Population Structure Using Mitochondrial DNA for the Conservation of Liobagrus geumgangensis (Siluriformes: Amblycipitidae), an Endemic Freshwater Fish in Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Genomic DNA Extraction
2.2. mtDNA Sequencing and Sequence Assembly
2.3. Sequence Analysis of Genetic Diversity and Structure in mtDNA
3. Results
3.1. Genetic Diversity
3.2. Population Genetic Structure
4. Discussion
5. Conservation Implications
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, S.H.; Yun, S.W.; Park, J.Y. A new species of torrent catfish, Liobagrus geumgangensis (Teleostei, Siluriformes, Amblycipitidae), from Korea. ZooKeys 2023, 1180, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, D.J.; Sutton, T.M. Seasonal movement patterns, habitat use, and home range of flathead catfish in the lower St. Joseph River, Michigan. N. Am. J. Fish. Manag. 2005, 25, 256–269. [Google Scholar] [CrossRef]
- Kadye, W.T.; Booth, A.J. Movement patterns and habitat selection of invasive A frican sharptooth catfish. J. Zool. 2013, 289, 41–51. [Google Scholar] [CrossRef]
- Fonseca, F.S.; Domingues, R.R.; Hallerman, E.M.; Hilsdorf, A.W. Genetic diversity of an imperiled Neotropical catfish and recommendations for its restoration. Front. Genet. 2017, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Chakona, A.; Gouws, G.; Kadye, W.T.; Mpopetsi, P.P.; Skelton, P.H. Probing hidden diversity to enhance conservation of the endangered narrow-range endemic Eastern Cape rocky, Sandelia bainsii (Castelnau 1861). Koedoe Afr. Protected Area Conserv. Sci. 2020, 62, a1627. [Google Scholar] [CrossRef]
- Kim, K.R.; Choi, H.K.; Lee, T.W.; Lee, H.J.; Yu, J.N. Population structure and genetic diversity of the spotted sleeper Odontobutis interrupta (Odontobutidae), a fish endemic to Korea. Diversity 2023, 15, 913. [Google Scholar] [CrossRef]
