Abstract
Intestinal bacterial communities play an important role in the growth and health of aquatic animal hosts and have drawn increasing attention. However, the role of the intestinal microbiota in the growth of freshwater prawns remains unclear. Here, the intestinal microbiota of freshwater prawns (Macrobrachium rosenbergii) at different life stages (one, two, and three months old) were investigated using 16S rRNA sequencing. The results showed that community richness and diversity increased with growth, which might be one of the reasons that the prawns maintained a fast growth rate before sexual maturation. Three core phyla were identified in the one-month-old group, namely, Firmicutes (79.24%), Proteobacteria (17.09%) and Actinobacteriota (2.01%). Five core phyla were identified in the two-month-old group, including Firmicutes (47.84%), Proteobacteria (44.22%), Actinobacteriota (1.83%), Acidobacteriota (1.66%) and Bacteroidota (1.24%), and the core phyla in the three-month-old group were similar, except for Acidobacterota, which was not identified. A total of 12 core genera were identified in all samples, and significant differences were observed in the relative abundance of gut microbiota between the three groups (p < 0.05). Exiguobacterium, Lactococcus and Shewanella were the three most significantly differentially abundant genera between stages. In addition, Candidatus Hepatoplasma was detected only in the two- and three-month-old prawn groups. This study provides information on the differences in the intestinal microbiota in different developmental stages, which contribute to adaptation to salinity in the early developmental stage and digestive ability to meet the growth needs of Macrobrachium rosenbergii.
Key Contribution:
The intestinal microbiota is different among different developmental stages of Macrobrachium rosenbergii, affecting adaptation to the environment and digestive ability to meet growth needs. In different stages of development, the composition of the intestinal microbial community may vary, and reflects the diet, digestive and absorption capacity in different developmental stages of Macrobrachium rosenbergii, especially around one month old.
1. Introduction
Freshwater prawns are extensively reared worldwide and annually provide more than 5 million tons of high-quality, high-protein animal food products for humans. The giant freshwater prawn (Macrobrachium rosenbergii) is the most important freshwater prawn species worldwide, especially in China [1,2]. Macrobrachium rosenbergii was introduced into China in 1976 and has become the most economically valuable farmed freshwater prawn species in China since the discovery that its larvae require brackish conditions for survival and the development of mass rearing techniques for hatchery production of prawn postlarvae [3]. The fast growth rate and adaptability of feed habits have also played a key role in the rapid development of the Macrobrachium rosenbergii industry. Previous studies have shown that the structure and composition of the intestinal microbiota are involved in host immune adaptations and health [4], such as Campylobacter abundance in the gut of chicken (0 to 7 D of age), which is associated with increased expression of the immune gene beta-defensin. The intestinal microbiota has been found to be involved in the growth and nutrient digestion of the host in humans and aquatic animals [5,6,7]; it is also highly associated with the development of the host [8]. For example, the intestinal microbial diversity of Macrobrachium rosenbergii decreased significantly with development in the zoeae larva stage from zoeae to postlarvae, but the dominant phyla of the microbial community were the same. A previous study has shown that the changing of the dominant phyla from embryonic stages to postlarval stages in Macrobrachium rosenbergii was attributed to the maternal and environmental microbe community, and promotes prawn growth and physiological health [9]. Although there is a close association between intestinal microbes and growth, the structure of the shrimp microbiota is affected by lifestyle, organ, developmental stage, diet, and health status [10]. For example, a higher bacterial diversity was observed in freshwater shrimp than in marine shrimp, but the microbiota composition of marine shrimp was significantly impacted by lifestyle, organ, and developmental stage [11,12,13]. Diet can also impact the host intestinal microbial community and affect host growth and health [14]. For this reason, the intestinal microbial community was treated as a key factor in evaluating the immune health of shrimp in Macrobrachium rosenbergii, Procambarus clarkii, Marsupenaeus japonicus and Litopenaeus vannamei [15].
