Abstract
Yellow catfish (Pelteobagrus fulvidraco) is an important freshwater fish species in aquaculture. However, as intensive farming has rapidly expanded, infectious diseases caused by various bacteria, such as Aeromonas hydrophila, have also increased. We conducted a transcriptomic analysis of head kidney from P. fulvidraco stimulated by A. hydrophila strain SHOU. A total of 43,249 unigenes with an average length of 1342 bp were obtained following assembly and annotation. By analyzing GO and KEGG enrichment, many differentially expressed genes (DEGs) and pathways related to immunity were identified. Out of the 973 DEGs that were identified, 553 were upregulated and 420 were downregulated. Moreover, KEGG enrichment analysis revealed that the innate immune system, including the TNF signaling pathway, NF-kappa B signaling pathway, and the Toll-like receptor signaling pathway involved in the defense, is activated against infectious response. Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis demonstrated that immune response genes were upregulated in response to A. hydrophila stimulation compared to the control. In conclusion, this study provides valuable insights into the immune defense mechanism of P. fulvidraco and sheds light on the host immune genes involved in the response to bacterial infection.
Keywords:
RNA-seq; bacterial infection; differential expressed genes; yellow catfish; immune signaling pathway Key Contribution:
This study contributes to a better understanding of the immune defense mechanism of P. fulvidraco, and also provides valuable insight into the host immune genes involved in the response to bacterial infection.
1. Introduction
Pelteobagrus fulvidraco is a freshwater teleost that is omnivorous. It is highly sought after in China and other Asian countries due to its high market value and delicious meat, making it the preferred choice for diverse cultures [1]. This species can be found in eastern Asia and is one of the important species of freshwater in China, with an impressive annual output exceeding 598,000 tons in 2022 [2]. Unfortunately, rapid development has led to an increase in the infectious diseases caused by various bacteria [3]. One of the most destructive pathogens, causing significant losses, is Aeromonas hydrophila, a Gram-negative bacterium, which is an oxidase-positive and facultative anaerobic. This is an opportunistic aquatic pathogen that produces various virulence factors, including hemolysins, aerolysins, adhesins, enterotoxins, phospholipase, and lipase [4], P. fulvidraco has become increasingly popular as a model fish for studying breeding technology, toxicology, immune response, and lipid metabolism in recent years [5,6,7,8,9]. However, the effect of A. hydrophila on P. fulvidraco has not been reported.
Fish immunity encompasses two essential components, namely the innate system and the adaptive system, which makes them the valuable models for studying the evolutionary history of immune systems [10]. The innate immune system is the first line of defense against pathogens and plays a crucial role in the immune response in fish. Various tissues, including the gills, skin, spleen, head kidney, and gut, are important sites of immune defense [11]. Unlike other animals, teleost fish possess a unique organ called the head kidney, which bears resemblance to the adrenal gland found in mammals. The head kidney contains lymphoid cells, which are responsible for cytokine production in the immune system, as well as endocrine cells that secrete cortisol, catecholamines, and thyroid hormones [12]. The close proximity of the immune and endocrine systems in the head kidney enables bidirectional signaling [13]. The head kidney in teleost fish is a separate organ that resembles the adrenal gland in mammals. It contains cytokine-producing immune system cells and endocrine cells that release cortisol, catecholamines, and thyroid hormones [14,15]. These immune cells from the head kidney of fishes are involved in recognizing and eliminating pathogens, as well as regulating immune responses [16,17]. The macrophages and neutrophils found in the head kidney of teleost fish are responsible for engulfing and destroying invading pathogens [18]. These immune cells located in the head kidney are responsible for producing cytokines, which are chemical messengers that regulate immune cell communication and response [19]. Meanwhile, the head kidney can also undergo changes in response to environmental factors or pathogen exposure [20].
Transcriptome analysis of the head kidney can provide valuable insights into the fish immune system, such as identifying differentially expressed genes or pathways involved in immune responses. Overall, the head kidney is a crucial organ of the fish immune system and plays a significant role in immune cell development, pathogen recognition, and immune responses [21,22]. The head kidney of fish is considered to be one of the model tissues for studying immunity. RNA-seq technology has several distinctive features, including its wide coverage of the genome, high sensitivity, unbiased expression analysis, and cost-effectiveness, and can provide a new method for transcriptome research [23]. Transcriptome analysis is a powerful tool for studying host–pathogen interactions and deciphering the genetic responses of hosts to pathogens, which can help to identify the genes involved in pathogen recognition, immune signaling pathways, and the production of defense molecules such as antimicrobial peptides. It can also reveal changes in gene expression patterns during different stages of infection, providing a temporal understanding of host–pathogen interactions. This allows for the identification of key genes and pathways involved in the immune response, providing valuable insights into disease mechanisms and potential therapeutic targets [24].
In this study, the head kidney of P. fulvidraco was used as a template to establish transcriptomic libraries injected with PBS and A. hydrophila. The differential expression genes (DEGs) in two libraries were compared. Subsequently, we focused on identifying the immune-related differentially expressed genes (DEGs) through GO enrichment and KEGG enrichment after enriching these DEGs. This research provides novel insights into the innate immune response of P. fulvidraco when stimulated by A. hydrophila.
2. Materials and Methods
2.1. Ethics Statement
All experiments conducted in the present study were approved by the Committee of the Yancheng Teachers University (No. YCTU-2020005).
2.2. Sampling Collection
Yellow catfish weighing approximately 50 g from Yancheng market in Jiangsu province, China, were bought and acclimated at a temperature of 24 °C for duration of two weeks. A total of ten fish were used, with five fish injected intraperitoneally with 100 μL PBS as a control, and the other five fish injected with A. hydrophila (1 × 107 CFU/mL) strain SHOU provided by Shanghai Ocean University. After 24 h of injection, the head kidney of each fish was collected, frozen in liquid nitrogen, and stored at −80 °C for further use.
