Abstract
Water quality is crucial for the ecological health of rivers. However, assessing environmental stressors in large river basins has been challenging due to limited biodiversity monitoring tools. Combining environmental DNA and water quality monitoring presents new possibilities for evaluating the impact of dissolved organic matter (DOM) on fish diversity. Case studies from the Jinshui River, Futou Lake, and Gan River in the Jinshui River Basin demonstrated that eDNA biomonitoring reached 84.62% OTU asymptote (176 OTUs) and 91.06% species asymptote (49 species). The Gan River had 1.21 and 1.26 times more fish OTUs than Futou Lake and the Jinshui River, with 20 overlapping species among the areas. We identified typical excitation-emission matrix (EEM) components of DOM and three PARAFAC fluorescent components: C1 (microbial humic-like), C2 (terrestrial humic-like), and C3 (tryptophan-like). Sequence diversity was positively correlated with EC, TDS, pH, NH3-N, DO, CODMn, biological index (BIX), and freshness index (β/α). Taxonomic diversity positively correlated with spectral slope ratio (SR) and C3. Functional diversity positively correlated with SR but negatively correlated with humification index (HIX). The combined eDNA and DOM monitoring approach shows promise for future assessments of fish biodiversity in river basin environments.
Keywords:
fish biodiversity; eDNA metabarcoding; dissolved organic matter (DOM); spatial pattern; river Key Contribution:
Combining eDNA and water quality monitoring in the Jinshui River Basin effectively assessed the impact of dissolved organic matter (DOM) on fish diversity, revealing important correlations and highlighting the approach’s potential for evaluating biodiversity in large river basins.
1. Introduction
Freshwater fish at higher trophic levels serve as key indicators of ecosystem health [1]. They significantly contribute to biomass production, regulate food webs and nutrient cycles, and maintain ecosystem stability [2]. The decline in fish biodiversity in China underscores the need for improved monitoring [3]. Essential Biodiversity Variables (EBVs) assess biodiversity [4]. These include genetic diversity (sequence diversity) [5], covering species diversity (taxonomic diversity) [6], and functional diversity [7]. Environmental DNA (eDNA) metabarcoding offers a reliable, accurate, and effective method for biodiversity monitoring [8], detecting species without direct observation [9].
eDNA offers a more sensitive and efficient method for monitoring aquatic species compared to traditional approaches [10]. Fish eDNA is effective for addressing ecological and environmental issues [11]. eDNA metabarcoding has been employed to assess fish diversity across different ecosystems, including rivers [12], estuaries [13], lakes [14], ponds [15], wetlands [16], coasts [17], and oceans [18]. eDNA monitoring also explores factors affecting freshwater fish diversity, such as sequencing diversity [19], taxonomic diversity [20], and functional diversity [21]. However, human activities [22], pollution [23], and water quality [24] pose serious threats to fish diversity. Fish can accumulate toxins and are sensitive to low pollutant levels [25]. In areas of uneven contamination, some organisms may migrate to avoid exposure [24], which can lead to biodiversity loss and impact ecosystem structure and function [26].
Pollutants in water bodies, such as PAHs, heavy metals, and trace organic contaminants, pose significant threats to the survival of fish. Dissolved organic matter (DOM), a complex and heterogeneous mixture of active organic species (e.g., polysaccharides, proteins, lignin) and diverse functional groups (e.g., aldehyde, amino, carboxyl, ester, hydroxyl, ketone, phenol), is distinguished by its widespread distribution, compositional variability, and high reactivity in natural waters [27]. Due to these characteristics, DOM directly impacts aquatic organisms [28] and interacts with substances like heavy metals [29], polycyclic aromatic hydrocarbons (PAHs) [30], and organic microcontaminants [31]. By influencing the fate and transport of these trace contaminants, DOM plays a critical role in mitigating their harmful effects, particularly the toxicity of metals [32] and PAHs [33], by altering their bioavailability and reducing their impact on fish. Specifically, PAHs induce immunotoxicity in fish [34], making them more susceptible to diseases [35]. However, PAHs bind strongly to organic matter, especially humic substances, which significantly reduces their uptake compared to freely dissolved forms [36]. Similarly, heavy metals such as Cu, Pb, and Cd primarily affect fish gills, disrupting ion transport and balance and often leading to mortality [37,38,39]. Here, DOM provides protective effects by accumulating on fish gills [40], altering the electrical properties of heavy metals, and regulating ion transport [41]. For example, DOM can protect fish from severe Cu toxicity [42] by complexing Cu ions and modifying the transport and permeability properties of fish gills [43]. Additionally, the toxicity of Cu and Pb can vary by up to two-fold depending on the type of DOM present [29]. Further evidence shows that DOM concentrations above 15 ppm carbon significantly reduce cadmium (Cd2+) accumulation in zebrafish (Danio rerio) eggs, underscoring its role in mitigating cadmium’s ecotoxicological impacts [44]. DOM also provides protective effects on fish gills under varying pH conditions. At low pH, DOM from the upper Rio Negro helps fish maintain gill function in ion-poor, acidic environments [45]. Notably, natural DOM may directly interact with gills to provide protection, which can override protective effects typically associated with calcium ions (Ca2+) [46]. At near-neutral pH, DOM from allochthonous sources enhances gill ionoregulation by altering the electrical properties of the gills [41]. Moreover, DOM reduces the bioaccumulation of organic microcontaminants such as benzo(k)fluoranthene [47], phenanthrene [48], and prometryn [49], with higher concentrations of humic acid leading to a significant decrease in their uptake by fish.
The decline in fish biodiversity in the Yangtze River Basin highlights the need for better monitoring [50]. While fish diversity in the Yangtze River Basin has been well studied [50,51], the Jinshui River Basin (Jinshui River, Futou Lake, and Gan River), a tributary, remains underexplored, particularly regarding the effects of DOM on fish community structure. We propose to use eDNA analysis to investigate how DOM properties and sources affect fish biodiversity in the Jinshui River Basin. By performing 12S rDNA high-throughput sequencing and analyzing ecological traits, this study aims to (a) explore the spatial characteristics of DOM; (b) compare eDNA results with traditional fishing methods; (c) assess fish diversity in terms of sequence, taxonomic, and functional aspects; and (d) elucidate the impacts of DOM on various facets of fish diversity. The findings will highlight DOM’s spatial impact on fish biodiversity, aiding river ecological protection efforts, particularly in the Jinshui River Basin.
2. Materials and Methods
2.1. eDNA Sample Collection
On 10–11 June 2023, we sampled ten sites across three sub-catchments of the Jinshui River Basin (29.7186–30.3138° N, 114.1313–114.3854° E). These sites comprised four sites along the Gan River, four around Futou Lake, and two within the Jinshui River (Figure 1 and Table S1). At each site, surface water was collected three times at a depth of 0.5 m using a 1 L water sampler. The sample were combined in a 5 L polyethylene bucket and kept at 4 °C. Each liter of water was filtered with 0.45 μm Mixed Cellulose Ester (MCE) microporous filter membranes (JinTeng, Tianjin, China) in triplicate, with concentrated samples sealed in 5 mL cryopreservation tubes (AXYGEN, Union City, CA, USA) and stored at −80 °C before environmental DNA extraction. A filter blank of 1 L DEPC water was used to control for contamination.
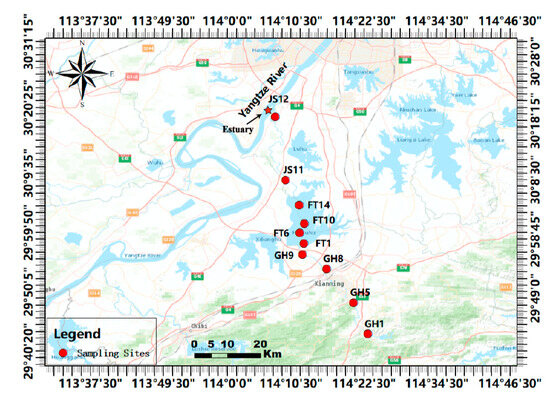
Figure 1.
Sampling sites selected in the Jinshui River Basin, China. The red circles indicate the sampling sites (see Supplementary Table S1 for details information).
2.2. eDNA Extraction, PCR Amplification, and Sequencing
eDNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, North Rhine–Westphalia, Germany) with a modified protocol [52]. Quality was assessed by 1.0% agarose gel electrophoresis and concentration measured with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Samples were stored at −80 °C.
The hypervariable region 12S rDNA gene of the fish mitochondrial was amplified using MiFish-U-F (5′-GTCGGTAAAACTCGTGCCAGC-3′) and MiFish-U-R (5′-CATAGTGGGGTATCTAATCCCAGTTTG-3′) primers [53] on a T100 Thermal Cycler PCR thermocycler (BIO-RAD, Hercules, CA, USA). The PCR mixture included 10 μL of 2 × Phanta Max Master Mix, 0.8 μL of each 5 μM primer, 10 ng of template DNA, and ddH2O to a final volume of 20 µL. PCR conditions were 94 °C for 5 min, 35 cycles of 98 °C for 20 s, 60 °C for 45 s, 72 °C for 30 s, and a final extension at 72 °C for 10 min. Products were extracted from a 2% agarose gel, purified with the PCR Clean-Up Kit (YuHua, Shanghai, China), and quantified with a Qubit 4.0 (Thermo Fisher Scientific, Waltham, MA, USA).
