Highlighting the Importance of Correct Sex Identification in Chondrichthyan Genomic Studies, Using the White Shark as an Example
Abstract
1. Introduction
2. Materials and Methods
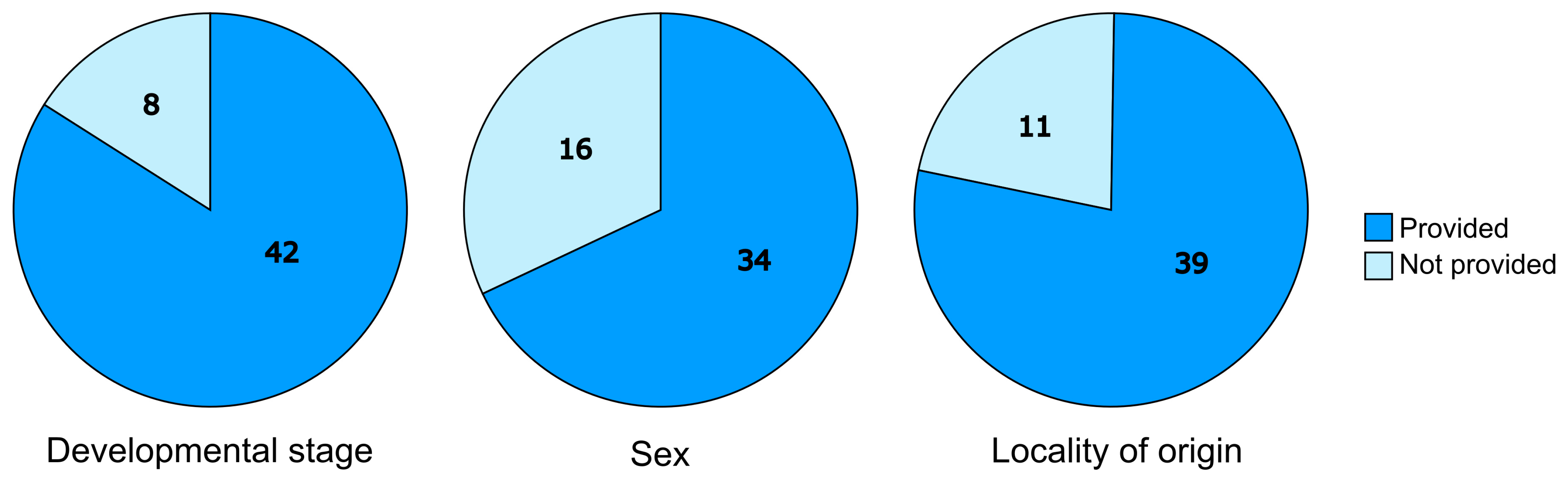
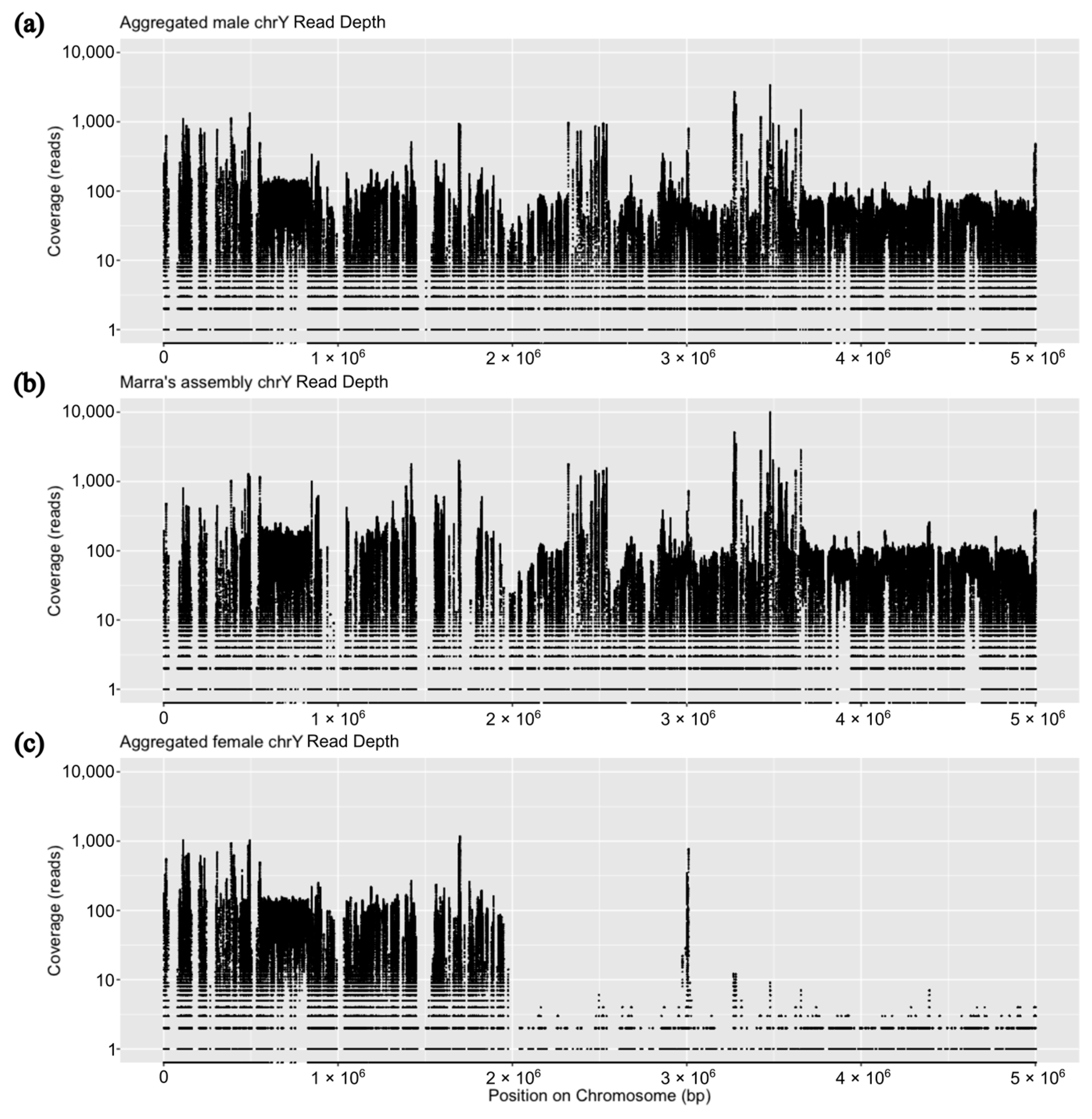
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiffer, P.H.; Kroiher, M.; Kraus, C.; Koutsovoulos, G.D.; Kumar, S.; Camps, J.I.R.; Nsah, N.A.; Stappert, D.; Morris, K.; Heger, P.; et al. The Genome of Romanomermis culicivorax: Revealing Fundamental Changes in the Core Developmental Genetic Toolkit in Nematoda. BMC Genom. 2013, 14, 923. [Google Scholar] [CrossRef] [PubMed]
- Hara, Y.; Yamaguchi, K.; Onimaru, K.; Kadota, M.; Koyanagi, M.; Keeley, S.D.; Tatsumi, K.; Tanaka, K.; Motone, F.; Kageyama, Y.; et al. Shark Genomes Provide Insights into Elasmobranch Evolution and the Origin of Vertebrates. Nat. Ecol. Evol. 2018, 2, 1761–1771. [Google Scholar] [CrossRef] [PubMed]
- Toh, H.; Yang, C.; Formenti, G.; Raja, K.; Yan, L.; Tracey, A.; Chow, W.; Howe, K.; Bergeron, L.A.; Zhang, G.; et al. A Haplotype-Resolved Genome Assembly of the Nile Rat Facilitates Exploration of the Genetic Basis of Diabetes. BMC Biol. 2022, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Gonzaga-Jauregui, C.; Lupski, J.R.; Gibbs, R.A. Human Genome Sequencing in Health and Disease. Annu. Rev. Med. 2012, 63, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shearwin-Whyatt, L.; Li, J.; Song, Z.; Hayakawa, T.; Stevens, D.; Fenelon, J.C.; Peel, E.; Cheng, Y.; Pajpach, F.; et al. Platypus and Echidna Genomes Reveal Mammalian Biology and Evolution. Nature 2021, 592, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, Z.; Bai, H.; Huang, Y.; Kang, N.; Ding, X.; Liu, J.; Luo, H.; Yang, C.; Chen, W.; et al. Evolutionary Analysis of a Complete Chicken Genome. Proc. Natl. Acad. Sci. USA 2023, 120, e2216641120. [Google Scholar] [CrossRef]
- Jebb, D.; Huang, Z.; Pippel, M.; Hughes, G.M.; Lavrichenko, K.; Devanna, P.; Winkler, S.; Jermiin, L.S.; Skirmuntt, E.C.; Katzourakis, A.; et al. Six Reference-Quality Genomes Reveal Evolution of Bat Adaptations. Nature 2020, 583, 578–584. [Google Scholar] [CrossRef]