2.3. RNA Sequencing
Novogene Experimental Department in Beijing, China, constructed the library and RNA-seq analysis. Trizol reagent (Sangon, Shanghai, China) was used to extract total RNA from the head kidney, following the manufacturer’s instructions. The RNA extraction process encompasses the following four methods: 1% agarose gel was used to monitor RNA degradation; the NanoPhotometer spectrophotometer (IMPLEN, Westlake Village, CA, USA) was used to determine the purity of the RNA; to measure the RNA concentration, the Qubit 2.0 Fluorometer with the Qubit RNA Assay Kit was utilized. Additionally, the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA) was employed to assess the integrity of the RNA with the RNA Nano 6000 Assay Kit.
After the sample was extracted, the mRNA was enriched by the oligo-dT-attached magnetic beads (Ambion, Foster City, CA, USA). Subsequently, we randomly divided the mRNA into short segments using a fragment buffer. Short segments of mRNA were used as templates. The first- and second-strand cDNAs were randomly synthesized using hexamer primers. PCR amplification was performed on the fragments with terminal repair and containing a single nucleotide A, following agarose gel electrophoresis. After undergoing purification, adaptor addition, and cDNA length selection, the library, which had an average length of 200 bp, was sequenced using the Illumina HiSeq 2000 system (Illumina, San Diego, CA, USA).
2.4. Transcriptome Assembly and Annotation
In the original reads, the sequence connector and primer sequence were removed, low-quality data were filtered out, and data quality was ensured [25]. Using Trinity v2.11.0 software, the clean reads were assembled into transcripts and the unigenes were obtained by extracting the longest transcript for each gene [26]. The unigenes were annotated using five databases: Gene Ontology (GO), evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG), Swissprot, KEGG Ortholog database (KO) and NCBI non-redundant protein sequences (Nr), using BLAST with a cutoff E-value of 10−5 [27].
2.5. Analysis of Differentially Expressed Genes
The expression levels of genes in each library were calculated using RSEM (RNA-seq by Expectation Maximization) based on the RPKM value [28]. The read count of each gene was obtained from the location results. Before analyzing differential gene expression, the read count of each sequence library was adjusted by a scale normalization factor using the edgeR package 3.0.8 [29]. A differential expression analysis of the two libraries was performed using the DEG-seq R package. The p-values were adjusted using q-values [30]. Differential expression was considered significant if the q-value was less than 0.005 [31,32]. The enrichment analysis of the GO and KEGG pathways was performed using the DEGs. A GO enrichment analysis of DEGs was carried out using the GOseq R package 1.10.0, which can account for gene length bias. The statistical enrichment of DEGs in KEGG pathways was tested using KOBAS software v2.0.12 [33,34,35].
2.6. qRT-PCR Validation
To verify the expression level of the most promising candidate immune genes, 11 genes were randomly selected from the differentially expressed genes. Specific primers were designed using Primer 5.0 (Table S1). For qRT-PCR, we used the synthesized cDNA obtained by using the TUREscript cDNA Synthesize Kit (Aidlab, Beijing, China) as the RNA template. The internal reference gene was β-actin. qRT-PCR was performed using the SYBR Green qPCR Mix Kit (Aidlab, Beijing, China) on the Mastercycle ep realplex (Eppendorf, Hamburg, Germany) instrument. The cycling conditions were as follows: 95 °C for 30 s, 40 cycles of 95 °C for 30 s, and 58 °C for 25 s. Three independent experiments were conducted with a final step of 72 °C for 25 s. The relative expression level of each gene was calculated using the 2−∆∆CT method [36]. The data were reported as the mean ± standard deviation (SEM). Each sample had three repeated measurements to ensure statistical reliability, and statistical significance was considered when p < 0.05.
3. Result and Discussion
3.1. Transcriptome Sequencing and Assembly
In this study, RNA-seq was used to investigate the immune mechanism of P. fulvidraco after A. hydrophila injection. A total of 43,835,966 clean reads were obtained in the treatment group and 53,492,522 clean reads in the control group. The results showed that Q20 and Q30 accounted for 91.13% and 92.43%, respectively, in the control group, compared to 96.93% and 96.42% in the treatment group. Additionally, the GC content in the treatment group and control group was 44.65% and 44.74% respectively. The transcripts were spliced into 120,492 transcripts with a mean length of 1575, N50 length of 2594, and N90 length of 644 (Table 1). Previous studies using RNA-seq confirmed that the data on the P. fulvidraco head kidney transcriptomes were accurately constructed, serving as a valuable genetic resource for further research [18,21,24]. These results indicated that the sequence data were of high quality and the transcriptome analysis results were reliable. Among these transcripts, 33,034 were in the range 300–500 bp, 26,918 were 501–1000 bp, 27,081 were 1–2k bp, and 33,459 transcripts were longer than 2k bp. The majority of the assembled transcripts were longer than 2k bp, as observed from the length distribution (Figure 1).

Table 1.
RNA-seq reads.

Figure 1.
Length distribution of the unigenes and transcripts.