Purified amplicons were pooled and sequenced in paired-end mode on an Illumina MiSeq 2500 (Illumina, San Diego, CA, USA) following Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China) protocols. Raw sequencing data were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA1132588).
2.3. Data Processing
The sequencing depth of 12S rRNA sequencing was determined by generating an average of 75,686 raw sequences per sample, a total of 227,579 raw sequences. The raw FASTQ files were first de-multiplexed using an in-house perl script, then quality-filtered with fastp version 0.19.6 [54] and merged using FLASH version 1.2.7 [55]. The quality filtering followed these criteria: (i) Reads were truncated at any site with an average quality score < 20 over a 50 bp sliding window, and shorter than 50 bp or containing ambiguous characters were discarded. (ii) Only overlapping sequences longer than 10 bp were assembled with a maximum mismatch ratio of 0.2 in the overlap region; unassembled reads were discarded. (iii) Samples were distinguished based on barcodes and primers, with exact barcode matching and up to 2 nucleotide mismatches allowed in primer matching. The optimized sequences were then clustered into operational taxonomic units (OTUs) using UPARSE 7.1 [56,57] with a 97% sequence similarity threshold. The most abundant sequence for each OTU was selected as a representative sequence. To account for sequencing depth effects on alpha and beta diversity measures, the 12S rRNA gene sequences from each sample were rarefied to 3009, achieving an average Good’s coverage of 99.09%.
2.4. Rarefaction Curve
In the Jinshui River Basin, rarefaction curves were created for fish species and OTUs identified through eDNA analysis using the R package iNEXT. These curves revealed differences between observed and extrapolated species and OTU rarity.
Historical fish catch data from May and November 2020 were collected using electric fishing and ground cage methods (details in Text S1). Taxonomic details were confirmed through fishbase.org. A Pearson correlation analysis compared fish species counts from traditional fishing and eDNA data. Fish status information in China was obtained from fishbase.org using the R package rfishbase.
2.5. Fish Sequence, Taxonomic, and Functional Diversity
To compare sequence diversity across areas, a paired Kruskal–Wallis test was used to assess differences based on average sequence variants within species [58]. Fish richness, calculated using the Chao index, represented taxonomic diversity. Functional richness (FRic) was determined by integrating traits such as migration patterns, trophic levels, and feeding types with the mFD package [52]. FRic was assessed using convex hull volumes in multidimensional trait space (details in Text S1). GLM regression and Pearson analysis examined the correlation between sequence, taxonomic, and functional diversity in each area.
2.6. Differences in Diversity Facets Across Areas
Principal coordinate analysis (PCoA) was used to investigate differences in fish sequence, taxonomic, and functional diversity in the Jinshui River Basin. Sequence and functional diversity were analyzed with Bray–Curtis dissimilarity, while taxonomic diversity was assessed using weighted UniFrac dissimilarity. PCoA for taxonomic diversity was conducted at the species level. ANOSIM analysis was used to evaluate differences between areas.
LOESS analysis (locally weighted scatterplot smoothing) examined correlations between diversity facets (sequence, taxonomic, functional) and distances from the Yangtze River. Distances to the Yangtze River Estuary (E114.127°, N30.331°) were calculated using the R package geosphere.
2.7. Spectroscopic Analysis
The water samples were collected simultaneously with the eDNA water samples, with three replicates taken at each site. Each replicate consisted of 500 mL of water, which was filtered within 24 h using 0.45 μm MCE filters. Ultraviolet–visible absorbance and excitation–emission matrices (EEMs) fluorescence spectroscopy were performed within a week of sample collection to analyze DOM. Absorbance was measured from 200 to 700 nm using a SPECORD 200 PLUS ultraviolet–visible spectrophotometer (Analytik Jena, Jena, Thuringia, Germany) with Milli-Q water as the reference. EEM fluorescence was analyzed with an F-7000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) using a 700 V xenon lamp. The value of specific ultraviolet absorbance at 254 nm (SUVA254) and spectral slope ratio (SR) was calculated from ultraviolet–visible absorption spectroscopic analysis. The biological index (BIX), fluorescence index (FI), humification index (HIX), and freshness index (β/α) were calculated from the EEMs of the dissolved organic matter (DOM) samples. See details in Text S1.
PARAFAC modeling in R with the staRdom package decomposed the EEM data into individual components. Component fluorescence intensity was based on maximum fluorescence (Fmax), and the number of components was determined by split-half validation. Identified components were compared with published data on the OpenFluor website: http://www.openfluor.org (accessed on 28 May 2024), using a Tucker congruence coefficient above 0.99 for similarity. See Text S1 for more details.
2.8. Evaluation the Impact of DOM and Physicochemical Parameters on Fish Diversity
The water samples were collected simultaneously with the eDNA water samples, with three replicates taken at each site. Each replicate consisted of 1 L of water. A comprehensive analysis was conducted, measuring a total of 14 physicochemical parameters (Table S3) at each sampling site, which included water factors measured using an HQ30d portable meter and nutrients analyzed through chemical methods, see details in Text S1.
To identify significant environmental factors associated with various fish diversity facets within the Jinshui River Basin, Mantel’s test was employed. For the analysis of fish structure at order level between sites, the R package vegan was utilized, employing redundancy analysis (RDA). Spearman’s correlation analysis was used to assess the relationships between environmental factors and fish species, with significance defined as p < 0.05 and |r| > 0.5. The network plot visualizations were generated using Gephi software (version 0.10.1, WebAtlas, Paris, France).
Statistical analyses were performed in R (version 4.4.0), and figures were created using the R packages ggplot2, ggpubr, and igraph.
3. Results
3.1. eDNA Annotation and Rarefaction Curve
In the Jinshui River Basin, comprising 10 sites, eDNA results reached 84.62% of OTUs asymptote (176 ASVs) and 91.06% of species asymptote (49 sp.) (Figure 2). In total, 9 orders, 21 families, 36 genera, and 45 fish species were detected in the Jinshui River Basin, with 8.89% being endemic, 15.56% introduced, and 75.55% native to China (Figure 3D and Table S2). Fish OTUs in the Gan River were 1.21 and 1.26 times richer than those in the Futou Lake and Jinshui River, respectively (Figure 3A). The highest number of fish species was found in the Gan River (40 species), followed by Futou Lake (31 species) and the Jinshui River (24 species), with 20 species overlapping among the three areas (Figure 3C).
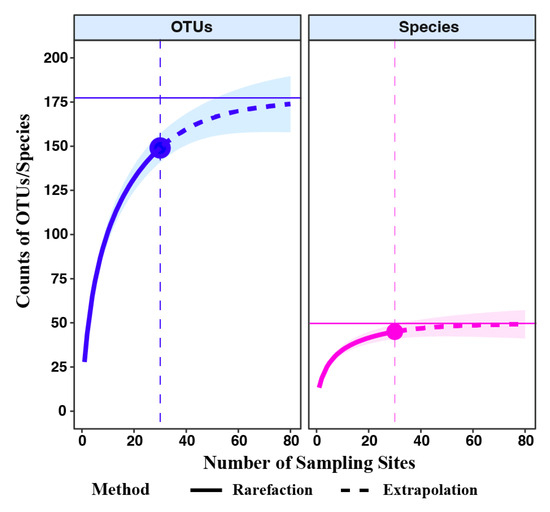
Figure 2.
Cumulative curve for fish OTUs and species based on observed numbers in the Jinshui River Basin.
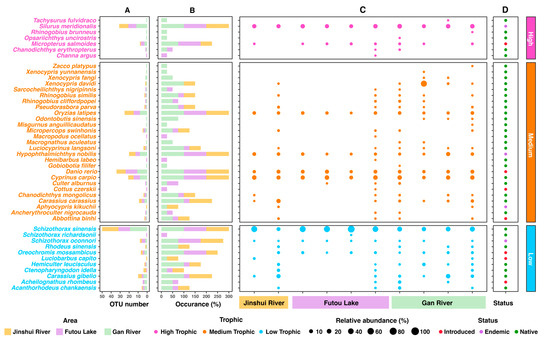
Figure 3.
Monitoring fish species in the Jinshui Basin using environmental DNA: the number of OTUs (A), occurrence (B), relative abundance (C), status in China (D).
3.2. Fish Trophic Level, Occurrence, and Dominant Species Based on eDNA Analysis
Fish trophic levels were categorized into three levels: high level (15.55%), medium level (60%), and low level (24.45%) of the total 45 fish species across 10 sites in the Jinshui River Basin (Figure 3). Their trophic levels and relationships within the food web were explored in the Jinshui River Basin (Figure S2B), revealing that detection signals of high-trophic fish increased significantly with greater distance between sampling sites and the Yangtze River Estuary (Figure S2C). Detection signals for high-trophic fish in the Gan River were higher, with their average occurrence being 1.46 times and 1.64 times greater than in the Jinshui River and Futou Lake, respectively (Figure 3B).