- Tan, M.; Redmond, A.K.; Dooley, H.; Nozu, R.; Sato, K.; Kuraku, S.; Koren, S.; Phillippy, A.M.; Dove, A.D.; Read, T. The Whale Shark Genome Reveals Patterns of Vertebrate Gene Family Evolution. eLife 2021, 10, e65394. [Google Scholar] [CrossRef]
- Formenti, G.; Theissinger, K.; Fernandes, C.; Bista, I.; Bombarely, A.; Bleidorn, C.; Ciofi, C.; Crottini, A.; Godoy, J.A.; Höglund, J.; et al. The Era of Reference Genomes in Conservation Genomics. Trends Ecol. Evol. 2022, 37, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Dussex, N.; van der Valk, T.; Morales, H.E.; Wheat, C.W.; Díez-del-Molino, D.; von Seth, J.; Foster, Y.; Kutschera, V.E.; Guschanski, K.; Rhie, A.; et al. Population Genomics of the Critically Endangered Kākāpō. Cell Genom. 2021, 1, 100002. [Google Scholar] [CrossRef]
- Morin, P.A.; Archer, F.I.; Avila, C.D.; Balacco, J.R.; Bukhman, Y.V.; Chow, W.; Fedrigo, O.; Formenti, G.; Fronczek, J.A.; Fungtammasan, A.; et al. Reference Genome and Demographic History of the Most Endangered Marine Mammal, the Vaquita. Mol. Ecol. Resour. 2021, 21, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Larivière, D.; Abueg, L.; Brajuka, N.; Gallardo-Alba, C.; Grüning, B.; Ko, B.J.; Ostrovsky, A.; Palmada-Flores, M.; Pickett, B.D.; Rabbani, K.; et al. Scalable, Accessible and Reproducible Reference Genome Assembly and Evaluation in Galaxy. Nat. Biotechnol. 2024, 42, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.-P.; Sun, C.-L.; Punt, A.E.; Liu, K.-M. Demographic Analysis of the Shortfin Mako Shark, Isurus oxyrinchus, in the Northwest Pacific Using a Two-Sex Stage-Based Matrix Model. ICES J. Mar. Sci. 2014, 71, 1604–1618. [Google Scholar] [CrossRef]
- Wilson Sayres, M.A. Genetic Diversity on the Sex Chromosomes. Genome Biol. Evol. 2018, 10, 1064–1078. [Google Scholar] [CrossRef] [PubMed]
- Phillips, N.M.; Devloo-Delva, F.; McCall, C.; Daly-Engel, T.S. Reviewing the Genetic Evidence for Sex-Biased Dispersal in Elasmobranchs. Rev. Fish Biol. Fish. 2021, 31, 821–841. [Google Scholar] [CrossRef]