Additionally, some studies have demonstrated how gut microbiota enhances host nutritional metabolism and immune function. For instance, bacterial communities can increase cellulose-degrading enzymes and confer upon their host optimal adaptation to its environment by modulating its metabolism [16]. A previous study has shown that nutrient competition is the primary mechanism through which the intestinal microbiota hampered the colonization of the pathogen Salmonella to prolong mice survival [17]. According to the present study, the composition of the intestinal microbial community is important for increasing digestion and absorption efficiency and improving growth and immunity response activity in shrimp. However, little is known about the changes in the intestinal bacterial community of Macrobrachium rosenbergii during the postlarval to adult period in normal culture.
Compared with marine shrimp, such as Litopenaeus vannamei, the intestinal microbiota of freshwater prawn, Macrobrachium rosenbergii, is poorly understood, particularly in the rearing stage, and ignores the biological interactions between intestinal bacteria and the host [7]. With the increasing economic importance of and attention being paid to disease issues in the worldwide production of Macrobrachium rosenbergii, the influence of the intestinal microbial community at the rearing stage has become a pressing issue. Studying the diversity of the intestinal microbiota composition of aquaculture species could help us to assess their health status and establish healthy rearing strategies. To understand that issue more deeply, our study performed Illumina PE250 high-throughput sequencing of 16S rRNA gene V3V4 amplicons to investigate the bacterial communities of the intestinal microbes of Macrobrachium rosenbergii at different rearing stages. The results will help us to understand the relationship between growth and bacterial communities in Macrobrachium rosenbergii, which will be beneficial for developing new dietary formulations, health assessments at the rearing stage, and exploration of microbial agents.
2. Materials and Methods
2.1. Sample Collection
Healthy juvenile Macrobrachium rosenbergii freshwater prawns of one month old (Y1), two months old (Y2), and three months old (Y3) with body weights of 1.97 ± 0.96 g, 20.73 ± 3.86 g and 21.85 ± 7.74 g, respectively, were collected from a farm pond in Xiangshan, Zhejiang Province, China. The prawns were cultured in a natural environment and fed twice a day with commercial diets (Tech-Bank Food Co, Ltd., Nanjing, China), and the initial rearing density was 30–50 k post larva per 0.0667 hectares of water with an average depth of 1.5 m. A total of 26 freshwater prawn individuals, 13 males and 13 females (3 males and 3 females in the Y1 stage, 5 males and 5 females in both the Y2 and Y3 stage), were randomly collected for sampling. The sampled prawns were first anesthetized by putting them on ice for 1 min. After anesthetizing, the complete intestinal tissues were immediately sampled using sterile equipment, frozen in liquid nitrogen, and stored at −80 °C until DNA extraction.
2.2. Genomic DNA (gDNA) Extraction and 16S rDNA Sequencing
Total gDNA extraction and sequencing were performed according to standard protocols. Briefly, gDNA from each sample was extracted using the CTAB method [18]. DNA concentration and purity were monitored on 1% agarose gels, and then the quality and quantity of DNA were assessed using a NanoDrop 2000 spectrophotometer (Bio–Rad Laboratories Inc., Hercules, CA, USA). The 16S rRNA gene fragments were amplified using a 341f/806r primer set targeting the V3 and V4 regions (341f: 5′–CCTAYGGGRBGCASCAG–3′, 806r: 5′–GGACTACNNGGGTATCTAAT–3′). All PCRs were carried out (98 °C for 1 min, followed by 30 cycles at 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 30 s, and finally elongation at 72 °C for 5 min) in a 15 μL reaction mixture containing Phusion® High-Fidelity PCR Master Mix (New England Biolabs, EvryCedex, France), 0.2 μM forward and reverse primers, and approximately 10 ng template DNA. The PCR products were detected using 2% agarose gel electrophoresis and mixed in ratios of equal density. Then, the mixed PCR products were purified with a Qiagen Gel Extraction Kit (Qiagen, Dusseldorf, Germany). Sequencing libraries were constructed using a TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations. The library quality was assessed using a Qubit@ 2.0 Fluorometer (Thermo Scientific, Waltham, MA, USA) and the Agilent Bioanalyzer 2100 system. The library was sequenced on an Illumina NovaSeq platform, and 250 bp paired-end reads were generated. All raw data generated in this study were deposited in the NCBI (National Center for Biotechnology Information), with the accession code PRJNA1007864.