Based on eDNA analysis, the dominant fish species can be clearly distinguished across the three areas (Figure 3). For instance, species such as Silurus meridionalis (Siluriformes order, high-trophic), Oryzias latipes (Beloniformes order, medium-trophic), Danio rerio (Cypriniformes order, medium-trophic), Hypophthalmichthys nobilis (Cypriniformes order, medium-trophic), Cyprinus carpio (Cypriniformes order, medium-trophic), and Schizothorax sinensis (Cypriniformes order, low-trophic) were present in all three areas with a 100% occurrence (Figure 3B) and high relative abundance (Figure 3C), indicating their dominance. However, species Channa argus (Anabantiformes order, high-trophic), Macropodus ocellatus (Anabantiformes order, medium-trophic), and Schizothorax richardsonii (Cypriniformes order, low-trophic) were only detected in the Futou Lake, each with a 25% occurrence (Figure 3B). In contrast, species Opsariichthys uncirostris (Cypriniformes order, high-trophic) and Odontobutis sinensis (Gobiiformes order, medium-trophic) were only detected in the Gan River, with occurrence of 25% and 75% (Figure 3B), respectively, illustrating that these species mainly inhabit this area. The evolutionary tree of fish can also be further explored based on the eDNA results obtained from the Jinshui River Basin (Figure S2A).
3.3. Diversity Facets: Variations Across Areas
Using eDNA analysis, distinct variations were revealed in the sequence, taxonomic, and functional diversity of fish in the Jinshui River Basin. The sequence diversity was significantly higher in Futou Lake compared to the Jinshui and Gan Rivers, whereas the Gan River exhibited significantly higher taxonomic and functional diversity than Futou Lake (Figure 4A, p < 0.05). The PCo1 axis effectively distinguished the sequencing diversity, taxonomic community structures, and functional structures among the Jinshui River, Futou Lake, and Gan River (ANOSIM, sequencing: Figure S1A, R = 0.4071, p = 0.001; taxonomic: Figure S1B, R = 0.2997, p = 0.001; functional: Figure S1C, R = 0.2354, p = 0.007).
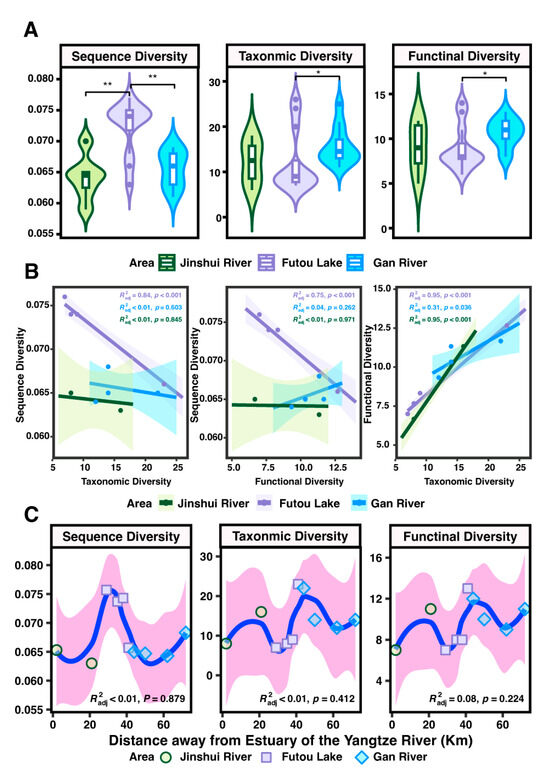
Figure 4.
Fish sequence, taxonomic, and functional diversity in the Jinshui River Basin. (A) Assessing diversity facets variation across river areas using the Wilcoxon Test. (B) Correlations between three diversity facets. (C) Trend of three facets of diversity with increasing distance from sampling sites to the Yangtze River Estuary using LOSSEN analysis with locally weighted scatterplot smoothing. “*” represents the degree of significance: “*” represents p < 0.05; “**” represents p < 0.01.
In Futou Lake, a negative correlation was observed between sequence diversity and taxonomic diversity (Pearson R = 0.84, p < 0.001), with similar trends observed between sequence and functional diversity (Figure 4B, R = −0.75, p < 0.001). In contrast, taxonomic and functional diversity showed a positive relationship in the Jinshui River (R = 0.95, p < 0.001), Futou Lake (R = 0.95, p < 0.001), and Gan River (R = 0.31, p =0.036) (Figure 4B).
Fish diversity (sequencing, taxonomic, and functional) varied with the distance from the Yangtze River Estuary in the three areas of the Jinshui River Basin (Figure 4C). With the increasing distance from Yangtze River Estuary, the sequence diversity increased from Jinshui River and reached the peak in Futou Lake, then sharply decreased in Gan River (R < 0.01, p = 0.897). Taxonomic diversity increased in the Jinshui River, reached their lowest point in Futou Lake, peaked at the confluence of the Jinshui River into Futou Lake, and then decreased again in the Gan River (R < 0.01, p = 0.412), and functional diversity exhibited similar trends (R = 0.08, p = 0.224).
3.4. The Composition, Fluorescence Characteristics, and PARAFAC Components of Dissolved Organic Matter (DOM)
Ultraviolet–visible absorbance spectroscopy was used to analyze the composition and variability characteristics of DOM (Figure 5 and Table S4). The highest average SUVA254 values were observed downstream of the Gan River, followed by the confluence of the Gan River and Futou Lake, indicating a higher abundance of aromatic substances in these areas compared to Futou Lake and Jinshui River. The mean SR value was also highest downstream of the Gan River, suggesting an enrichment of low-molecular-weight compounds, followed by the Jinshui River and Futou Lake.
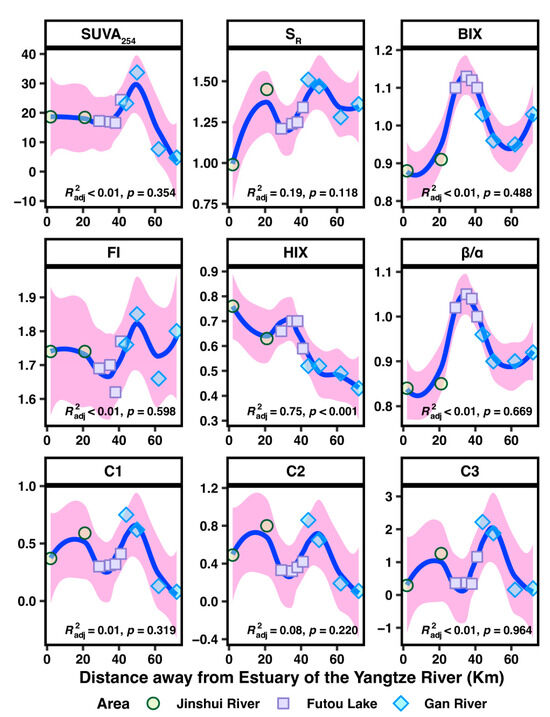
Figure 5.
Correlations between composition, fluorescence characteristics, and EEM-PARAFAC components of dissolved organic matter (DOM) and the Yangtze River Estuary distances in the Jinshui River Basin: A LOESS analysis (locally weighted scatterplot smoothing).
To explore the sources contributing to DOM, fluorescence characteristics such as the BIX, β/α, FI, and HIX were calculated (Figure 5 and Table S4). The highest average values BIX and β/α were found in Futou Lake, followed by the Gan River and Jinshui River. This suggests that Futou Lake had recently produced, labile DOM, while the Jinshui River exhibited older, more recalcitrant, and less autogenic DOM. The range of FI values indicated that DOM in these areas derives from both microbial and terrestrial sources, with DOM in the Gan River primarily from microorganisms and in Futou Lake mainly from terrestrial sources. The HIX value was highest in the Jinshui River and lowest in the Gan River, indicating that humification and molecular complexity decreased with increasing distance from the Yangtze River Estuary.
In the Jinshui River Basin, typical EEM components of DOM were identified, and three PARAFAC fluorescent components were established through split-half validation (Figures S6 and S7). The values of Component 1, Component 2, and Component 3 exhibited similar trends with increasing distance from the Yangtze River Estuary (Figure 5 and Table S4). These include two humic-like (Component 1 and Component 2) and one protein-like (Component 3) (Figure S4, Tables S4 and S5). Component 1 (C1), with its peak excitation at 260/290 nm and emission at 390 nm, closely resembles Peak M (ex300/em390), which is identified as a microbial humic-like component [59]. Component 2 (C2), characterized by Ex/Em wavelengths of 260/445 nm, is classified as a blend of traditional humic-like peaks, specifically peak A (ex260/em450), indicative of a terrestrial humic-like component representative of terrestrial environments [60]. Component 3 (C3) had excitation and emission peaks at 275 nm and 330 nm, respectively, typical of protein-like (tryptophan-like) compounds [61]. The C1/C3 ratio was positively correlated with C2/C3 (Figure S8, Pearson R = 0.97, p < 0.001). C3 reflects the characteristics of DOM in downstream of Jinshui River; C1, C2, and C3 reflect the characteristics of DOM in Futou Lake; and C1 and C2 reflect the characteristics of DOM in Gan River.
3.5. Impact Evaluation of DOM
In the Jinshui River Basin, fish sequence diversity was most strongly associated with physicochemical parameters and DOM (Figure 6). The physicochemical parameters significantly related to sequence diversity were pH (p < 0.01), total dissolved solids (TDS, p < 0.01), electrical conductivity (EC, p < 0.01), ammonia nitrogen (NH3-N, p < 0.01), permanganate index (CODMn, p < 0.05), and dissolved oxygen (DO, p < 0.05), as well as fluorescence parameters, including β/α (p < 0.01) and BIX (p < 0.05). Taxonomic diversity had a strong correlation with SR (p < 0.01), followed by C3 (p < 0.05) and pH (p < 0.05). Functional diversity was least influenced by environmental factors and strongly correlated only with SR (p < 0.01) and HIX (p < 0.05).