3.2. Assembly and Functional Classification of Unigenes
To obtain comprehensive gene function information, we annotated single genes using seven different databases including NR, NT, Pfam, KOG/COG, Swiss-prot, KEGG and GO. Further assembly of unigenes resulted in 43,249 unigenes with a total length of 58,057,882 and a mean length of 1342 (Table 2). Among these unigenes, 12,399 were in the range 300–500 bp, 13,772 were 500–1000 bp, 8126 were 1–2k bp, and 8985 unigenes were longer than 2k bp (Table 2). The majority of the unigenes had lengths between 500 and 1000 bp, with the remaining ones being evenly distributed (Figure 1). In at least one database, 67.89% of the unigenes were annotated, while 13.93% were annotated in all seven databases, as indicated in Table 3. 25,239 unigenes in NR (58.35%), 21,274 unigenes in NT (49.18%), 12,792 unigenes in KO (29.57%), 17,536 unigenes in SwissProt (40.54%), 17,793 unigenes in Pfam (41.14%), 17,793 unigenes in GO (41.14%), and 8427 unigenes in KOG (19.48%) were successfully annotated (Figure 2). Figure 3 also shows more details in the seven databases. Among these unigenes, 31.1% had an evalue exceeding 1 × 10−30, and 90.2% exhibited over 60% similarity with the annotated findings in the NCBI database. Furthermore, P. fulvidraco had the highest number of hits to the Lctalurus punctatus (72.1%), followed by Pygocentrus nattereri (4.8%), Astyanax mexicanus (3.9%), Cyprinus carpio (1.6%), and Danio rerio (1.2%) (Figure 4). These results were similar to those obtained from the fourfinger threadfin fish [18].

Table 2.
Assembled transcripts and unigenes obtained from P. fulvidraco head kidney transcriptome analysis.

Table 3.
A summary of unigenes was annotated in seven databases.

Figure 2.
Annotated unigenes in seven databases.
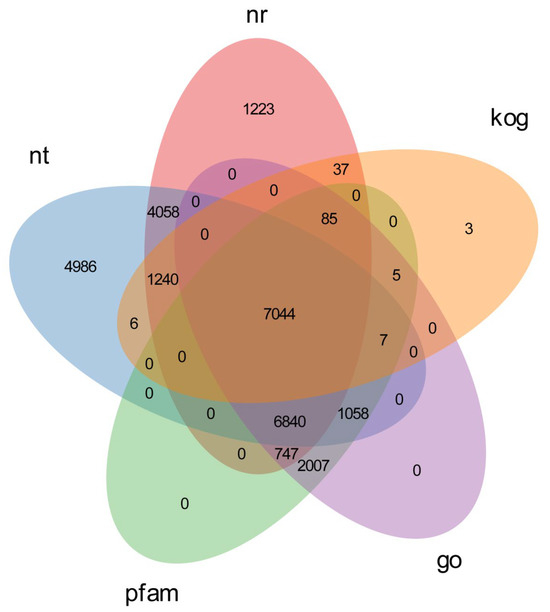
Figure 3.
Distribution of unigenes annotated in the Nr database.
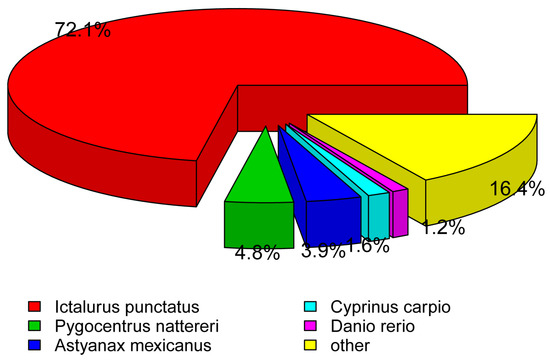
Figure 4.
Species distribution of the best hits for each unigene.
3.3. GO and KEGG Pathways Analysis
To obtain a deeper understanding of the biological importance of a specific gene, we utilized GO, KOG, and KEGG to determine its functional categories. After annotating the genes using GO, they were then categorized into three levels based on their biological process, cellular component, and molecular function, referred to as BP, CC, and MF, respectively. Fifty-six functional terms were assigned to a total of 17,793 unigenes (41.14%) that were successfully annotated in the GO database. The unigenes in the BP category were mainly associated with the cellular process, metabolic process, and single-organism process. The CC category was mostly represented by terms related to cell and cell parts. In the MF category, unigenes involved in binding and catalytic activity were highly represented (Figure 5). The results were similar to recent studies of blunt snout bream [37] and naked carp [38].
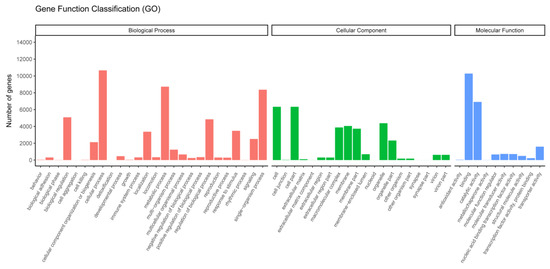
Figure 5.
Gene ontology (GO) annotation of the assembled unigenes.
A total of 26 groups were formed to categorize 8427 genes in the KOG analysis. The highest number of unigenes was found in the signal transduction mechanisms category (T), followed by general function prediction only (R), and posttranslational modification, protein turnover, and chaperones (O). The unnamed protein and cell motility categories had the lowest number of unigenes. Several unigenes (V) were assigned to the cluster of defense mechanisms, suggesting their involvement in immune defense in P. fulvidraco (Figure 6).
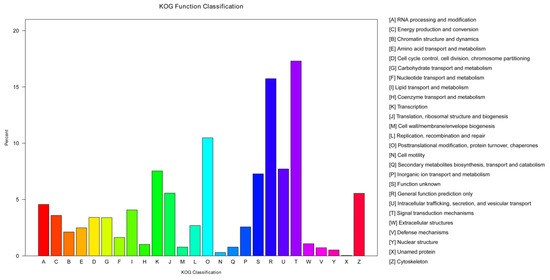
Figure 6.