- Maxwell, S.L.; Fuller, R.A.; Brooks, T.M.; Watson, J.E. Biodiversity: The ravages of guns, nets and bulldozers. Nature 2016, 536, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Markert, J.A.; Champlin, D.M.; Gutjahr-Gobell, R.; Grear, J.S.; Kuhn, A.; McGreevy, T.J.; Roth, A.; Bagley, M.J.; Nacci, D.E. Population genetic diversity and fitness in multiple environments. BMC Evol. Biol. 2010, 10, 205. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Brauer, C.J.; Beheregaray, L.B. Recent and rapid anthropogenic habitat fragmentation increases extinction risk for freshwater biodiversity. Evol. Appl. 2020, 13, 2857–2869. [Google Scholar] [CrossRef] [PubMed]
- Gurevitch, J.; Padilla, D.K. Are invasive species a major cause of extinctions? Trends Ecol. Evol. 2004, 19, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R. Conservation genetics. Annu. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef]
- Baltazar-Soares, M.; de Araújo Lima, A.R.; Silva, G. Targeted sequencing of mitochondrial genes reveals signatures of molecular adaptation in a nearly panmictic small pelagic fish species. Genes 2021, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.Y.; Wang, J.J.; Ren, J.F.; Li, F.; Wu, J.X.; Zhou, J.J.; Li, J.; Yang, J.; Lin, H.D.; Yang, J.; et al. Historical landscape evolution shaped the phylogeography and population history of the cyprinid fishes of Acrossocheilus (Cypriniformes: Cyprinidae) according to mitochondrial DNA in Zhejiang Province, China. Diversity 2023, 15, 425. [Google Scholar] [CrossRef]
- Ríos, N.; Casanova, A.; Hermida, M.; Pardo, B.G.; Martínez, P.; Bouza, C.; García, G. Population genomics in Rhamdia quelen (Heptapteridae, Siluriformes) reveals deep divergence and adaptation in the Neotropical region. Genes 2020, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Baharum, S.N.; Nurdalila, A.A. Application of 16s rDNA and cytochrome b ribosomal markers in studies of lineage and fish populations structure of aquatic species. Mol. Biol. Rep. 2012, 39, 5225–5232. [Google Scholar] [CrossRef] [PubMed]