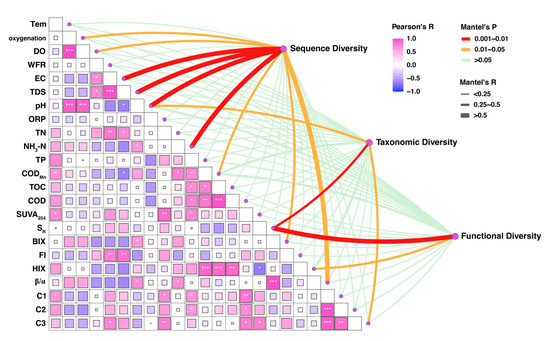
Figure 6.
Impact of dissolved organic matter (DOM) optical properties, PARAFAC components, and physicochemical parameters in the Jinshui River Basin on fish sequence, taxonomic, and functional diversity. “*” represents the degree of significance: “*” represents p < 0.05; “**” represents p < 0.01; “***” represents p < 0.001.
The correlation between physicochemical parameters and DOM with fish relative abundance at both the order and species levels was analyzed using redundancy analysis (RDA) (Figure S9) and co-occurrence network analysis (Figure S10), respectively. The Centrarchiformes order showed a positive relationship with the BIX, the β/α ratio, and DO. The Gobiiformes and Cichliformes order were both positively correlated with the SR, C3, and fluorescence index (FI). Ammonia nitrogen (NH3-N) and total organic carbon (TOC) showed negatively correlation with fish distribution. The Cypriniformes order showed positive correlations with the HIX, SUVA254, TOC, total nitrogen (TN), and NH3-N (Figure S9). The relative abundance of Carassius carassius was positively correlated with pH, DO, oxygenation, and CODMn. Similarly, the relative abundance of Carassius gibelio showed positive correlations with these parameters, as well as with NH3-N. The number of fish species associated with SR was the largest, followed by C3, C1, and HIX (Figure S10).
4. Discussion
4.1. Impact Evaluation of Physicochemical Parameters and DOM on Fish Diversity Facets
Significant environmental differences might could explain the distinct sequence variants, species composition, and functional diversity recovered by eDNA in the Jinshui River Basin. In this study, we found more correlations between environmental factors and sequence diversity compared to the other two diversity facets (Figure 6). This observation aligns with a previous study indicating that fish sequence diversity in the Yalujiang Estuary (YLJK) in the Yellow Sea of China was more strongly associated with environmental factors than taxonomic and functional diversity facets [52]. In Futou lake, the chlorophyll-a concentrations showed the similar trends with DO and pH (Figure S3), a pattern supported by a previous study that found a positive correlation between chlorophyll-a levels, DO, and pH [62]. NH3-N, as the primary excretory product of fish [63], is toxic to fish by inducing homeostasis disruption and toxicity through shared mechanisms [64]. Regions with evaluated NH3-N and CODMn level can promote phytoplankton growth, leading to increased chlorophyll-a levels; as phytoplankton photosynthesize, they release oxygen, thereby raising DO and pH levels [65]. However, abundant nutrients can also lead to water eutrophication and oxygen depletion [66]. Oxygen is a key limiting factor for many fish species, with low levels potentially diminishing effective population sizes and thereby reducing genetic diversity [67]. Higher DO levels support a greater variety of fish species [68], reduce physiological stress [69], and promote the growth of plants and algae that sustain robust food webs [70], all while indicating good water quality and maintaining habitat integrity [71]. A previous study found that the β/α ratio and BIX were highly correlated [72], and a similar correlation was observed across the Jinshui River Basin in this study (Figure S5C). In Futou Lake, the highest BIX and β/α values were observed compared to other areas (Figure 5), indicating high levels of autochthonous (microbial-derived) and recently produced DOM [72]. Healthy microbial activity, as indicated by high BIX, supports primary producers [73], which in turn support higher trophic levels, including fish [74]. Bacterioplankton that utilize DOM provide extra energy to zooplankton, which are then consumed by planktivorous fish [75].
Notably, the similar trends in taxonomic and functional diversity with increasing distance from the Yangtze River Estuary (Figure 4C) align with the significant positive correlations observed between these facets (Figure 4B) as discovered in earlier research [52]. These results indicate that eDNA is a powerful tool for mapping the spatial distribution of fish diversity in rivers and reflecting the relationship between different diversity facets [52,76].
High SR values represent a higher presence of low molecular weight, diminished aromaticity and photobleaching [77], higher algal contribution, and enhanced biological degradation [72]. Previous studies have recorded SR values in various aquatic environments, with an average value of 0.99–1.12 in Yongding River Basin [78] and 0.70–2.40 in lake waters [79]. In this study, initial SR values varied from 0.99 to 1.51, suggesting that DOM was primarily derived from autochthonous origins. In the downstream of the Gan River to the entrance of Futou Lake, SR values, along with taxonomic and functional diversity, were the highest (Figure 5), suggesting that taxonomy and functional diversity of fish were positively correlated with algal contribution and biological degradation.
Furthermore, the highest C3 was found downstream of Gan River, which might be caused by high temperature and may have accelerated phytoplankton growth and led to an increase in protein-like substances [80]; then, the phytoplankton could be consumed by grazing fish directly or indirectly (e.g., phytoplankton–zooplankton–fish model) [81]. A higher degree of DOM humification indicates that fewer nutrients in the water are available for cyanobacteria absorption and utilization, which can promote the shift from an algae-dominated system to one dominated by macrophytes [82]. These results indicating that functional diversity is significantly influenced by the increasing humification and molecular complexity of DOM. Humic substances (HSs) can impact fish by lowering ammonia and nitrite toxicity in zebrafish (Danio rerio) embryos [83], reducing iron toxicity in brown trout (Salmo trutta) [84], and decreasing the uptake of mercury, cadmium, and zinc in chinook salmon (Oncorhynchus tshawytscha) eggs [85]. Another study found that HS-like and tryptophan-like components can accumulate in the blood of tilapia [86]. These findings may explain why we observed that humic-like and tryptophan-like substances positively affected fish taxonomic and functional diversity.
4.2. Factors Determining River Basin Fish Community Structures
At the order level, RDA analysis revealed that DO was the main factor driving the fish community structure, followed by temperature (Figure S9A). Oxygen levels strongly influenced the spatial distribution of fish species [67]. DO and temperature significantly affected both fish biomass and species richness [87]. The concentration of NH3-N was negatively correlated with fish distribution. This finding is in agreement with a prior study that revealed a negative correlation between NH3-N and CODMn levels and fish distribution in the middle and downstream regions of the Yangtze River Basin, indicating considerable water pollution in these zones [51]. SR showed a positive correlation with the relative abundance of Gobiiformes (Figure S9B), indicating that Gobiiformes might be mainly influenced by the aromaticity of DOM. The relative abundance of Centrarchiformes was positively associated with BIX and β/α values (Figure S9B), suggesting that Centrarchiformes primarily inhabited waters characterized by autochthonous (microbial-derived) and recently produced DOM. Additionally, higher HIX levels, indicating strong humification and fewer nutrients for cyanobacteria, promote the transition to a macrophyte-dominated water body [82]. The positive correlation between HIX and Cypriniformes (Figure S9B) might be attributed to the dominance of Cyprinus carpio in the Jinshui River Basin (Figure 3). As omnivores, these fish usually forage in shallow waters where water hyacinth and rooted macrophytes are abundant [88].
The variation in fish communities was also assessed at the species level. We observed that pH had a positive correlation with five fish species, whereas CODMn showed a positive correlation with four fish species and a negative correlation with one species (Figure S10A). A previous study revealed that pH had a significant impact on the fish community [89]. Another study found that DOM derived from planktonic algae leads to increased CODMn levels along the middle route of the South-to-North Water Diversion Project (SNWD) in North China [90]. This is consistent with our results, where biological activity and the presence of recently produced autochthonous DOM in Futou Lake were associated with the highest CODMn levels. Among the DOM optical properties and PARAFAC components, SR had the strongest correlation with fish species, being associated with eight species (positively: seven species; negatively: one species). Similarly, microbial humic-like substances (C1) were positively correlated with eight species, while protein-like (tryptophan-like) substances (C3) showed a positive correlation with seven species (Figure S10B). These results indicate that these fish primarily inhabit waters characterized by a greater algal contribution, less humified organic matter, and more biodegradable organic matter (C3: tryptophan-like substances).