KOG annotation of the assembled unigenes. A specific functional cluster’s number of unigenes is shown on the y-axis. The x-axis lists categories indicated by letters in the column on the right.
KEGG offers a comprehensive database to effectively examine gene functions and establish connections between genomic data and advanced functional information [39]. In the present study, 12,235 genes were divided into five categories. Within the cellular processes, transport and catalism accounted for the most genes, followed by cell community. Signal transduction had the highest number of genes in the environmental information process. Folding, sorting, and graduation had the highest representation in genetic information processing, followed by translation. Metabolism was the largest category, with carbohydrate, lipid, and the amino acid metabolism being the top three pathways. The immune system and endocrine system were the largest two systems in organismal systems (Table S2; Figure 7).
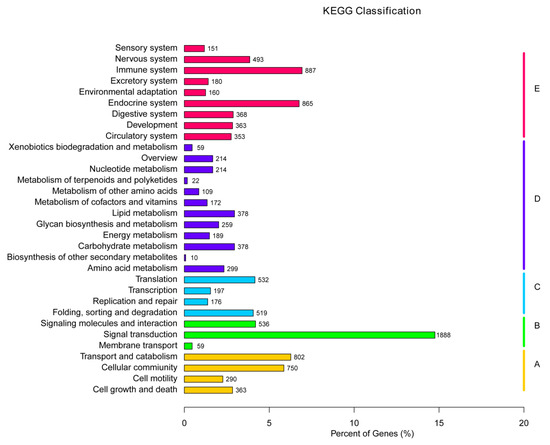
Figure 7.
KEGG annotation of assembled unigenes. Different colored bars represent different groups.
3.4. Enrichment Analysis of GO and KEGG Pathways on the Basis of DEGs
The identification of DEGs between the injection of A. hydrophila and control groups revealed a total of 973 unigenes in the head kidney showed significant differential expression following A. hydrophila injection. Among these unigenes, 553 were significantly upregulated and 420 were significantly downregulated (Figure 8). Pathway significant enrichment can be used to determine the most significant biochemical metabolic pathway and signal transduction pathway involved in the differential expression of genes upon which organisms depend to carry out their biological functions. DEGs, in the analysis of Gene Ontology (GO), can be categorized into Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). In the CC category, the highest proportion is attributed to the extracellular region, while, in the MF category, cytokine receptor binding is the most prevalent. KEGG is a major public database for pathways [39]. Following the mapping to the KEGG database, it was determined that the annotation of the 973 DEGs was successful, and they were allocated to a total of 231 pathways. Among these pathways, Figure 9 displays the top 20 most abundant DEGs associated with P. fulvidraco infection. Some of the genes involved in the immune defense system include those related to the NF-κB signaling pathway, Toll-like receptor signaling pathway, TNF signaling pathway, and cytokine–cytokine receptor interaction. The NF-kappa B signaling pathway is an important signal molecule involved in common biological immune defense [40]. The NF-κB family of transcription factors regulates numerous genes involved in diverse cellular processes, including cell proliferation, differentiation, genome stability, and both the innate and adaptive immune responses [41]. The responsibility for pathogen recognition and the initiation of innate immune responses lies with Toll-like receptors (TLRs), which are well-studied pattern recognition receptors [42]. TLRs have played a crucial role in connecting the initial recognition of pathogens by innate immune cells to the activation of the adaptive immune response [43]. Cytokines play a critical role in regulating and mobilizing cells involved in innate and adaptive inflammatory host defenses, as well as cell growth, differentiation, cell death, angiogenesis, and processes aimed at restoring homeostasis [44]. These soluble extracellular proteins or glycoproteins serve as crucial intercellular regulators [45]. Cytokines are released by different cells in the body, typically as a response to an activating stimulus [46]. These cytokines initiate responses by attaching to specific receptors on the cell surface of target cells. Based on their structure, cytokines can be categorized into various families, while their receptors can also be grouped in a similar manner [47]. The TNF signaling pathway has a critical function in diverse physiological and pathological processes, such as cell growth, specialization, programmed cell death, the regulation of immune response, and the triggering of inflammation [48]. Since it was discovered twenty years ago, TNF has been known as an anti-cancer agent while also having a multifunctional impact on lipid metabolism, coagulation, insulin resistance, and endothelial function [49]. Cells can receive both survival and death signals from members of the TNFR superfamily [50]. Tumor necrosis factor alpha (TNF-α) is a versatile cytokine that plays a crucial role in regulating numerous signaling pathways associated with inflammation, immunity, apoptosis (cell death), anti-apoptosis (cell survival), and even the development of tumors [51]. These signaling pathways play a crucial role in enhancing the immune response to foreign pathogens by activating a range of signal transduction pathways.
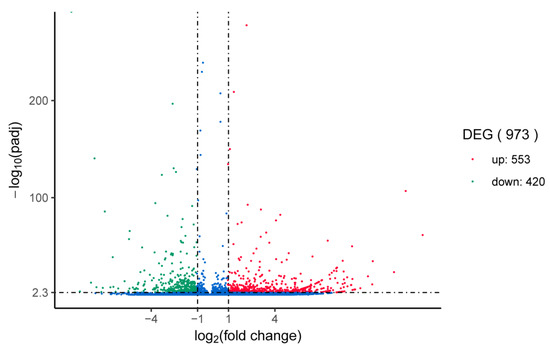
Figure 8.
Volcano analysis of different expression genes (DEGs) in head kidney of yellow catfish compared to control at 24 h post injection. The volcano plot displays the expression levels of each unigene, with upregulation shown as up and downregulation as down. The X-axis represents the fold change of the gene between groups, while the Y-axis displays the LogP of the unigene.