- Wu, J.; Liu, F.; Jiao, J.; Luo, H.; Fan, S.; Liu, J.; Wang, H.; Cui, N.; Zhao, N.; Qu, Q.; et al. Comparative Genomics Illuminates Karyotype and Sex Chromosome Evolution of Sharks. Cell Genom. 2024, 4, 100607. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Uno, Y.; Kadota, M.; Nishimura, O.; Nozu, R.; Murakumo, K.; Matsumoto, R.; Sato, K.; Kuraku, S. Elasmobranch Genome Sequencing Reveals Evolutionary Trends of Vertebrate Karyotype Organization. Genome Res. 2023, 33, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Vicoso, B. Molecular and Evolutionary Dynamics of Animal Sex-Chromosome Turnover. Nat. Ecol. Evol. 2019, 3, 1632–1641. [Google Scholar] [CrossRef]
- Pinto, B.J.; Keating, S.E.; Nielsen, S.V.; Scantlebury, D.P.; Daza, J.D.; Gamble, T. Chromosome-Level Genome Assembly Reveals Dynamic Sex Chromosomes in Neotropical Leaf-Litter Geckos (Sphaerodactylidae: Sphaerodactylus). J. Hered. 2022, 113, 272–287. [Google Scholar] [CrossRef] [PubMed]
- Donahue, C.S.C.; William, H. A Karyotypic Study of Three Species of Rajiformes (Chondrichthyes, Pisces). Can. J. Genet. Cytol. 1974, 16, 203–211. [Google Scholar] [CrossRef] [PubMed]
- de Souza Valentim, F.C.; Porto, J.I.R.; Feldberg, E. Chromosomal Characterization of Amazonian Freshwater Stingrays with Evidence for New Karyomorphs and XX/XY Sex Chromosomes. Genet. Mol. Biol. 2019, 42, 578–593. [Google Scholar] [CrossRef]
- Maddock, M.B.; Schwartz, F.J. Elasmobranch Cytogenetics: Methods and Sex Chromosomes. Bull. Mar. Sci. 1996, 58, 147–155. [Google Scholar]
- Uno, Y.; Nozu, R.; Kiyatake, I.; Higashiguchi, N.; Sodeyama, S.; Murakumo, K.; Sato, K.; Kuraku, S. Cell Culture-Based Karyotyping of Orectolobiform Sharks for Chromosome-Scale Genome Analysis. Commun. Biol. 2020, 3, 652. [Google Scholar] [CrossRef] [PubMed]
- Marra, N.J.; Stanhope, M.J.; Jue, N.K.; Wang, M.; Sun, Q.; Pavinski Bitar, P.; Richards, V.P.; Komissarov, A.; Rayko, M.; Kliver, S.; et al. White Shark Genome Reveals Ancient Elasmobranch Adaptations Associated with Wound Healing and the Maintenance of Genome Stability. Proc. Natl. Acad. Sci. USA 2019, 116, 4446–4455. [Google Scholar] [CrossRef] [PubMed]