5. Conclusions
Using eDNA metabarcoding analysis, our study demonstrated how DOM sources impact fish biodiversity in the Jinshui River Basin. We detected 45 species through eDNA method. eDNA results from 10 sites reached 84.62% OTUs asymptote (176 ASVs) and 91.06% species asymptote (49 species). The Gan River had 1.21 and 1.26 times more fish OTUs than Futou Lake and the Jinshui River, respectively. It also had the highest number of fish species (40), followed by Futou Lake (31) and the Jinshui River (24), with 20 species overlapping among the areas. Sequence diversity was highest in Futou Lake, while taxonomic and functional diversity were highest in the Gan River. There was a positive relationship between taxonomic and functional diversity in the Jinshui River Basin. We identified typical EEM components of DOM and three PARAFAC fluorescent components: C1 (microbial humic-like), C2 (terrestrial humic-like), and C3 (protein-like, tryptophan-like). Sequence diversity in Futou Lake was notably high and positively correlated with various water quality parameters (dissolved oxygen, electrical conductivity, total dissolved solids, pH, and ammonia nitrogen), as well as β/α ratio and BIX, which represent autochthonous (microbial-derived) and recently produced DOM. Both taxonomic and functional diversity exhibited similar patterns as the distance from the Yangtze River Estuary increased, indicating a positive relationship with SR, which reflects algal contributions and biodegradable DOM. Taxonomic diversity was positively linked with high C3 (represents DOM bioavailability), whereas functional diversity negatively correlated with HIX (represents humification). Humic-like substance (C1 and C2) also positively affects fish taxonomic and functional diversity. The combined eDNA and DOM monitoring approach holds promise for future assessments of fish biodiversity in river basin environments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes9120489/s1, Text S1: Supplementary Methods and Results; Table S1: Information about sampling sites in the Jinshui River Basin; Table S2: Information on 45 species fish in the Jinshui River Basin; Table S3: Physicochemcal parameters in the Jinshui River Basin; Table S4: Optical properties in the Jinshui River Basin; Table S5: Analysis of PARAFAC component descriptions and wavelength positions, alongside comparisons with analogous components identified in the OpenFluor database; Table S6: Topological properties of co-occurring networks; Figure S1: Principal co-ordinates analysis (PCoA) on fish sequence diversity (A), taxonomic composition of communities (B), and functional composition (C) in the Jinshui River Basin. Sequence diversity and functional structure were analyzed using Bray–Curtis dissimilarity, while taxonomic diversity was assessed using weighted UniFrac dissimilarity. Differences between areas were evaluated through ANOSIM analysis to identify variations; Figure S2: Fish evolutionary tree (A) and food web relationships in Jinshui River, Futou Lake, and Gan River (C) within Jinshui River Basin (B) based on eDNA data in the Jinshui River Basin; Figure S3: Correlations between physicochemical parameters and the Yangtze River Estuary distances in the Jinshui Basin: A LOESS Analysis (Locally weighted scatterplot smoothing). Tem represents Temperature (°C); Oxygenation represents dissolved oxygen percentage; DO represents dissolved oxygen (mg/L); WFR represents water flow rate (m/s); EC represents electrical conductivity (μS/cm); TDS represents total dissolved solids (mg/L); ORP represents oxidation-reduction potential (mV); NH3-N represents ammonia nitrogen (mg/L); TP represents represents total phosphorus (mg/L); IMn represents permanganate index (mg/L); TOC represents total organic carbon (mg/L); TOD represents total oxygen demand (mg/L); Chl-a represents chlorophyl-a; Figure S4: Contour plots of dissolved organic matter (DOM) in the Jinshui River Basin using excitation–emission matrix fluorescence spectroscopy (EEM) combined with parallel factor analysis (PARAFAC); Figure S5: Biplot of (A) biological index (BIX)–fluorescence index (FI), (B) humification index (HIX)–biological index (BIX), (C) freshness index (β/α)–biological index (BIX), (D) humification index (HIX)–fluorescence index (FI), (E) freshness index (β/α)–fluorescence index (FI), and (F) freshness index (β/α)–humification index (HIX). The likelihood that each point falls within the highest density region, as defined by the specified probs; Figure S6: Comparisons of excitation and emission spectra, split into halves, based on the three-component PARAFAC model; Figure S7: Spectral comparisons: 3-Component PARAFAC Model versus OpenFluor Database. The similarity score was 0.99 for both the excitation and emission of Component 1 and Component 3, and it was 0.98 for the excitation and 0.99 for the emission of Component 2; Figure S8: Discriminating sampling sites using C1/C3 ratios versus C2/C3 ratios; Figure S9: Redundancy analysis (RDA) of fish species at order level with physicochemical parameters (A), DOM optical properties and PARAFAC components (B) in Jinshui Basin, respectively. Fish species at order level are indicated by the pink arrows; physicochemical parameters, optical properties, and PARAFAC components are indicated by blue arrows; Figure S10: Network analysis of fish species with physicochemical parameters (A) and DOM optical properties and PARAFAC components (B). The criteria were p < 0.05 and r >|0.5|; red and green lines represent the positive and negative correlations between fish species and environmental factors, respectively; the thickness of the edges is proportional to the strength of the correlation; the color of nodes represents the fish species at order level. ([91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110] are cited in the Supplementary Materials)
Author Contributions
S.C.: Data curation, Formal analysis, Visualization, Writing—original draft, Writing—review and editing. J.Z.: Formal analysis. H.X.: Formal analysis. Q.Y.: Writing—review and editing. J.L.: Formal analysis. L.Z.: Resources, Funding acquisition. N.L.: Writing—review and editing. Y.W.: Supervision, Methodology, Writing—review and editing. F.M.: Supervision, Funding acquisition, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2021YFC3200100) and Joint Research Project II on Ecological Environment Protection and Restoration of the Yangtze River (2022-LHYJ-02-0506-09).
Institutional Review Board Statement
Since this study used a non-invasive method, eDNA metabarcoding, to obtain fish diversity information through water sample collection rather than direct fish capture, it does not involve ethical concerns.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Whitfield, A.K.; Elliott, M. Fishes as Indicators of Environmental and Ecological Changes within Estuaries: A Review of Progress and Some Suggestions for the Future. J. Fish Biol. 2002, 61, 229–250. [Google Scholar] [CrossRef]
- Villéger, S.; Brosse, S.; Mouchet, M.; Mouillot, D.; Vanni, M.J. Functional Ecology of Fish: Current Approaches and Future Challenges. Aquat. Sci. 2017, 79, 783–801. [Google Scholar] [CrossRef]
- Xiang, T.; Dong, X.; Ju, T.; Shi, L.; Grenouillet, G. Anthropogenic Activities and Environmental Filtering Have Reshaped Freshwater Fish Biodiversity Patterns in China over the Past 120 Years. J. Environ. Manag. 2023, 344, 118374. [Google Scholar] [CrossRef]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential Biodiversity Variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef]
- Paz-Vinas, I.; Loot, G.; Stevens, V.M.; Blanchet, S. Evolutionary Processes Driving Spatial Patterns of Intraspecific Genetic Diversity in River Ecosystems. Mol. Ecol. 2015, 24, 4586–4604. [Google Scholar] [CrossRef]
- Iknayan, K.J.; Tingley, M.W.; Furnas, B.J.; Beissinger, S.R. Detecting Diversity: Emerging Methods to Estimate Species Diversity. Trends Ecol. Evol. 2014, 29, 97–106. [Google Scholar] [CrossRef]