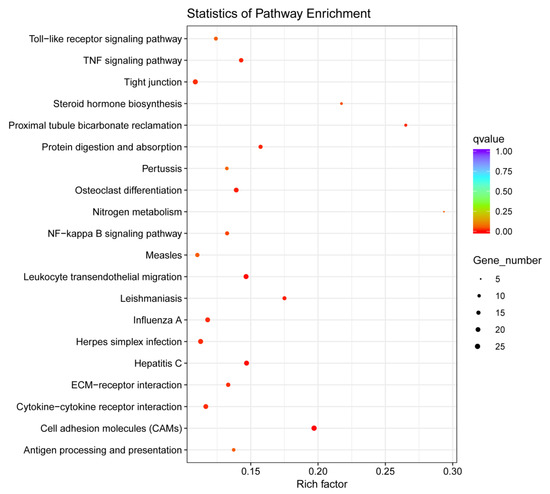
Figure 9.
KEGG pathway enrichment analysis of DEGs between groups. The point size shows the number of DEGs enriched in the KEGG pathway. The point color shows different q-values, as indicated on the right.
3.5. Validation of RNA-Seq Results Using RT-qPCR
To further confirm the differential expression of genes (DEGs), qRT-PCR analysis was conducted to randomly select seven genes (Figure 10). The results indicated that most of the immune-related genes were upregulated compared to the control group, which confirmed the reliability of the transcriptomic data. Among them, the C3 complement component, Galectin-3, Calpain-9, and E3 ubiquitin–protein ligase showed significant increases compared to the control group, followed by the C-type lectin, Claudin-7, and Interferon-induced protein 2. The complement system, which is crucial for innate immunity, is composed of a complex protein network [52]. C3 complement component (C3) plays a critical role in the complement cascade and immune defense by participating in complement activation [53]. The mRNA expression levels of C3 in Oreochromis niloticus were significantly upregulated in the liver, spleen, and head kidney after challenge with Streptococcus agalactiae and A. hydrophila [54]. Similarly, Ctenopharyngodon idella showed significantly upregulated C3 mRNA expression levels in the gill, liver, spleen, intestine, trunk kidney, and head kidney after infection with A. hydrophila [55]. Galectins (Gals) belong to the β-galactoside binding protein family and are involved in various immune responses during pathogenic infections [56]. Larimichthys crocea Gal-3 expression was significantly upregulated after infection with Pseudomonas plecoglossicida [57]. In C. idella, the liver and spleen showed an increase in Gal-3 expression when exposed to grass carp reovirus (GCRV), lipopolysaccharide (LPS), and polyinosinic:polycytidylic acid (poly I:C) challenge [58]. Calpains are calcium-dependent neutral proteases that play important roles in multiple cellular functions [59]. Calpain-9-like protein was identified in the skin mucus of C. idella after A. hydrophila infection [60]. E3 ubiquitin–protein ligases are essential for the selection of proteins targeted for degradation through the ubiquitin proteasome pathway, and they are involved in various cellular processes, including signal transduction, transcription, immune response, cell cycle, and apoptosis [61]. The ubiquitin E3 ligase TRIM10 acts as a positive regulator of STING signaling and promotes STING aggregation and activation in the Golgi apparatus during cGAS-STING-mediated antiviral and antitumor immunity [62]. C-type lectins (CTLs) are carbohydrate-binding proteins that function as pattern recognition receptors involved in phagocytosis and the elimination of pathogens. They play a crucial role in antigen presentation and initiation of the adaptive immune response [63]. The liver, spleen, head kidney, and blood showed an upregulation in the expression of P. fulvidraco CTL following the challenge with lipopolysaccharide and polyriboinosinic polyribocytidylic acid [9]. Following Cryptocaryon irritans challenge, the head kidney and liver of Lates calcarifer experienced an induction in the mRNA transcript levels of CTL [64]. Similarly, the mRNA transcript levels of Misgurnus anguillicaudatus CTL were significantly upregulated after immune challenge with A. hydrophila [65]. Interferons induce the expression of interferon-inducible proteins, which play a crucial role in regulating antiviral, cell growth, immune modulatory, and anti-tumor functions [66,67]. These proteins play various roles within cells, such as controlling RNA and protein metabolism, facilitating growth and differentiation, regulating apoptosis, and transmitting signals. Therefore, the qRT-PCR analysis results corroborated the reliability of the RNA-seq data.
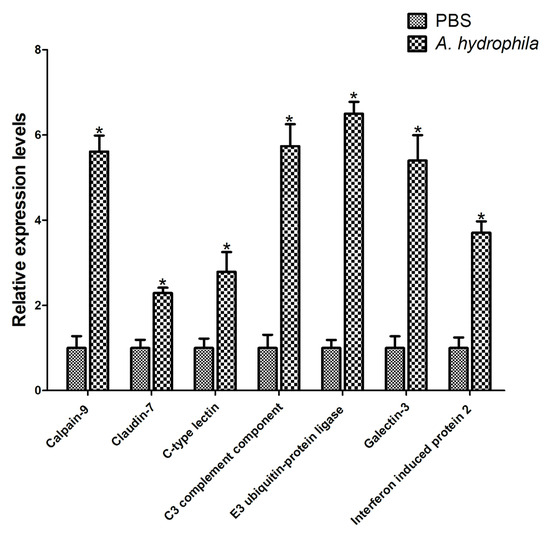
Figure 10.
The relative expression profiles of randomly chosen genes involved in the immune response were analyzed. PBS injections were used as the control groups. The gene expression level in the control group was designated as 1.0. The data were presented as the average fold change (means ± SE, n = 3) compared to the untreated group. The values were marked with asterisks (* p < 0.05) to indicate significant differences from the control at the corresponding time point.