2.3. Data Analysis
Paired-end reads were assigned to samples based on their unique barcodes and were truncated by cutting off the barcodes and primer sequences, and they were merged using FLASH [19]. Quality filtering of the raw tags was performed using fastp (Version 0.20.0) software to obtain high-quality clean tags. The clean tags were compared with the reference Silva database (https://www.arbsilva.de/, accessed on 20 August 2023) using the software Vsearch (Version 2.15.0) to detect chimera sequences, and then the chimera sequences were removed to obtain the effective tags [20]. For the effective tags obtained previously, denoising was performed with DADA2 or the deblur module in QIIME2 software (Version QIIME2-202006) to obtain initial ASVs (amplicon sequence variants) (default: DADA2), and then ASVs with an abundance of less than 5 were filtered out [21]. Species annotation was performed using QIIME2 software, and the annotation database was the Silva database. The absolute abundance of ASVs was normalized using a standard sequence number corresponding to the sample with the least sequences. The processing parameters in the above steps were all default values. Subsequent analyses of alpha diversity and beta diversity were all performed based on the output normalized data. The rarefied ASV count data were used for alpha diversity analysis. In order to analyze the diversity, richness and uniformity of the communities in the sample, alpha diversity was calculated from six indices in QIIME2, including Observed_otus, Chao1, Shannon, Simpson, Dominance, and Good’s coverage. Rarefaction curves were generated based on these three metrics’ indication of the diversity estimated by the Shannon index.
2.4. Phylogenetic Distance and Community Distribution
In order to study the phylogenetic relationship of each ASV and the differences in the dominant species among different samples or groups, multiple sequence alignment was performed using QIIME2 software. Weighted and unweighted UniFrac distances were calculated using QIIME2, and were used to construct a phylogenetic tree in ClustalW. The three-dimensional PCoA (Principal Coordinate Analysis) results were displayed using the QIIME2 package, while the two-dimensional PCoA results were displayed using the ade4 package and ggplot2 package in R software (Version 2.15.3).
Furthermore, to study the functions of the communities in the samples and determine the different functions of the communities in the different groups, PICRUSt2 software (http://huttenhower.sph.harvard.edu/picrust/, accessed on 27 August 2023) (Version 2.1.2-b) was used for functional annotation analysis.
2.5. Statistical Analysis
To find the significance of the differences in community structure between the groups, the adonis and anosim functions in QIIME2 software were used. R software (Version 3.5.3) was utilized for MetaStat and Wilcoxon analyses to determine the significantly different relative species abundances of each taxonomic level (phylum, class, order, family, genus). To identify the biomarkers, LEfSe software (http://huttenhower.sph.harvard.edu/lefse/, accessed on 27 August 2023)) (Version 1.0) was used to perform an LEfSe analysis (LDA score threshold: 4). A Kruskal–Wallis test was performed to find out the different functions of the communities in the samples and in the different groups; PICRUSt2 software (Version 2.1.2-b) was used for function annotation analysis.
3. Results
3.1. OTU Analysis and Alpha Diversity
A total of 412,939, 753,650 and 733,101 qualified reads were obtained from the gut contents of Y1, Y2 and Y3, respectively, with an average of 73,065 qualified reads in each sample. All qualified reads were dereplicated to obtain final amplicon sequence variants (ASVs) using the DADA2 method, with the same similarity values reaching 100%, and the observed ASV curves of all samples and the species accumulation approached a saturation plateau (Figure 1A,B). The results indicated that sequencing depth and sampling amount were sufficient to cover most of the microbial community. A total of 5124 unique ASVs were detected, and 881, 3176 and 2372 unique ASVs were detected in the Y1, Y2 and Y3 groups, respectively; 149 unique ASVs were shared by the three groups (Figure 1C). In addition, the average ASV numbers obtained from each sample in the Y1, Y2 and Y3 groups were 233 ± 80, 506 ± 249 and 436 ± 129, respectively. All ASVs were annotated at different taxonomic levels, and 39 phylum-level taxa were identified in all samples to understand the species composition of the microbial community.