- Jarzyna, M.A.; Jetz, W. Detecting the Multiple Facets of Biodiversity. Trends Ecol. Evol. 2016, 31, 527–538. [Google Scholar] [CrossRef]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A.; et al. Reliable, Verifiable and Efficient Monitoring of Biodiversity via Metabarcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Yu, D.W.; De Bruyn, M. Environmental DNA for Wildlife Biology and Biodiversity Monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis—A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of Fish eDNA and Its Applications in Ecology and Environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef] [PubMed]
- Sales, N.G.; Wangensteen, O.S.; Carvalho, D.C.; Deiner, K.; Præbel, K.; Coscia, I.; McDevitt, A.D.; Mariani, S. Space-Time Dynamics in Monitoring Neotropical Fish Communities Using eDNA Metabarcoding. Sci. Total Environ. 2021, 754, 142096. [Google Scholar] [CrossRef]
- Van Driessche, C.; Everts, T.; Neyrinck, S.; Halfmaerten, D.; Verschelde, P.; Breine, J.; Bonte, D.; Brys, R. Environmental DNA Metabarcoding Reflects Spatiotemporal Fish Community Shifts in the Scheldt Estuary. Sci. Total Environ. 2024, 934, 173242. [Google Scholar] [CrossRef]
- Hänfling, B.; Lawson Handley, L.; Read, D.S.; Hahn, C.; Li, J.; Nichols, P.; Blackman, R.C.; Oliver, A.; Winfield, I.J. Environmental DNA Metabarcoding of Lake Fish Communities Reflects Long-term Data from Established Survey Methods. Mol. Ecol. 2016, 25, 3101–3119. [Google Scholar] [CrossRef]
- Harper, L.R.; Buxton, A.S.; Rees, H.C.; Bruce, K.; Brys, R.; Halfmaerten, D.; Read, D.S.; Watson, H.V.; Sayer, C.D.; Jones, E.P.; et al. Prospects and Challenges of Environmental DNA (eDNA) Monitoring in Freshwater Ponds. Hydrobiologia 2019, 826, 25–41. [Google Scholar] [CrossRef]
- Zou, K.; Chen, J.; Ruan, H.; Li, Z.; Guo, W.; Li, M.; Liu, L. eDNA Metabarcoding as a Promising Conservation Tool for Monitoring Fish Diversity in a Coastal Wetland of the Pearl River Estuary Compared to Bottom Trawling. Sci. Total Environ. 2020, 702, 134704. [Google Scholar] [CrossRef]
- Rey, A.; Viard, F.; Lizé, A.; Corre, E.; Valentini, A.; Thiriet, P. Coastal Rocky Reef Fish Monitoring in the Context of the Marine Strategy Framework Directive: Environmental DNA Metabarcoding Complements Underwater Visual Census. Ocean. Coast. Manag. 2023, 241, 106625. [Google Scholar] [CrossRef]
- Fraija-Fernández, N.; Bouquieaux, M.-C.; Rey, A.; Mendibil, I.; Cotano, U.; Irigoien, X.; Santos, M.; Rodríguez-Ezpeleta, N. Marine Water Environmental DNA Metabarcoding Provides a Comprehensive Fish Diversity Assessment and Reveals Spatial Patterns in a Large Oceanic Area. Ecol. Evol. 2020, 10, 7560–7584. [Google Scholar] [CrossRef]
- Tsuji, S.; Maruyama, A.; Miya, M.; Ushio, M.; Sato, H.; Minamoto, T.; Yamanaka, H. Environmental DNA Analysis Shows High Potential as a Tool for Estimating Intraspecific Genetic Diversity in a Wild Fish Population. Mol. Ecol. Resour. 2020, 20, 1248–1258. [Google Scholar] [CrossRef]
- Zhang, S.; Lu, Q.; Wang, Y.; Wang, X.; Zhao, J.; Yao, M. Assessment of Fish Communities Using Environmental DNA: Effect of Spatial Sampling Design in Lentic Systems of Different Sizes. Mol. Ecol. Resour. 2020, 20, 242–255. [Google Scholar] [CrossRef]
- Marques, V.; Castagné, P.; Polanco, A.; Borrero-Pérez, G.H.; Hocdé, R.; Guérin, P.; Juhel, J.; Velez, L.; Loiseau, N.; Letessier, T.B.; et al. Use of Environmental DNA in Assessment of Fish Functional and Phylogenetic Diversity. Conserv. Biol. 2021, 35, 1944–1956. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Logez, M.; Xu, J.; Tao, S.; Villéger, S.; Brosse, S. Human Impacts on Global Freshwater Fish Biodiversity. Science 2021, 371, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, X.; Zhao, G.; Zuo, C.; Alofs, K.; Wang, R. Pathways Linking Watershed Development and Riparian Quality to Stream Water Quality and Fish Communities: Insights from 233 Subbasins of the Great Lakes Region. Water Res. 2024, 261, 121964. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, R.S.; Silva, G.C.; Bazzan, T.; De La Torre, F.; Nebo, C.; Siqueira-Silva, D.H.; Cardoso-Silva, S.; Pompêo, M.L.M.; De Paiva, T.C.B.; Da Silva, F.T.; et al. Connections among Land Use, Water Quality, Biodiversity of Aquatic Invertebrates, and Fish Behavior in Amazon Rivers. Toxics 2022, 10, 182. [Google Scholar] [CrossRef]
- Topić Popović, N.; Strunjak-Perović, I.; Barišić, J.; Kepec, S.; Jadan, M.; Beer-Ljubić, B.; Matijatko, V.; Palić, D.; Klobučar, G.; Babić, S.; et al. Native Prussian Carp (Carassius gibelio) Health Status, Biochemical and Histological Responses to Treated Wastewaters. Environ. Pollut. 2016, 218, 689–701. [Google Scholar] [CrossRef]
- Araújo, C.V.M.; Laissaoui, A.; Silva, D.C.V.R.; Ramos-Rodríguez, E.; González-Ortegón, E.; Espíndola, E.L.G.; Baldó, F.; Mena, F.; Parra, G.; Blasco, J.; et al. Not Only Toxic but Repellent: What Can Organisms’ Responses Tell Us about Contamination and What Are the Ecological Consequences When They Flee from an Environment? Toxics 2020, 8, 118. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, Y.; Wang, X.C.; Wang, Y.; Dzakpasu, M. Characterization and Biogeochemical Implications of Dissolved Organic Matter in Aquatic Environments. J. Environ. Manag. 2021, 294, 113041. [Google Scholar] [CrossRef]
- Wood, C.M.; Al-Reasi, H.A.; Smith, D.S. The Two Faces of DOC. Aquat. Toxicol. 2011, 105, 3–8. [Google Scholar] [CrossRef]
- Schwartz, M.L.; Curtis, P.J.; Playle, R.C. Influence of Natural Organic Matter Source on Acute Copper, Lead, and Cadmium Toxicity to Rainbow Trout (Oncorhynchus mykiss). Environ. Toxicol. Chem. 2004, 23, 2889–2899. [Google Scholar] [CrossRef]
- Cornelissen, G.; Okkenhaug, G.; Breedveld, G.D.; Sørlie, J.-E. Transport of Polycyclic Aromatic Hydrocarbons and Polychlorinated Biphenyls in a Landfill: A Novel Equilibrium Passive Sampler to Determine Free and Total Dissolved Concentrations in Leachate Water. J. Hydrol. 2009, 369, 253–259. [Google Scholar] [CrossRef]
- Artifon, V.; Zanardi-Lamardo, E.; Fillmann, G. Aquatic Organic Matter: Classification and Interaction with Organic Microcontaminants. Sci. Total Environ. 2019, 649, 1620–1635. [Google Scholar] [CrossRef] [PubMed]
- Baken, S.; Degryse, F.; Verheyen, L.; Merckx, R.; Smolders, E. Metal Complexation Properties of Freshwater Dissolved Organic Matter Are Explained by Its Aromaticity and by Anthropogenic Ligands. Environ. Sci. Technol. 2011, 45, 2584–2590. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, Z.; Wu, K.; Lan, J.; Li, T.; Yuan, D. Speciation, Distribution and Migration Pathways of Polycyclic Aromatic Hydrocarbons in a Typical Underground River System in Southwest China. J. Hydrol. 2021, 596, 125690. [Google Scholar] [CrossRef]
- Reynaud, S.; Deschaux, P. The Effects of Polycyclic Aromatic Hydrocarbons on the Immune System of Fish: A Review. Aquat. Toxicol. 2006, 77, 229–238. [Google Scholar] [CrossRef]
- Honda, M.; Suzuki, N. Toxicities of Polycyclic Aromatic Hydrocarbons for Aquatic Animals. Int. J. Environ. Res. Public Health 2020, 17, 1363. [Google Scholar] [CrossRef]
- Moeckel, C.; Monteith, D.T.; Llewellyn, N.R.; Henrys, P.A.; Pereira, M.G. Relationship between the Concentrations of Dissolved Organic Matter and Polycyclic Aromatic Hydrocarbons in a Typical U.K. Upland Stream. Environ. Sci. Technol. 2014, 48, 130–138. [Google Scholar] [CrossRef]
- Giles, M.A. Electrolyte and Water Balance in Plasma and Urine of Rainbow Trout (Salmo gairdneri) during Chronic Exposure to Cadmium. Can. J. Fish. Aquat. Sci. 1984, 41, 1678–1685. [Google Scholar] [CrossRef]
- Laurén, D.J.; McDonald, D.G. Acclimation to Copper by Rainbow Trout, Salmo gairdneri: Physiology. Can. J. Fish. Aquat. Sci. 1987, 44, 99–104. [Google Scholar] [CrossRef]
- Hollis, L.; McGeer, J.C.; McDonald, D.G.; Wood, C.M. Cadmium Accumulation, Gill Cd Binding, Acclimation, and Physiological Effects during Long Term Sublethal Cd Exposure in Rainbow Trout. Aquat. Toxicol. 1999, 46, 101–119. [Google Scholar] [CrossRef]
