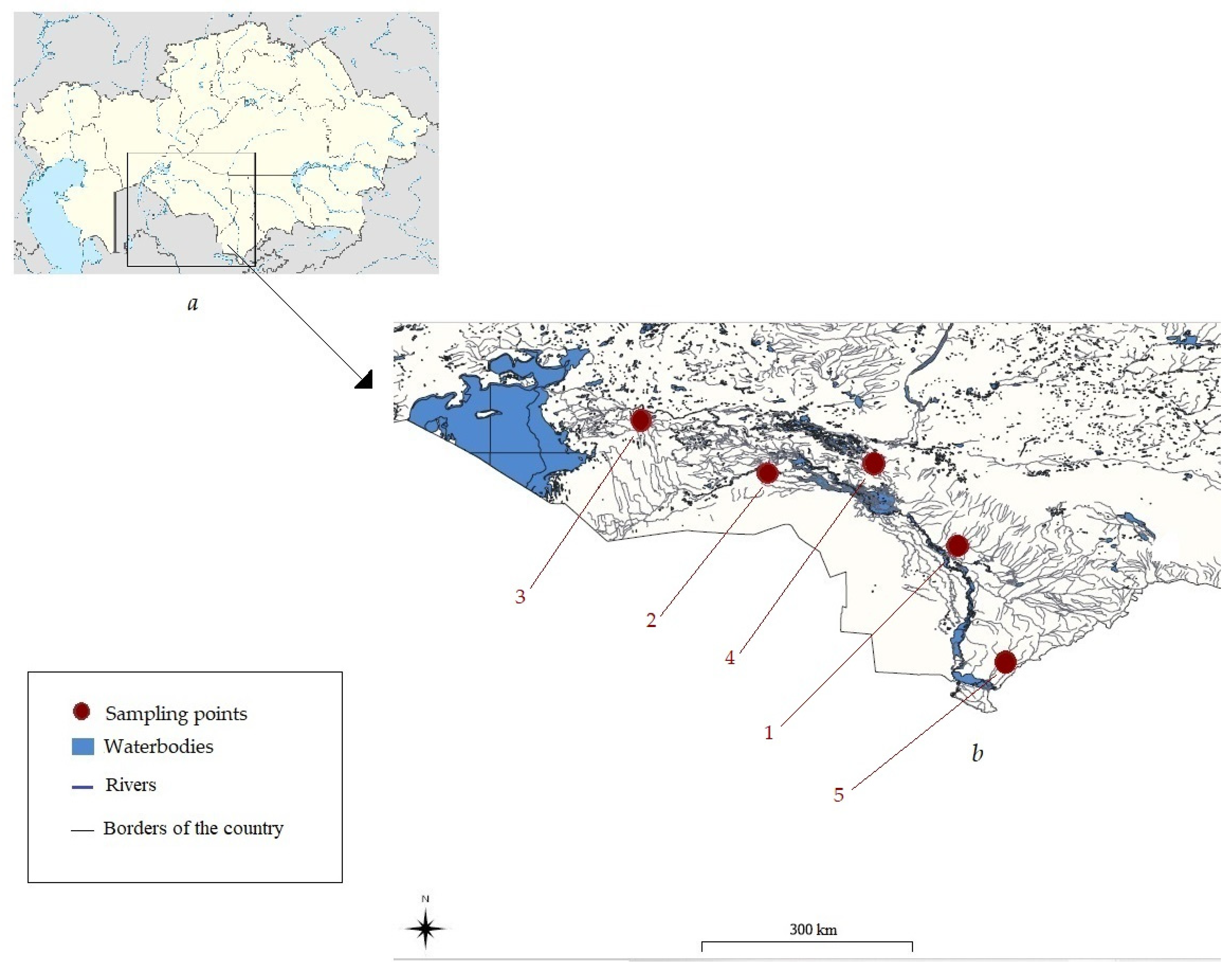

| N° | Location | Species | Number of Sampled Individuals (n = 81) | Latitude | Longitude |
|---|---|---|---|---|---|
| 1 | Bairykum village | L. brachycephalus | 7 | 42.0764 | 68.4753 |
| 2 | Kyzylorda region | L. brachycephalus | 21 | 45.757381 | 62.322459 |
| L. conocephalus | 2 | ||||
| 3 | Basykara dam | L. brachycephalus | 25 | 45.7583156 | 62.3254401 |
| 4 | Rice check | L. brachycephalus | 22 | 45.21043 | 64.12054 |
| L. conocephalus | 2 | ||||
| 5 | Badam River | L. conocephalus | 2 | 42.3088933 | 69.5388738 |
| Locus Name | Primer Sequence (5′–3′) | Amplicon Size | Repeat Motif | Tail of Primer | Fluorescent Marker |
|---|---|---|---|---|---|
| M1287 | F:TAATTAGCAACAGGCCCGCA R: TGCGTTCCCGTGTTTGAATG | 170 | (AG)10 | T3 | PET |
| M1417 | F:CCAAGTCTCGCTATCCTCGG R:AAGAGGAGTGATGACAGCGC | 114 | (CCG)5 | CAG | NED |
| M1182 | F:GCTCTCGTTCCAGTCCAGAC R: AGCATCTGGCCATGATGGAG | 193 | (AATC)7 | CAG | VIC |
| M2237 | F:GAAGGTCACGTGGTTGTCCA R: AGGGAATTGGATGCAGCTCC | 91 | (AG)12 | CAG | PET |
| M2108 | F:GCTGCGGATTGGTCAAGAAC R: GCTCTTCTCCTCTCATCCGC | 91 | (AG)14 | M13 | NED |
| M2044 | F:TATGCAGCTTCCACCCACTG R:GTTCACGCTGTTTGCTGGAG | 103 | (AC)10 | M13 | NED |
| M2164 | F:GGCGTTGTTGAGCCAATCAG R: TGACTTTGGCAGGACGTGTT | 91 | (AGC)5 | M13 | VIC |
| M2306 | F:CAGTCCCAGACTCTTCCAGC R:CCGGTGTGCGATCCAATCTA | 302 | (ATC)5 | CAG | NED |
| M3230 | F:ATTGAGGATCCCGAGGCTCT R:CGATAAGCCCGTGAGACGTT | 149 | (AGG)5 | T3 | PET |
| M3264 | F:TGGTCATGCATGCGGTACAT R:AAGGTCACTGAAGTGCTGCA | 159 | (ACAG)6 | T3 | NED |
| M3318 | F:AGTGAAAGCATGTCCAGGCA R:GGAACTGGCCGTGAAATGG | 217 | (AG)18 | CAG | VIC |
| M3444 | F:ATGACTCAGGTGAAGCAGGC R:CCGCTCCTGCTTGACTTCAT | 223 | (AGC)7 | CAG | VIC |