- Rhie, A.; McCarthy, S.A.; Fedrigo, O.; Damas, J.; Formenti, G.; Koren, S.; Uliano-Silva, M.; Chow, W.; Fungtammasan, A.; Kim, J.; et al. Towards Complete and Error-Free Genome Assemblies of All Vertebrate Species. Nature 2021, 592, 737–746. [Google Scholar] [CrossRef]
- Wagner, I.; Smolina, I.; Koop, M.E.L.; Bal, T.; Lizano, A.M.; Choo, L.Q.; Hofreiter, M.; Gennari, E.; de Sabata, E.; Shivji, M.S.; et al. Genome Analysis Reveals Three Distinct Lineages of the Cosmopolitan White Shark. Curr. Biol. 2024, 34, 3582–3590.e4. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly: Sebastopol, CA, USA, 2020; ISBN 978-1-4919-7516-9. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Wickham, H. Elegant Graphics for Data Analysis (Ggplot2). Appl. Spat. Data Anal. R. 2009, 784, 785. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Devloo-Delva, F.; Gosselin, T.; Butcher, P.A.; Grewe, P.M.; Huveneers, C.; Thomson, R.B.; Werry, J.M.; Feutry, P. An R-Based Tool for Identifying Sex-Linked Markers from Restriction Site-Associated DNA Sequencing with Applications to Elasmobranch Conservation. Conserv. Genet. Resour. 2024, 16, 11–16. [Google Scholar] [CrossRef]
- Hendon, J.M.; Koester, D.M.; Hoffmayer, E.R.; Driggers, W.B.; Cicia, A.M. Occurrence of an Intersexual Blacktip Shark in the Northern Gulf of Mexico, with Notes on the Standardization of Classifications for This Condition in Elasmobranchs. Mar. Coast. Fish. 2013, 5, 174–180. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, M.; Jiang, C.; Yang, S.; Xiao, J. Occurrence of an Intersexual Pacific Spadenose Shark Scoliodon macrorhynchos from the Southern Taiwan Strait. Mar. Coast. Fish. 2017, 9, 573–576. [Google Scholar] [CrossRef]
- Perrin, N. Sex Reversal: A Fountain of Youth for Sex Chromosomes? Evolution 2009, 63, 3043–3049. [Google Scholar] [CrossRef]