- Campbell, P.G.; Twiss, M.R.; Wilkinson, K.J. Accumulation of Natural Organic Matter on the Surfaces of Living Cells: Implications for the Interaction of Toxic Solutes with Aquatic Biota. Can. J. Fish. Aquat. Sci. 1997, 54, 2543–2554. [Google Scholar] [CrossRef]
- Galvez, F.; Donini, A.; Playle, R.C.; Smith, D.S.; O’Donnell, M.J.; Wood, C.M. A Matter of Potential Concern: Natural Organic Matter Alters the Electrical Properties of Fish Gills. Environ. Sci. Technol. 2008, 42, 9385–9390. [Google Scholar] [CrossRef] [PubMed]
- Crémazy, A.; Braz-Mota, S.; Brix, K.V.; Duarte, R.M.; Val, A.L.; Wood, C.M. Investigating the Mechanisms of Dissolved Organic Matter Protection against Copper Toxicity in Fish of Amazon’s Black Waters. Sci. Total Environ. 2022, 843, 157032. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, A.Y.O.; Playle, R.C.; Val, A.L.; Wood, C.M. Physiological Action of Dissolved Organic Matter in Rainbow Trout in the Presence and Absence of Copper: Sodium Uptake Kinetics and Unidirectional Flux Rates in Hard and Softwater. Aquat. Toxicol. 2004, 70, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Burnison, B.K.; Meinelt, T.; Playle, R.; Pietrock, M.; Wienke, A.; Steinberg, C.E.W. Cadmium Accumulation in Zebrafish (Danio rerio) Eggs Is Modulated by Dissolved Organic Matter (DOM). Aquat. Toxicol. 2006, 79, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Duarte, R.M.; Smith, D.S.; Val, A.L.; Wood, C.M. Dissolved Organic Carbon from the Upper Rio Negro Protects Zebrafish (Danio rerio) against Ionoregulatory Disturbances Caused by Low pH Exposure. Sci. Rep. 2016, 6, 20377. [Google Scholar] [CrossRef]
- Wood, C.M.; Matsuo, A.Y.O.; Wilson, R.W.; Gonzalez, R.J.; Patrick, M.L.; Playle, R.C.; Luis Val, A. Protection by Natural Blackwater against Disturbances in Ion Fluxes Caused by Low pH Exposure in Freshwater Stingrays Endemic to the Rio Negro. Physiol. Biochem. Zool. 2003, 76, 12–27. [Google Scholar] [CrossRef]
- Chen, S.; Ke, R.; Zha, J.; Wang, Z.; Khan, S.U. Influence of Humic Acid on Bioavailability and Toxicity of Benzo[k]Fluoranthene to Japanese Medaka. Environ. Sci. Technol. 2008, 42, 9431–9436. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, C.; Cheng, P.; He, X.; Zhu, Y.; Zhang, Y. Influences of Humic Acid on the Bioavailability of Phenanthrene and Alkyl Phenanthrenes to Early Life Stages of Marine Medaka (Oryzias melastigma). Environ. Pollut. 2016, 210, 211–216. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhu, L. Effect of Humic Acid on Prometryn Bioaccumulation and the Induction of Oxidative Stress in Zebrafish (Danio rerio). RSC Adv. 2016, 6, 16790–16797. [Google Scholar] [CrossRef]
- Ye, S.; Li, Z.; Zhang, T.; Liu, J.; Xie, S. Assessing Fish Distribution and Threats to Fish Biodiversity in the Yangtze River Basin, China. Ichthyol. Res. 2014, 61, 183–188. [Google Scholar] [CrossRef]
- Qian, M.-M.; Wang, Z.-Y.; Zhou, Q.; Wang, J.; Shao, Y.; Qiao, Q.; Fan, J.-T.; Yan, Z.-G. Environmental DNA Unveiling the Fish Community Structure and Diversity Features in the Yangtze River Basin. Environ. Res. 2023, 239, 117198. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhang, J.; Wang, Z.; Lin, J.; Huang, X.; Liu, W.; Li, H.; Pellissier, L.; Zhang, X. Holistic Impact Evaluation of Human Activities on the Coastal Fish Biodiversity in the Chinese Coastal Environment. Environ. Sci. Technol. 2022, 56, 6574–6583. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a Set of Universal PCR Primers for Metabarcoding Environmental DNA from Fishes: Detection of More than 230 Subtropical Marine Species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Manel, S.; Guerin, P.-E.; Mouillot, D.; Blanchet, S.; Velez, L.; Albouy, C.; Pellissier, L. Global Determinants of Freshwater and Marine Fish Genetic Diversity. Nat. Commun. 2020, 11, 692. [Google Scholar] [CrossRef]
- Murphy, K.R.; Hambly, A.; Singh, S.; Henderson, R.K.; Baker, A.; Stuetz, R.; Khan, S.J. Organic Matter Fluorescence in Municipal Water Recycling Schemes: Toward a Unified PARAFAC Model. Environ. Sci. 2011, 45, 2909–2916. [Google Scholar] [CrossRef]
- Jørgensen, L. Global Trends in the Fluorescence Characteristics and Distribution of Marine Dissolved Organic Matter. Mar. Chem. 2011, 126, 139–148. [Google Scholar] [CrossRef]
- Yu, H.; Liang, H.; Qu, F.; Han, Z.; Shao, S.; Chang, H.; Li, G. Impact of Dataset Diversity on Accuracy and Sensitivity of Parallel Factor Analysis Model of Dissolved Organic Matter Fluorescence Excitation-Emission Matrix. Sci. Rep. 2015, 5, 10207. [Google Scholar] [CrossRef] [PubMed]
- Zang, C.; Huang, S.; Wu, M.; Du, S.; Scholz, M.; Gao, F.; Lin, C.; Guo, Y.; Dong, Y. Comparison of Relationships Between pH, Dissolved Oxygen and Chlorophyll a for Aquaculture and Non-Aquaculture Waters. Water Air Soil Pollut. 2011, 219, 157–174. [Google Scholar] [CrossRef]
- Whiles, M.R.; Huryn, A.D.; Taylor, B.W.; Reeve, J.D. Influence of Handling Stress and Fasting on Estimates of Ammonium Excretion by Tadpoles and Fish: Recommendations for Designing Excretion Experiments. Limnol. Oceanogr. Methods 2009, 7, 1–7. [Google Scholar] [CrossRef]
- Edwards, T.M.; Puglis, H.J.; Kent, D.B.; Durán, J.L.; Bradshaw, L.M.; Farag, A.M. Ammonia and Aquatic Ecosystems—A Review of Global Sources, Biogeochemical Cycling, and Effects on Fish. Sci. Total Environ. 2024, 907, 167911. [Google Scholar] [CrossRef]
- Fu, X.; Zheng, M.; Su, J.; Xi, B.; Wei, D.; Wang, X. Spatiotemporal Patterns and Threshold of Chlorophyll-a in Lake Taihu Based on Microcystins. Environ. Sci. Pollut. Res. 2023, 30, 49327–49338. [Google Scholar] [CrossRef]
- Howarth, R.; Chan, F.; Conley, D.J.; Garnier, J.; Doney, S.C.; Marino, R.; Billen, G. Coupled Biogeochemical Cycles: Eutrophication and Hypoxia in Temperate Estuaries and Coastal Marine Ecosystems. Front. Ecol. Environ. 2011, 9, 18–26. [Google Scholar] [CrossRef]
- Fourtune, L.; Paz-Vinas, I.; Loot, G.; Prunier, J.G.; Blanchet, S. Lessons from the Fish: A Multi-Species Analysis Reveals Common Processes Underlying Similar Species-Genetic Diversity Correlations. Freshw. Biol. 2016, 61, 1830–1845. [Google Scholar] [CrossRef]
- Wang, L.; Lyons, J.; Kanehl, P.; Bannerman, R. Impacts of Urbanization on Stream Habitat and Fish Across Multiple Spatial Scales. Environ. Manag. 2001, 28, 255–266. [Google Scholar] [CrossRef]
- Rahel, F.J.; Olden, J.D. Assessing the Effects of Climate Change on Aquatic Invasive Species. Conserv. Biol. 2008, 22, 521–533. [Google Scholar] [CrossRef]
- López Muñoz, I.; Bernard, O. Modeling the Influence of Temperature, Light Intensity and Oxygen Concentration on Microalgal Growth Rate. Processes 2021, 9, 496. [Google Scholar] [CrossRef]
- Matthews, W.J.; Marsh-Matthews, E. Effects of Drought on Fish across Axes of Space, Time and Ecological Complexity. Freshw. Biol. 2003, 48, 1232–1253. [Google Scholar] [CrossRef]
- Hansen, A.M.; Kraus, T.E.C.; Pellerin, B.A.; Fleck, J.A.; Downing, B.D.; Bergamaschi, B.A. Optical Properties of Dissolved Organic Matter (DOM): Effects of Biological and Photolytic Degradation. Limnol. Oceanogr. 2016, 61, 1015–1032. [Google Scholar] [CrossRef]
- Hu, A.; Choi, M.; Tanentzap, A.J.; Liu, J.; Jang, K.-S.; Lennon, J.T.; Liu, Y.; Soininen, J.; Lu, X.; Zhang, Y.; et al. Ecological Networks of Dissolved Organic Matter and Microorganisms under Global Change. Nat. Commun. 2022, 13, 3600. [Google Scholar] [CrossRef] [PubMed]
- Malzahn, A.M.; Aberle, N.; Clemmesen, C.; Boersma, M. Nutrient Limitation of Primary Producers Affects Planktivorous Fish Condition. Limnol. Oceanogr. 2007, 52, 2062–2071. [Google Scholar] [CrossRef]
- Boulion, V.V. Relationship between Phytoplankton, Heterotrophic Plankton, and Planktivorous Fish Productions in Different Water Bodies. Dokl. Biol. Sci. 2023, 513, S5–S9. [Google Scholar] [CrossRef]
- Suárez-Castro, A.F.; Raymundo, M.; Bimler, M.; Mayfield, M.M. Using Multi-Scale Spatially Explicit Frameworks to Understand the Relationship between Functional Diversity and Species Richness. Ecography 2022, 2022, e05844. [Google Scholar] [CrossRef]