| M4211 | F:CTAGACGAGCAGCACTGGAG R:CATTAGACAGCCGAGCCCTT | 109 | (AC)11 | CAG | PET |
| M4455 | F:TGTATGACGCTGGTTGGAGC R:ATGATACGATCCCAGCGCTG | 110 | (AC)11 | M13 | NED |
| M4215 | F:CGAGCCGATCTCTGTCTGTG R:CCCAAACCCAAGAAAGTGCG | 91 | (AATC)10 | M13 | PET |
| M4138 | F:CTGGCTGTCAACCTGTGGAA R:CTCCAGAGTCCGTACCTGGA | 153 | (AG)10 | CAG | VIC |
| M4474 | F:AACACTGACCATGTGACGCA R:CCAACTTCTGGTCCGGCATA | 238 | (AC)11 | T3 | PET |
| Sampling Points | Species | Na | Ne | Ho | He | Gis |
|---|---|---|---|---|---|---|
| Bairykum v. | L. brachycephalus | 3.267 | 2.283 | 0.518 | 0.497 | −0.042 |
| Kyzylorda r. | L. brachycephalus | 4.533 | 2.653 | 0.578 | 0.546 | −0.058 |
| L. conocephalus | ||||||
| Basykara d. | L. brachycephalus | 5.067 | 2.707 | 0.540 | 0.539 | −0.001 |
| Rice check | L. brachycephalus | 4.667 | 2.659 | 0.557 | 0.552 | −0.008 |
| L. conocephalus | ||||||
| Badam R. | L. conocephalus | 1.867 | 1.812 | 0.433 | 0.428 | −0.014 |
| Locus | Na | Ne | Ho | Hs | Ht | H’t | Gis |
|---|---|---|---|---|---|---|---|
| M1287 | 11.000 | 2.946 | 0.827 | 0.699 | 0.730 | 0.738 | −0.184 |
| M1417 | 3.000 | 2.640 | 0.879 | 0.650 | 0.646 | 0.645 | −0.353 |
| M1182 | 5.000 | 2.678 | 0.821 | 0.660 | 0.695 | 0.704 | −0.243 |
| M2237 | 5.000 | 1.409 | 0.186 | 0.320 | 0.321 | 0.321 | 0.420 |
| M2108 | 3.000 | 1.057 | 0.037 | 0.057 | 0.055 | 0.055 | 0.341 |
| M2044 | 12.000 | 5.093 | 0.911 | 0.842 | 0.860 | 0.864 | −0.082 |
| M2164 | 3.000 | 1.062 | 0.041 | 0.064 | 0.061 | 0.061 | 0.357 |
| M2306 | 2.000 | 1.584 | 0.242 | 0.390 | 0.476 | 0.497 | 0.380 |
| M3230 | 2.000 | 1.996 | 0.654 | 0.514 | 0.503 | 0.500 | −0.273 |
| M3264 | 6.000 | 2.403 | 0.758 | 0.602 | 0.591 | 0.588 | −0.259 |