- Rodrigues, N.; Studer, T.; Dufresnes, C.; Perrin, N. Sex-Chromosome Recombination in Common Frogs Brings Water to the Fountain-of-Youth. Mol. Biol. Evol. 2018, 35, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.; Badawy, G.M.I.; Wallace, B.M.N. Amphibian Sex Determination and Sex Reversal. Cell. Mol. Life Sci. CMLS 1999, 55, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Charlesworth, D.; Qi, J.; Wu, R.; Chen, M.; Wang, Z.; Xu, L.; Fu, H.; Zhang, X.; Chen, X.; et al. Independent Evolution of Sex Chromosomes and Male Pregnancy–Related Genes in Two Seahorse Species. Mol. Biol. Evol. 2023, 40, msac279. [Google Scholar] [CrossRef] [PubMed]
- Baroiller, J.-F.; D’Cotta, H. The Reversible Sex of Gonochoristic Fish: Insights and Consequences. Sex. Dev. 2016, 10, 242–266. [Google Scholar] [CrossRef]
- Valdivieso, A.; Wilson, C.A.; Amores, A.; da Silva Rodrigues, M.; Nóbrega, R.H.; Ribas, L.; Postlethwait, J.H.; Piferrer, F. Environmentally-Induced Sex Reversal in Fish with Chromosomal vs. Polygenic Sex Determination. Environ. Res. 2022, 213, 113549. [Google Scholar] [CrossRef] [PubMed]
- Yano, K.; Tanaka, S. Hermaphroditism in the Lantern Shark Etmopterus unicolor (Squalidae, Chondrichthyes). Jpn. J. Ichthyol. 1989, 36, 338–345. [Google Scholar] [CrossRef]
- Palmer, D.H.; Rogers, T.F.; Dean, R.; Wright, A.E. How to Identify Sex Chromosomes and Their Turnover. Mol. Ecol. 2019, 28, 4709–4724. [Google Scholar] [CrossRef]
- Gamble, T.; Coryell, J.; Ezaz, T.; Lynch, J.; Scantlebury, D.P.; Zarkower, D. Restriction Site-Associated DNA Sequencing (RAD-Seq) Reveals an Extraordinary Number of Transitions among Gecko Sex-Determining Systems. Mol. Biol. Evol. 2015, 32, 1296–1309. [Google Scholar] [CrossRef]
- Quattro, J.M.; Driggers, W.B.I.; Grady, J.M.; Ulrich, G.F.; Roberts, M.A. Sphyrna gilberti Sp. Nov., a New Hammerhead Shark (Carcharhiniformes, Sphyrnidae) from the Western Atlantic Ocean. Zootaxa 2013, 3702, 159–178. [Google Scholar] [CrossRef]
- Johnson, G.J.; Buckworth, R.C.; Lee, H.; Morgan, J.A.T.; Ovenden, J.R.; McMahon, C.R. A Novel Field Method to Distinguish between Cryptic Carcharhinid Sharks, Australian Blacktip Shark Carcharhinus tilstoni and Common Blacktip Shark C. limbatus, despite the Presence of Hybrids. J. Fish Biol. 2016, 90, 39–60. [Google Scholar] [CrossRef]