4. Conclusions
In this study, we constructed and sequenced the head kidney response of Pelteobagrus fulvidraco to A. hydrophila injection. Illumina sequencing technology was utilized for sequence analysis, resulting in the identification of 973 DEGs. Among these, 553 were upregulated and 420 were downregulated. Furthermore, genes related to immune defense systems, including the TNF signaling pathway, NF-kappa B signaling pathway, Toll-like receptor signaling pathway, and cytokine–cytokine receptor interaction, were found, suggesting that these pathways play a role in the immune response of P. fulvidraco to A. hydrophila injection. These pathways are involved in the recognition of pathogens, inflammatory responses, and the activation of immune cells. Overall, these findings contribute to our understanding of the immune response of P. fulvidraco and shed light on the genes and pathways involved in their defense against A. hydrophila injection. These data could be valuable for fish health management, disease prevention, and potential interventions to enhance the immune response of fish species against bacterial infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9030100/s1, Table S1. Primers used in this study, Table S2. KEGG annotation of assembled unigenes.
Author Contributions
Conceptualization, Q.L.; methodology, S.J.; software, S.J.; validation, S.J., Y.L. and T.W.; formal analysis, S.J.; investigation, R.M. and Q.L.; resources, C.H.; data curation, S.J. and Q.L.; writing—original draft preparation, S.J.; writing—review and editing, Q.L.; supervision, Q.L.; project administration, Q.L.; funding acquisition, Q.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Research General Program of Jiangsu Provincial Higher Education Institutions (22KJA240002, 21KJA240003), the Fishery High-Quality Development Research Project of Yancheng, China (YCSCYJ2021022), the Science and Technology Vice-President Project of Jiangsu Province (FZ2023), the National Natural Science Foundation of China (32370556, 32070526 and 32270487), and the National Key R&D Program of China (2019YFD0900404-05). This study was sponsored by the Qing Lan Project of Jiangsu Province, and the “Outstanding Young Talents” of YCTU.
Institutional Review Board Statement
The Committee of the Yancheng Teachers University approved the animal protocols, and all experiments were performed under the applicable standards, with access No. YCTU-2020005 (16 March 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated for this study can be found in the GenBank accession no. PRJNA904403.
Conflicts of Interest
The authors declare no competing interests.
References
- Gong, G.; Dan, C.; Xiao, S.; Guo, W.; Huang, P.; Xiong, Y.; Wu, J.; He, Y.; Zhang, J.; Li, X.; et al. Chromosomal-level assembly of yellow catfish genome using third-generation DNA sequencing and Hi-C analysis. Gigascience 2018, 7, giy120. [Google Scholar] [CrossRef]
- Bureau of Fisheries of Ministry of Agriculture and Rural Affairs of the People’s Republic of China. China Fisheries Statistical Yearbook of 2022; China Agricultural Press: Beijing, China, 2022.
- You, S.-L.; Jiang, X.-X.; Zhang, G.-R.; Ji, W.; Ma, X.-F.; Zhou, X.; Wei, K.-J. Molecular Characterization of Nine TRAF Genes in Yellow Catfish (Pelteobagrus fulvidraco) and Their Expression Profiling in Response to Edwardsiella ictaluri Infection. Int. J. Mol. Sci. 2023, 24, 8363. [Google Scholar] [CrossRef]
- Harikrishnan, R.; Balasundaram, C. Modern trends in Aeromonas hydrophila disease management with fish. Rev. Fish. Sci. 2005, 13, 281–320. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, F.; Ou, M.; Liu, H.; Luo, Q.; Fei, S.; Zhao, J.; Chen, K.; Zhao, Q.; Li, K. Effects of Myostatin b Knockout on Offspring Body Length and Skeleton in Yellow Catfish (Pelteobagrus fulvidraco). Biology 2023, 12, 1331. [Google Scholar] [CrossRef]
- Tang, X.H.; Ye, X.T.; Wu, Q.C.; Tang, Y.Y.; Zhang, D.Z.; Liu, Q.N.; Tang, B.P.; Zhu, H.R. Molecular characterization and expression analysis of a novel C-type lectin (CTL) gene in yellow catfish Pelteobagrus fulvidraco. Aquacult. Rep. 2021, 20, 100640. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, D.-Y.; Guo, W.; Gong, G.; Chen, Z.-X.; Tang, Q.; Mei, J. Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco). Biology 2022, 11, 1818. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Zhang, D.G.; Lei, X.J.; Tan, X.Y.; Song, C.C.; Zheng, H.; Luo, Z. Effects of dietary selenium and oxidized fish oils on intestinal lipid metabolism and antioxidant responses of yellow catfish Pelteobagrus fulvidraco. Antioxidants 2022, 11, 1904. [Google Scholar] [CrossRef]
- Jiang, S.; Lei, Y.; Li, Y.; Sun, W.; Wang, T.; Ma, R.; Liu, Q.; Tang, B. Molecular Identification and Expression Analysis of an Intelectin Gene in the Yellow Catfish Pelteobagrus fulvidraco (Siluriformes: Bagridae). Fishes 2023, 8, 492. [Google Scholar] [CrossRef]