| M3444 | 8.000 | 2.885 | 0.824 | 0.675 | 0.676 | 0.676 | −0.221 |
| M4211 | 4.000 | 1.514 | 0.255 | 0.357 | 0.374 | 0.378 | 0.288 |
| M4215 | 9.000 | 3.030 | 0.237 | 0.721 | 0.811 | 0.833 | 0.672 |
| M4138 | 5.000 | 2.145 | 0.715 | 0.550 | 0.542 | 0.540 | −0.300 |
| M4474 | 12.000 | 2.430 | 0.490 | 0.618 | 0.738 | 0.768 | 0.207 |
| Mean | 6.000 | 2.325 | 0.525 | 0.515 | 0.539 | 0.545 | −0.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adyrbekova, K.; Perea, S.; Doadrio, I. Development and Characterization of Fifteen Polymorphic Microsatellite Loci for Rare and Endangered Species within Luciobarbus Heckel, 1843 Genus in the Aral Basin and Their Conservation Application. Fishes 2024, 9, 169. https://doi.org/10.3390/fishes9050169
Adyrbekova K, Perea S, Doadrio I. Development and Characterization of Fifteen Polymorphic Microsatellite Loci for Rare and Endangered Species within Luciobarbus Heckel, 1843 Genus in the Aral Basin and Their Conservation Application. Fishes. 2024; 9(5):169. https://doi.org/10.3390/fishes9050169
Chicago/Turabian StyleAdyrbekova, Kamila, Silvia Perea, and Ignacio Doadrio. 2024. "Development and Characterization of Fifteen Polymorphic Microsatellite Loci for Rare and Endangered Species within Luciobarbus Heckel, 1843 Genus in the Aral Basin and Their Conservation Application" Fishes 9, no. 5: 169. https://doi.org/10.3390/fishes9050169
APA StyleAdyrbekova, K., Perea, S., & Doadrio, I. (2024). Development and Characterization of Fifteen Polymorphic Microsatellite Loci for Rare and Endangered Species within Luciobarbus Heckel, 1843 Genus in the Aral Basin and Their Conservation Application. Fishes, 9(5), 169. https://doi.org/10.3390/fishes9050169