- Bórquez, J.; Valdovinos, C.; Brante, A. Genetic structure and diversity in the freshwater gastropod Chilina dombeiana in the Biobío River, Chile. Conserv. Genet. 2020, 21, 1023–1036. [Google Scholar] [CrossRef]
- Höglund, J.; Larsson, J.K.; Corrales, C.; Santafé, G.; Baines, D.; Segelbacher, G. Genetic structure among black grouse in Britain: Implications for designing conservation units. Anim. Conserv. 2011, 14, 400–408. [Google Scholar] [CrossRef]
- Kim, K.R.; Kwak, Y.H.; Sung, M.S.; Cho, S.J.; Bang, I.C. Population structure and genetic diversity of the endangered fish black shinner Pseudopungtungia nigra (Cyprinidae) in Korea: A wild and restoration population. Sci. Rep. 2023, 13, 9692. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Mega, K.S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Mishmar, D.; Ruiz-Pesini, E.; Golik, P.; Macaulay, V.; Clark, A.G.; Hosseini, S.; Brandon, M.; Easley, K.; Chen, E.; Brown, M.D. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 2003, 100, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Pesini, E.; Mishmar, D.; Brandon, M.; Procaccio, V.; Wallace, D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 2004, 303, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Huang, Y.; Zeng, Q.; Li, B.; Peng, Z. Genetic diversity and population history among geographic populations of Silurus asotus in different water systems in China based on mtDNA Cytb gene sequences. J. Fish. China 2017, 41, 1489–1499. [Google Scholar]