- Zhu, L.; Nie, L.; Zhu, G.; Xiang, L.X.; Shao, J.Z. Advances in research of fish immune-relevant genes: A comparative overview of innate and adaptive immunity in teleosts. Dev. Comp. Immunol. 2013, 39, 39–62. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Boahen, A.; Hu, D.; Adams, M.J.; Ma, B. Bidirectional crosstalk between the peripheral nervous system and lymphoid tissues/organs. Front. Immunol. 2023, 14, 1254054. [Google Scholar] [CrossRef] [PubMed]
- Smorodinskaya, S.; Kochetkov, N.; Gavrilin, K.; Nikiforov-Nikishin, D.; Reznikova, D.; Vatlin, A.; Klimuk, A.; Odorskaya, M.; Nikiforov-Nikishin, A.; Ponomarev, A.; et al. The Effects of Acute Bisphenol A Toxicity on the Hematological Parameters, Hematopoiesis, and Kidney Histology of Zebrafish (Danio rerio). Animals 2023, 13, 3685. [Google Scholar] [CrossRef] [PubMed]
- Geven, E.J.; Klaren, P.H. The teleost head kidney: Integrating thyroid and immune signalling. Dev. Comp. Immunol. 2017, 66, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Biasini, L.; Zamperin, G.; Pascoli, F.; Abbadi, M.; Buratin, A.; Marsella, A.; Panzarin, V.; Toffan, A. Transcriptome Profiling of Oncorhynchus mykiss Infected with Low or Highly Pathogenic Viral Hemorrhagic Septicemia Virus (VHSV). Microorganisms 2024, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Rauta, P.R.; Nayak, B.; Das, S. Immune system and immune responses in fish and their role in comparative immunity study: A model for higher organisms. Immunol. Lett. 2012, 148, 23–33. [Google Scholar] [CrossRef]
- Leal, Y.; Valenzuela-Muñoz, V.; Casuso, A.; Benavente, B.P.; Gallardo-Escárate, C. Comparative Transcriptomics in Atlantic Salmon Head Kidney and SHK-1 Cell Line Exposed to the Sea Louse Cr-Cathepsin. Genes 2023, 14, 905. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, S.; Wang, P.-C.; Chen, S.-C. Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum). Int. J. Mol. Sci. 2023, 24, 3832. [Google Scholar] [CrossRef]
- Gao, J.; Guo, H.-Y.; Liu, M.-J.; Zhu, K.-C.; Liu, B.; Liu, B.-S.; Zhang, N.; Jiang, S.-G.; Zhang, D.-C. Transcriptome Analysis of the Immune Process of Golden Pompano (Trachinotus ovatus) Infected with Streptococcus agalactiae. Fishes 2023, 8, 52. [Google Scholar] [CrossRef]
- Ghosh, D.; Datta, S.; Bhattacharya, S.; Mazumder, S. Long-term exposure to arsenic affects head kidney and impairs humoral immune responses of Clarias batrachus. Aquat. Toxicol. 2007, 81, 79–89. [Google Scholar] [CrossRef]
- Jiang, S.H.; Wu, L.X.; Cai, Y.T.; Ma, R.T.; Zhang, H.B.; Zhang, D.Z.; Tang, B.P.; Liu, Q.N.; Dai, L.S. Differentially expressed genes in head kidney of Pelteobagrus fulvidraco following Vibrio cholerae challenge. Front. Immunol. 2023, 13, 1039956. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.S.V.; Power, D.M.; Guerreiro, P.M.; Louro, B.; Chen, L.; Canário, A.V.M. Transcriptomic Down-Regulation of Immune System Components in Barrier and Hematopoietic Tissues after Lipopolysaccharide Injection in Antarctic Notothenia coriiceps. Fishes 2022, 7, 171. [Google Scholar] [CrossRef]
- Liu, Q.N.; Tang, Y.Y.; Zhou, M.J.; Luo, S.; Li, Y.T.; Wang, G.; Zhang, D.Z.; Yang, H.; Tang, B.P.; He, W.F. Differentially expressed genes involved in immune pathways from yellowhead catfish (Tachysurus fulvidraco) after poly (I:C) challenge. Int. J. Biol. Macromol. 2021, 183, 340–345. [Google Scholar] [CrossRef]
- Kim, J.; Cho, M.; Lim, J.; Choi, H.; Hong, S. Pathogenic Mechanism of a Highly Virulent Infectious Hematopoietic Necrosis Virus in Head Kidney of Rainbow Trout (Oncorhynchus mykiss) Analyzed by RNA-Seq Transcriptome Profiling. Viruses 2022, 14, 859. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar] [CrossRef]
- Storey, J.D. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tran, N.T.; Gao, Z.X.; Zhao, H.H.; Yi, S.K.; Chen, B.X.; Zhao, Y.H.; Lin, L.; Liu, X.Q.; Wang, W.M. Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish Shellfish Immunol. 2015, 45, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Zhang, C.; Zhang, R.; Zhao, K. Transcriptome profiling analysis of naked carp (Gymnocypris przewalskii) provides insights into the immune-related genes in highland fish. Fish Shellfish Immunol. 2015, 46, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef]
- Chandel, N.S.; Trzyna, W.C.; McClintock, D.S.; Schumacker, P.T. Role of oxidants in NF-kappa B activation and TNF-alpha gene transcription induced by hypoxia and endotoxin. J. Immunol. 2000, 165, 1013–1021. [Google Scholar] [CrossRef]
- Beutler, B. Toll-like receptors: How they work and what they do. Curr. Opin. Hematol. 2002, 9, 2–10. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Leng, T.; Akther, H.D.; Hackstein, C.; Powell, K.; King, T.; Friedrich, M.; McCuaig, S.; Neyazi, M.; Arancibia-Cárcamo, C.V.; Hagel, J.; et al. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 2019, 28, 3077–3091. [Google Scholar] [CrossRef] [PubMed]