- Lesturgie, P.; Denton, J.; Yang, L.; Corrigan, S.; Kneebone, J.; Laso-Jadart, R.; Lynghammar, A.; Fedrigo, O.; Mona, S.; Naylor, G.J.P. Short-term Evolutionary Implications of an Introgressed Size-Determining Supergene in a Vulnerable Population. Nat. Commun. 2024. [Google Scholar]
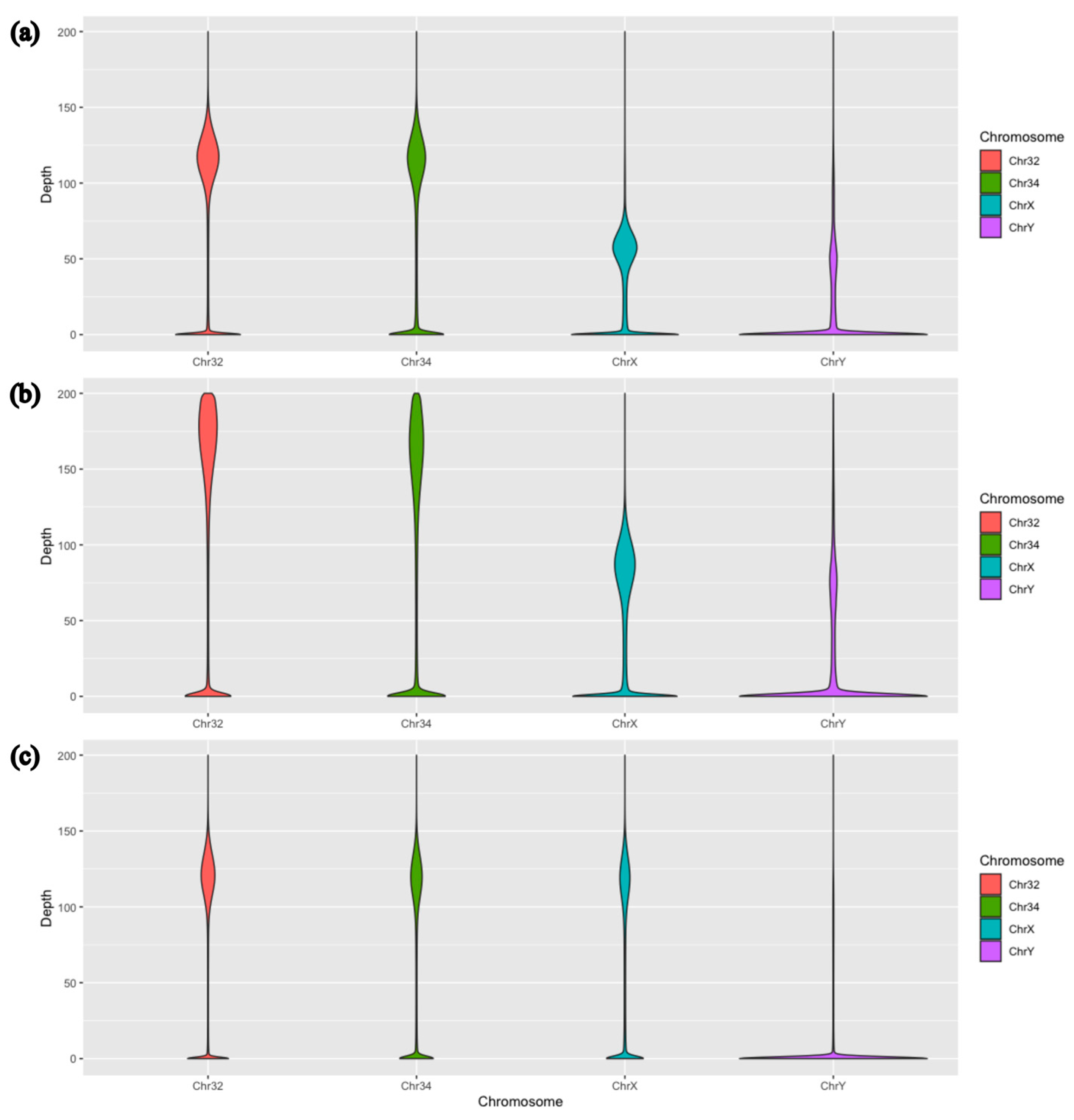
| Average Sequencing Depth | |||||
|---|---|---|---|---|---|
| Chr 1 | Chr 32 | Chr 34 | Chr X | Chr Y | |
| Marra et al. | 163.597 | 138.003 | 118.415 | 58.770 | 34.778 |
| Marra et al. (*) | 127.009 | 107.139 | 91.932 | 45.626 | 27.000 |
| Aggregated males | 107.164 | 93.507 | 83.988 | 41.034 | 23.439 |
| Aggregated females | 111.161 | 96.347 | 86.014 | 80.575 | 10.273 |
| χ2 Test p-Values | ||||
|---|---|---|---|---|
| Male Expected Ratio 2:1 | Female Expected Ratio 1:1 | |||
| Chr 32: Chr X | Chr 34: Chr X | Chr 32: Chr X | Chr 34: Chr X | |
| Marra et al. | 0.302 | 0.963 | * 1.620 × 10–8 | * 7.435 × 10–6 |
| Aggregated males | 0.486 | 0.903 | * 6.072 × 10–6 | * 1.222 × 10–4 |
| Aggregated females | * 5.711 × 10–4 | * 3.853 × 10–5 | 0.236 | 0.674 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-H.; Yang, L.; Naylor, G.J.P. Highlighting the Importance of Correct Sex Identification in Chondrichthyan Genomic Studies, Using the White Shark as an Example. Fishes 2024, 9, 520. https://doi.org/10.3390/fishes9120520
Lee S-H, Yang L, Naylor GJP. Highlighting the Importance of Correct Sex Identification in Chondrichthyan Genomic Studies, Using the White Shark as an Example. Fishes. 2024; 9(12):520. https://doi.org/10.3390/fishes9120520
Chicago/Turabian StyleLee, Szu-Hsuan, Lei Yang, and Gavin J. P. Naylor. 2024. "Highlighting the Importance of Correct Sex Identification in Chondrichthyan Genomic Studies, Using the White Shark as an Example" Fishes 9, no. 12: 520. https://doi.org/10.3390/fishes9120520
APA StyleLee, S.-H., Yang, L., & Naylor, G. J. P. (2024). Highlighting the Importance of Correct Sex Identification in Chondrichthyan Genomic Studies, Using the White Shark as an Example. Fishes, 9(12), 520. https://doi.org/10.3390/fishes9120520