- Frankham, R.; Ballou, J.D.; Eldridge, M.D.B.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the probability of outbreeding depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.J.; Coelho, H.; Collares-Pereira, M.J.; Coelho, M.M. Mitochondrial DNA variation in the highly endangered cyprinid fish Anaecypris hispanica: Importance for conservation. Heredity 2001, 87, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Mamuris, Z.; Stoumboudi, M.T.; Stamatis, C.; Barbieri, R.; Moutou, K.A. Genetic variation in populations of the endangered fish Ladigesocypris ghigii and its implications for conservation. Freshw. Biol. 2005, 50, 1441–1453. [Google Scholar] [CrossRef]
- Chan, J.; Li, W.; Hu, X.; Liu, Y.; Xu, Q. Genetic diversity and population structure analysis of Qinghai-Tibetan Plateau schizothoracine fish (Gymnocypris dobula) based on mtDNA D-loop sequences. Biochem. Syst. Ecol. 2016, 69, 152–160. [Google Scholar] [CrossRef]
- Hedrick, P.W.; Miller, P.S. Conservation genetics: Techniques and fundamentals. Ecol. Appl. 1992, 2, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Matocq, M.D.; Villablanca, F.X. Low genetic diversity in an endangered species: Recent or historic pattern? Biol. Conserv. 2001, 98, 61–68. [Google Scholar] [CrossRef]
- Choi, N.H.; Seo, W.I.; Kim, C.C.; Park, C.K.; Heo, S.J.; Yoon, S.M.; Han, K.; Lee, W.K.; Lee, W. Spawning behavior and Early Life Histoty of the Liobagrus mediadiposalis in the Korean Endemic Species. Korean J. Fish. Aquat. Sci. 2008, 41, 478–484. [Google Scholar]
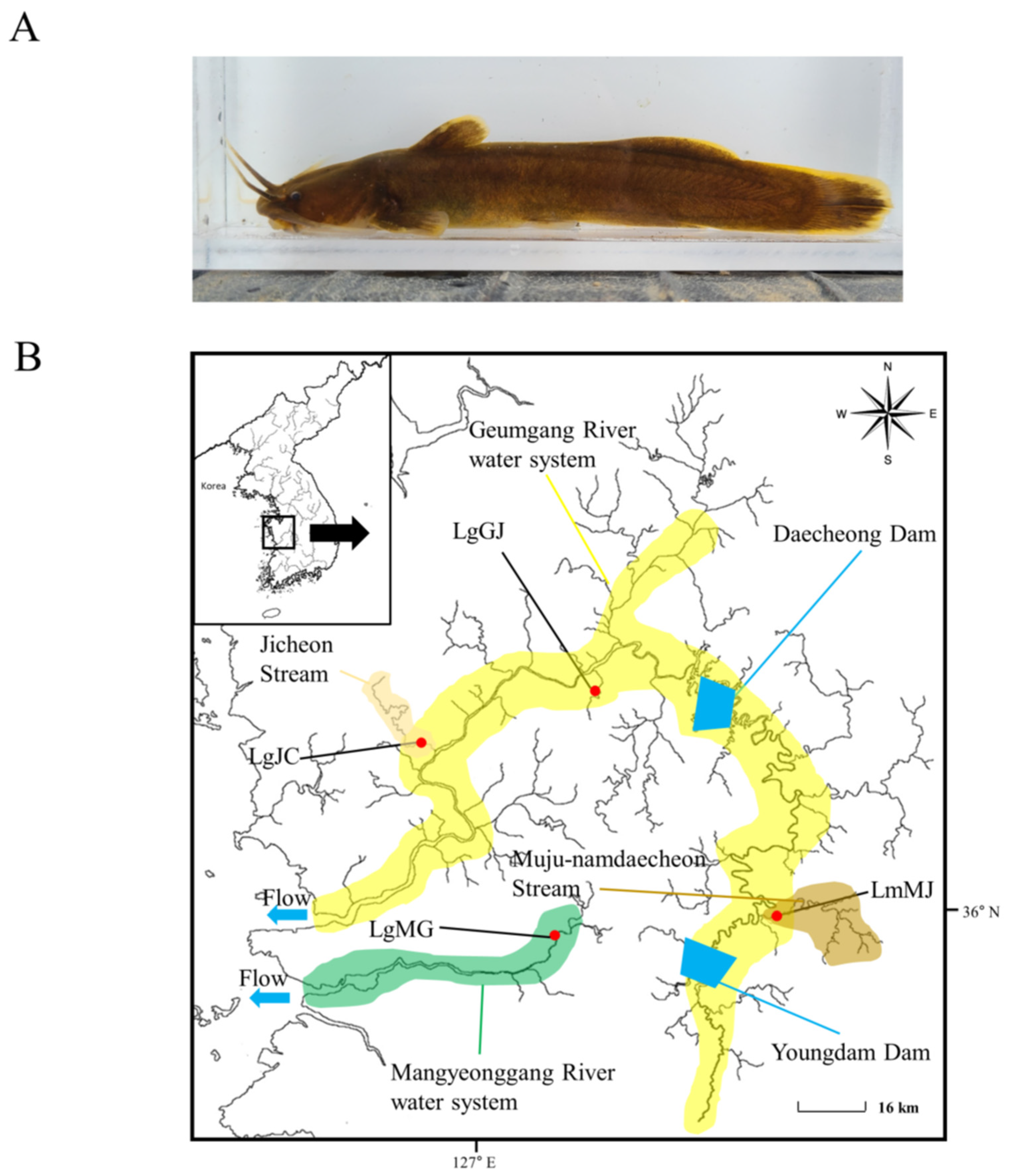
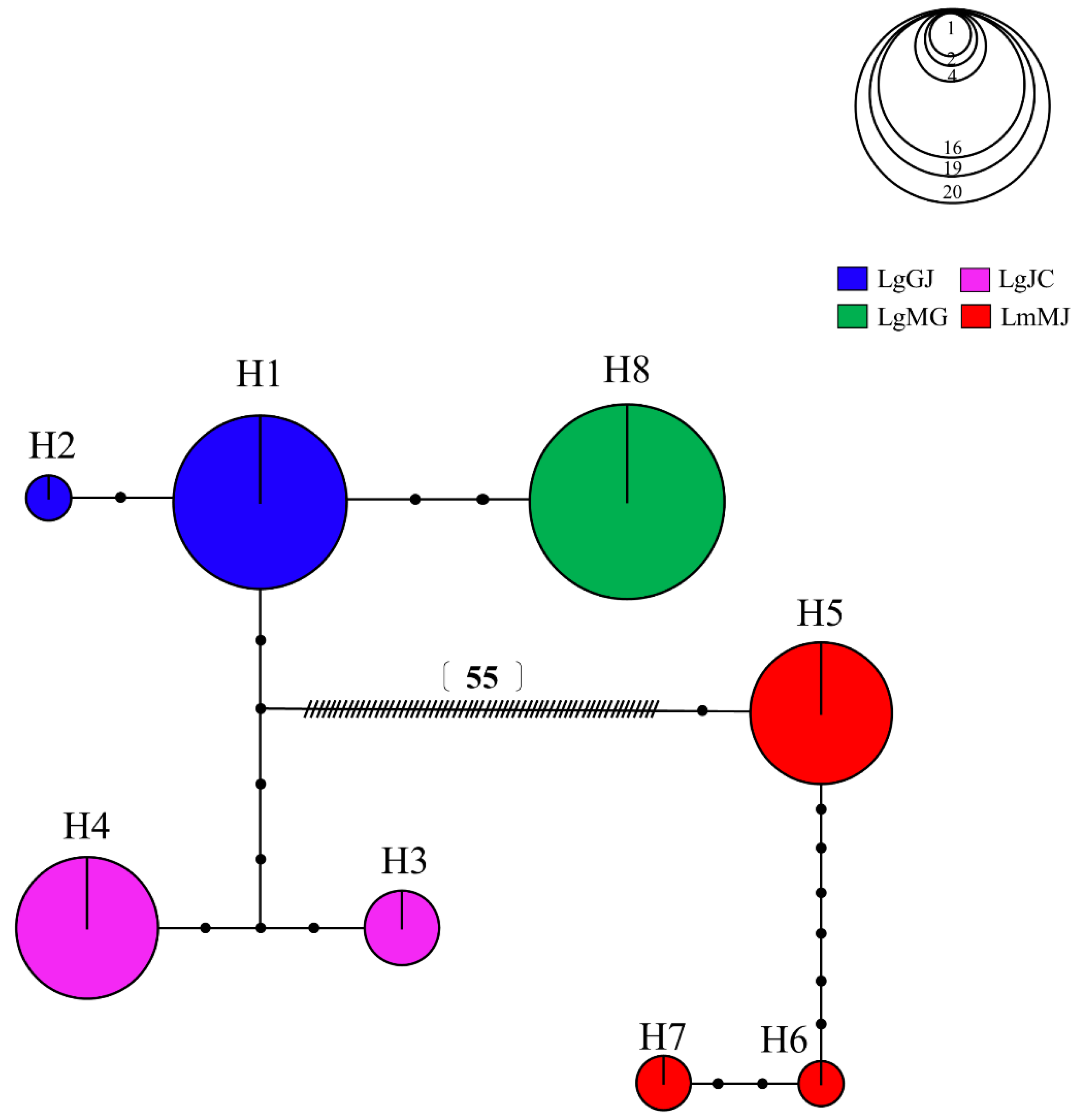
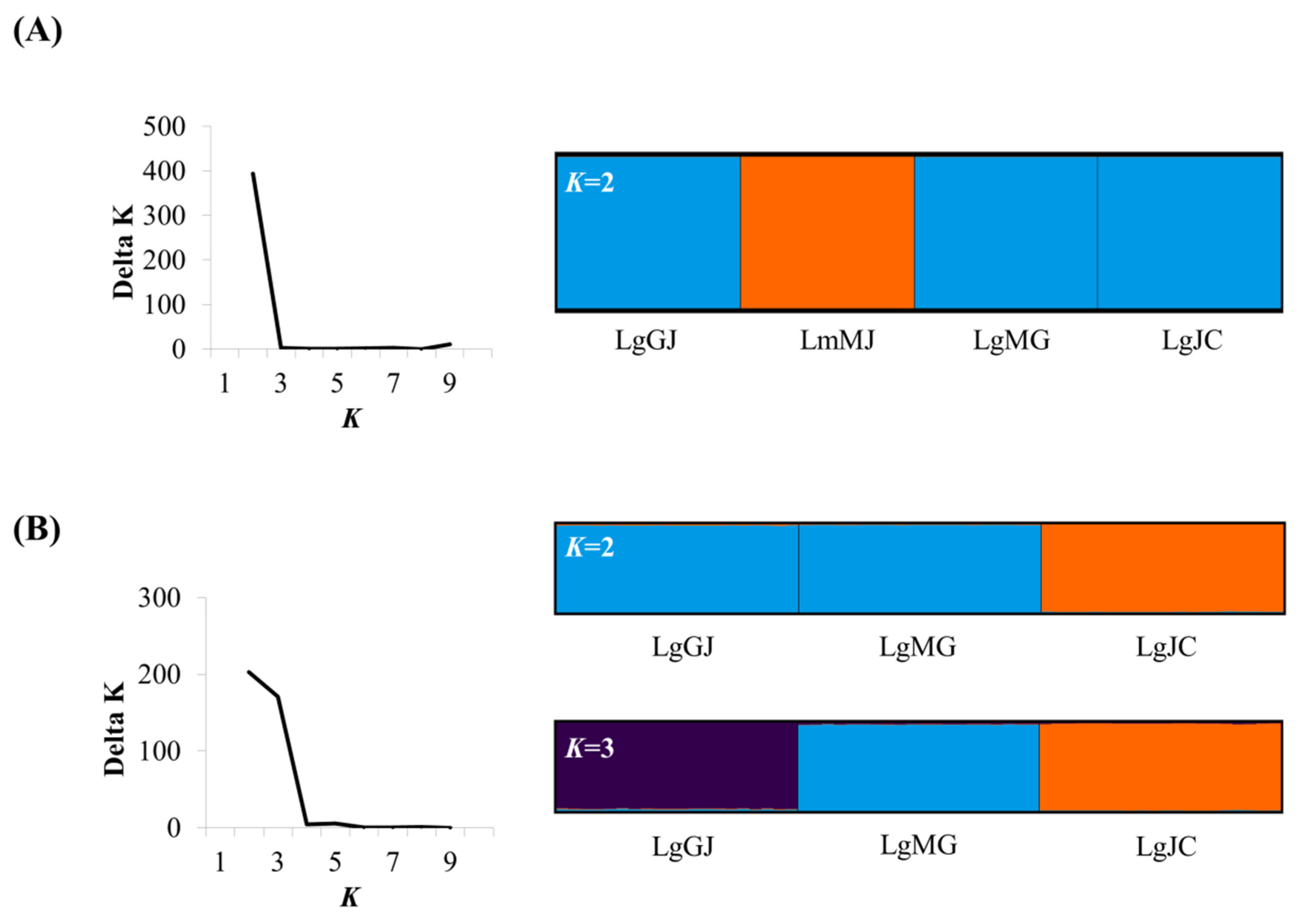
| Species | Population Code | Water System | N | h | Hd | Nucleotide Diversity (π) | D | F |
|---|---|---|---|---|---|---|---|---|
| L. geumgangensis | LgJC | Geumgang River | 20 | 2 | 0.337 | 0.00066 | 0.45727 | 1.985 |
| L. geumgangensis | LgGJ | Geumgang River | 20 | 2 | 0.100 | 0.00010 | −1.16439 | −0.879 |