- Vuletić, A.; Jovanić, I.; Jurišić, V.; Milovanović, Z.; Nikolić, S.; Spurnić, I.; Konjević, G. IL-2 and IL-15 induced NKG2D, CD158a and CD158b expression on T, NKT- like and NK cell lymphocyte subsets from regional lymph nodes of melanoma patients. Pathol. Oncol. Res. 2020, 26, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Vesely, M.D. Getting Under the Skin: Targeting Cutaneous Autoimmune Disease. Yale J. Biol. Med. 2020, 93, 197–206. [Google Scholar]
- Cárcamo-Martínez, Á.; Mallon, B.; Anjani, Q.K.; Domínguez-Robles, J.; Utomo, E.; Vora, L.K.; Tekko, I.A.; Larrañeta, E.; Donnelly, R.F. Enhancing intradermal delivery of tofacitinib citrate: Comparison between powder-loaded hollow microneedle arrays and dissolving microneedle arrays. Int. J. Pharm. 2021, 593, 120152. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-C.; Yeh, W.-C.; Ohashi, P.S. LPS/TLR4 Signal Transduction Pathway. Cytokine 2008, 42, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Blaser, H.; Dostert, C.; Mak, T.W.; Brenner, D. TNF and ROS Crosstalk in Inflammation. Trends Cell Biol. 2016, 26, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, S.; Huang, P.; Zhang, H.; Yu, C. Development of a prognostic model based on an immunogenomic landscape analysis of medulloblastoma. Biosci. Rep. 2021, 41, BSR20202907. [Google Scholar] [CrossRef]
- Zhu, M.; Song, Y.; Martínez-Cuesta, M.C.; Peláez, C.; Li, E.; Requena, T.; Wang, H.; Sun, Y. Immunological Activity and Gut Microbiota Modulation of Pectin from Kiwano (Cucumis metuliferus) Peels. Foods 2022, 11, 1632. [Google Scholar] [CrossRef]
- Carpanini, S.M.; Torvell, M.; Morgan, B.P. Therapeutic Inhibition of the Complement System in Diseases of the Central Nervous System. Front. Immunol. 2019, 10, 362. [Google Scholar] [CrossRef] [PubMed]
- Santana-Coelho, D.; Lugo, J.N. Hippocampal Upregulation of Complement Component C3 in Response to Lipopolysaccharide Stimuli in a Model of Fragile-X Syndrome. Curr. Issues Mol. Biol. 2023, 45, 9306–9315. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Mu, L.; Qiu, L.; Chen, N.; Li, J.; Zeng, Q.; Yin, X.; Ye, J. Complement C3 Regulates Inflammatory Response and Monocyte/Macrophage Phagocytosis of Streptococcus agalactiae in a Teleost Fish. Int. J. Mol. Sci. 2022, 23, 15586. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Shen, Y.; Wang, S.; Xu, X.; Dang, Y.; Zhang, M.; Li, L.; Zhang, J.; Wang, R.; Li, J. Complement component 3 (C3): An important role in grass carp (Ctenopharyngodon idella) experimentally exposed to Aeromonas hydrophila. Fish Shellfish Immunol. 2019, 88, 189–197. [Google Scholar] [CrossRef]
- Rabinovic, G.A. Role of galectins in inflammatory and immunomodulatory processes. Biochim. Biophys. Acta 2002, 1572, 274–284. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, B.; Li, W.; Han, F. Molecular Characterization of Galectin-3 in Large Yellow Croaker Larimichthys crocea Functioning in Antibacterial Activity. Int. J. Mol. Sci. 2023, 24, 11539. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, R.; Chu, P.; Chen, L.; Li, Y.; He, L.; Li, Y.; Liao, L.; Zhu, Z.; Wang, Y. Characterization and expression of galectin-3 in grass carp (Ctenopharyngodon idella). Dev. Comp. Immunol. 2020, 104, 103567. [Google Scholar] [CrossRef]
- Ono, Y.; Sorimachi, H. Calpains—An elaborate proteolytic system. Biochim. Biophys. Acta BBA Proteins Proteom. 2012, 1824, 224–236. [Google Scholar] [CrossRef]
- Ali, S.; Dawar, F.U.; Ullah, W.; Hassan, M.; Ullah, K.; Zhao, Z. Proteomic map of the differentially expressed proteins in the skin of Ctenopharyngodon idella against Aeromonas hydrophila infection. Fish Shellfish Immunol. Rep. 2023, 5, 100122. [Google Scholar] [CrossRef]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef]
- Kong, L.; Sui, C.; Chen, T.; Zhang, L.; Zhao, W.; Zheng, Y.; Liu, B.; Cheng, X.; Gao, C. The ubiquitin E3 ligase TRIM10 promotes STING aggregation and activation in the Golgi apparatus. Cell Rep. 2023, 42, 112306. [Google Scholar] [CrossRef]
- Ng, T.B.; Fai Cheung, R.C.; Wing Ng, C.C.; Fang, E.F.; Wong, J.H. A review of fish lectins. Curr. Protein Pept. Sci. 2015, 16, 337–351. [Google Scholar] [CrossRef]
- Mohd-Shaharuddin, N.; Mohd-Adnan, A.; Kua, B.C.; Nathan, S. Expression profile of immune-related genes in Lates calcarifer infected by Cryptocaryon irritans. Fish Shellfish Immunol. 2013, 34, 762–769. [Google Scholar] [CrossRef]
- Zhang, X.W.; Yang, C.H.; Zhang, H.Q.; Pan, X.T.; Jin, Z.Y.; Zhang, H.W.; Xia, X.H. A C-type lectin with antibacterial activity in weather loach, Misgurnus anguillicaudatus. J. Fish Dis. 2020, 43, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Langevin, C.; Aleksejeva, E.; Passoni, G.; Palha, N.; Levraud, J.P.; Boudinot, P. The antiviral innate immune response in fish: Evolution and conservation of the IFN system. J. Mol. Biol. 2013, 425, 4904–4920. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. Teleost fish interferons and their role in immunity. Dev. Comp. Immunol. 2011, 35, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).