- Helms, J.R.; Stubbins, A.; Ritchie, J.D.; Minor, E.C.; Kieber, D.J.; Mopper, K. Absorption Spectral Slopes and Slope Ratios as Indicators of Molecular Weight, Source, and Photobleaching of Chromophoric Dissolved Organic Matter. Limnol. Oceanogr. 2008, 53, 955–969. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, S.; Mu, E.; Zhao, Y.; Cheng, L.; Zhu, Y.; Yuan, Y.; Wang, Y.; Ding, A. Characterizing the Spatiotemporal Distribution of Dissolved Organic Matter (DOM) in the Yongding River Basin: Insights from Flow Regulation. J. Environ. Manag. 2023, 325, 116476. [Google Scholar] [CrossRef]
- Zhang, Y.; Van Dijk, M.A.; Liu, M.; Zhu, G.; Qin, B. The Contribution of Phytoplankton Degradation to Chromophoric Dissolved Organic Matter (CDOM) in Eutrophic Shallow Lakes: Field and Experimental Evidence. Water Res. 2009, 43, 4685–4697. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, Y.; Tian, Y.; Hou, Z.; He, K.; Fu, L.; Xu, H. Impact of Land Use on the DOM Composition in Different Seasons in a Subtropical River Flowing through a Region Undergoing Rapid Urbanization. J. Clean. Prod. 2019, 212, 1224–1231. [Google Scholar] [CrossRef]
- Ahoutou, M.K.; Yao, E.K.; Djeha, R.Y.; Kone, M.; Tambosco, K.; Duval, C.; Hamlaoui, S.; Bernard, C.; Bouvy, M.; Marie, B.; et al. Impacts of Nutrient Loading and Fish Grazing on the Phytoplankton Community and Cyanotoxin Production in a Shallow Tropical Lake: Results from Mesocosm Experiments. MicrobiologyOpen 2022, 11, e1278. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wu, X.; Zhi, G.; Yang, Y.; Wu, L.; Zhang, Y.; Zheng, B.; Qadeer, A.; Zheng, J.; Deng, W.; et al. Fluorescence Characteristics of DOM and Its Influence on Water Quality of Rivers and Lakes in the Dianchi Lake Basin. Ecol. Indic. 2022, 142, 109088. [Google Scholar] [CrossRef]
- Meinelt, T.; Kroupova, H.; Stüber, A.; Rennert, B.; Wienke, A.; Steinberg, C.E.W. Can Dissolved Aquatic Humic Substances Reduce the Toxicity of Ammonia and Nitrite in Recirculating Aquaculture Systems? Aquaculture 2010, 306, 378–383. [Google Scholar] [CrossRef]
- Peuranen, S.; Vuorinen, P.J.; Vuorinen, M.; Hollender, A. The Effects of Iron, Humic Acids and Low pH on the Gills and Physiology of Brown Trout (Salmo trutta). Ann. Zool. Fenn. 1994, 31, 389–396. [Google Scholar]
- Hammock, D.; Huang, C.C.; Mort, G.; Swinehart, J.H. The Effect of Humic Acid on the Uptake of Mercury(II), Cadmium(II), and Zinc(II) by Chinook Salmon (Oncorhynchus tshawytscha) Eggs. Arch. Environ. Contam. Toxicol. 2003, 44, 83–88. [Google Scholar] [CrossRef]
- Yamin, G.; Borisover, M.; Cohen, E.; Van Rijn, J. Accumulation of Humic-like and Proteinaceous Dissolved Organic Matter in Zero-Discharge Aquaculture Systems as Revealed by Fluorescence EEM Spectroscopy. Water Res. 2017, 108, 412–421. [Google Scholar] [CrossRef]
- Duque, G.; Gamboa-García, D.E.; Molina, A.; Cogua, P. Effect of Water Quality Variation on Fish Assemblages in an Anthropogenically Impacted Tropical Estuary, Colombian Pacific. Environ. Sci. Pollut. Res. 2020, 27, 25740–25753. [Google Scholar] [CrossRef]
- Ramírez-Herrejón, J.; Moncayo-Estrada, R.; Balart, E.; García Camacho, L.; Vital Rodríguez, B.; Alvarado Villanueva, R.; Ortega Murillo, R.; Caraveo-Patiño, J. Trophic Interrelations between Introduced Common Carp, Cyprinus Carpio (Actinopterygii: Cypriniformes: Cyprinidae), and Fish Community in a Eutrophic Shallow Lake. Acta Ichthyol. Piscat. 2014, 44, 45–58. [Google Scholar] [CrossRef]
- Wu, J.; Mao, R.; Li, M.; Xia, J.; Song, J.; Cheng, D.; Sun, H. Assessment of Aquatic Ecological Health Based on Determination of Biological Community Variability of Fish and Macroinvertebrates in the Weihe River Basin, China. J. Environ. Manag. 2020, 267, 110651. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Lei, P.; Xin, X.; Zhang, A.; Yin, W. Evidence on the Causes of the Rising Levels of CODMn along the Middle Route of the South-to-North Diversion Project in China: The Role of Algal Dissolved Organic Matter. J. Environ. Sci. 2022, 113, 281–290. [Google Scholar] [CrossRef]
- Weishaar, J.L.; Aiken, G.R.; Bergamaschi, B.A.; Fram, M.S.; Fujii, R.; Mopper, K. Evaluation of Specific Ultraviolet Absorbance as an Indicator of the Chemical Composition and Reactivity of Dissolved Organic Carbon. Environ. Sci. Technol. 2003, 37, 4702–4708. [Google Scholar] [CrossRef] [PubMed]
- Huguet, A.; Vacher, L.; Relexans, S.; Saubusse, S.; Froidefond, J.M.; Parlanti, E. Properties of Fluorescent Dissolved Organic Matter in the Gironde Estuary. Org. Geochem. 2009, 40, 706–719. [Google Scholar] [CrossRef]
- Parlanti, E.; Wörz, K.; Geoffroy, L.; Lamotte, M. Dissolved Organic Matter Fluorescence Spectroscopy as a Tool to Estimate Biological Activity in a Coastal Zone Submitted to Anthropogenic Inputs. Org. Geochem. 2000, 31, 1765–1781. [Google Scholar] [CrossRef]
- Cory, R.M.; McKnight, D.M. Fluorescence Spectroscopy Reveals Ubiquitous Presence of Oxidized and Reduced Quinones in Dissolved Organic Matter. Environ. Sci. Technol. 2005, 39, 8142–8149. [Google Scholar] [CrossRef]
- McKnight, D.M.; Boyer, E.W.; Westerhoff, P.K.; Doran, P.T.; Kulbe, T.; Andersen, D.T. Spectrofluorometric Characterization of Dissolved Organic Matter for Indication of Precursor Organic Material and Aromaticity. Limnol. Oceanogr. 2001, 46, 38–48. [Google Scholar] [CrossRef]
- Ohno, T. Fluorescence Inner-Filtering Correction for Determining the Humification Index of Dissolved Organic Matter. Environ. Sci. Technol. 2002, 36, 742–746. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Revell, L.J. Phytools: An R Package for Phylogenetic Comparative Biology (and Other Things). Methods Ecol. Evol. 2012, 3, 217–223. [Google Scholar] [CrossRef]
- Poelen, J.H.; Simons, J.D.; Mungall, C.J. Global Biotic Interactions: An Open Infrastructure to Share and Analyze Species-Interaction Datasets. Ecol. Inform. 2014, 24, 148–159. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal 2005, Complex Systems, 1695. Available online: https://igraph.org (accessed on 28 May 2024).
- Murphy, K.; Stedmon, C.; Wenig, P.; Bro, R. OpenFluor—A Spectral Database of Auto-Fluorescence by Organic Compounds in the Environment. Anal. Methods 2014, 6, 658. [Google Scholar] [CrossRef]
- Wang, S. Suspect Screening to Support Source Identification and Risk Assessment of Organic Micropollutants in the Aquatic Environment of a Sub-Saharan African Urban Center. Water Res. 2022, 220, 118706. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Spectral and Isotopic Characteristics of Particulate Organic Matter in a Subtropical Estuary under the Influences of Human Disturbance. J. Mar. Syst. 2020, 203, 103264. [Google Scholar] [CrossRef]
- Krylov, I.N.; Drozdova, A.N.; Labutin, T.A. Albatross R Package to Study PARAFAC Components of DOM Fluorescence from Mixing Zones of Arctic Shelf Seas. Chemom. Intell. Lab. Syst. 2020, 207, 104176. [Google Scholar] [CrossRef]
- Eder, A. Pathways and Composition of Dissolved Organic Carbon in a Small Agricultural Catchment during Base Flow Conditions. Ecohydrol. Hydrobiol. 2022, 22, 96–112. [Google Scholar] [CrossRef]
- Lee, D. Characteristics of Intracellular Algogenic Organic Matter and Its Reactivity with Hydroxyl Radicals. Water Res. 2018, 144, 13–25. [Google Scholar] [CrossRef]
- Murphy, K.R.; Ruiz, G.M.; Dunsmuir, W.T.M.; Waite, T.D. Optimized Parameters for Fluorescence-Based Verification of Ballast Water Exchange by Ships. Environ. Sci. Technol. 2006, 40, 2357–2362. [Google Scholar] [CrossRef]
- Stedmon, C.; Thomas, D.; Papadimitriou, S.; Granskog, M.; Dieckmann, G. Using Fluorescence to Characterize Dissolved Organic Matter in Antarctic Sea Ice Brines. J. Geophys. Res. 2011, 116, G03027. [Google Scholar] [CrossRef]
- Du, Y. Composition of Dissolved Organic Matter Controls Interactions with La and Al Ions: Implications for Phosphorus Immobilization in Eutrophic Lakes. Environ. Pollut. 2019, 248, 36–47. [Google Scholar] [CrossRef]
- Brogi, S.R.; Derrien, M.; Hur, J. In-Depth Assessment of the Effect of Sodium Azide on the Optical Properties of Dissolved Organic Matter. J. Fluoresc. 2019, 29, 877–885. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).