| L. geumgangensis | LgMG | Mangyeongang River | 20 | 1 | 0.000 | 0.00000 | - | - |
| L. geumgangensis | LgJC, LgGJ | Geumgang River | 40 | 4 | 0.619 | 0.00271 | 1.90551 | 4.441 |
| L. geumgangensis | LgMG | Mangyeongang River | 20 | 1 | 0.000 | 0.00000 | - | - |
| L. mediadiposalis | LmMJ | Geumgang River | 19 | 3 | 0.292 | 0.00231 | −0.28784 | 3.737 |
| L. geumgangensis | LgJC, LgGJ, LgMG | Geumgang and Mangyeongang River | 60 | 5 | 0.725 | 0.00319 | 1.88005 | 4.910 |
| LgJC | LgGJ | LgMG | LmMJ | |
|---|---|---|---|---|
| LgJC | - | 0.000 | 0.000 | 0.000 |
| LgGJ | 0.923 | - | 0.000 | 0.000 |
| LgMG | 0.952 | 0.976 | - | 0.000 |
| LmMJ | 0.976 | 0.980 | 0.981 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-R.; Sung, M.-S.; Kim, K.-S. Population Structure Using Mitochondrial DNA for the Conservation of Liobagrus geumgangensis (Siluriformes: Amblycipitidae), an Endemic Freshwater Fish in Korea. Fishes 2024, 9, 153. https://doi.org/10.3390/fishes9050153
Kim K-R, Sung M-S, Kim K-S. Population Structure Using Mitochondrial DNA for the Conservation of Liobagrus geumgangensis (Siluriformes: Amblycipitidae), an Endemic Freshwater Fish in Korea. Fishes. 2024; 9(5):153. https://doi.org/10.3390/fishes9050153
Chicago/Turabian StyleKim, Kang-Rae, Mu-Sung Sung, and Keun-Sik Kim. 2024. "Population Structure Using Mitochondrial DNA for the Conservation of Liobagrus geumgangensis (Siluriformes: Amblycipitidae), an Endemic Freshwater Fish in Korea" Fishes 9, no. 5: 153. https://doi.org/10.3390/fishes9050153
APA StyleKim, K.-R., Sung, M.-S., & Kim, K.-S. (2024). Population Structure Using Mitochondrial DNA for the Conservation of Liobagrus geumgangensis (Siluriformes: Amblycipitidae), an Endemic Freshwater Fish in Korea. Fishes, 9(5), 153. https://doi.org/10.3390/fishes9050153