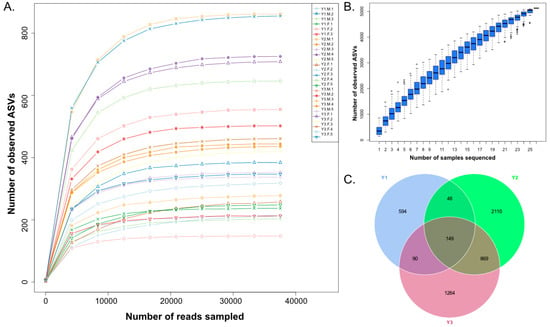
Figure 1.
Observed ASV analysis, species accumulation analysis, and ASV Venn analysis of the Y1, Y2, and Y3 groups. (A) The number of observed ASV curves in each sample; (B) The species accumulation number in different samples; (C) Venn diagram analysis showing the unique and shared ASVs of the Y1, Y2 and Y3 groups.
Alpha diversity indices, such as Chao1 (community richness indices), Simpson, and Shannon indices, were calculated to evaluate the microbial community richness and diversity of the Y1, Y2, and Y3 groups. The completeness of sequencing in all samples was estimated by the index of Good’s coverage, and almost all reached 100% (two samples reached 99.9% in the Y2 group). The microbial community richness indices of Chao1 and observed ASV in the Y1 group were significantly lower than those in the Y2 and Y3 groups (p < 0.01) (shown in Table 1). The Shannon index in the Y1 group was lower than that in the Y2 and Y3 groups, but the difference was not statistically significant (p > 0.05). The Simpson and dominance indices were also not different among the three groups. There was no significant difference in all richness and diversity indices between the Y2 and Y3 groups.

Table 1.
Statistics of richness and diversity indices of microbial community in samples.
3.2. Beta Diversity
In the beta diversity analysis, Bray–Curtis was used to compare the pairwise distance between samples, and a heatmap (Figure 2A) was used to visualize the results. From the results of the distance comparison, the samples within the Y1 group were significantly lower than those samples within the Y2 and Y3 groups. A higher diversity was investigated in the Y2 and Y3 groups. Principal coordinate analysis (PCoA) was conducted to compare the similarities and differences in the microbial community compositions of all samples (Figure 2B). The results showed that PC1, PC2 and PC3 were equal to 40.00%, 19.05%, and 15.59%, respectively. According to the results of the distance comparison, the Y1, Y2 and Y3 groups could be divided into three clusters. The clustering results showed that the Y1 group gathered in one branch, and the Y2 and Y3 groups gathered in another two branches, but individuals in the Y2 and Y3 groups did not show a clear clustering relationship (Figure 2C). Interestingly, males and females in the Y1 group showed an obvious clustering relationship, but those in the Y2 and Y3 groups did not.
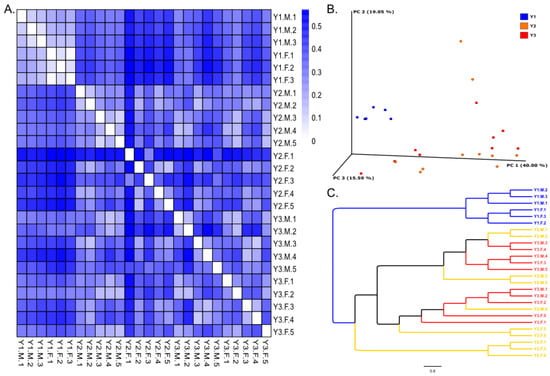
Figure 2.
Beta diversity analysis of samples in the three groups. (A) Heatmap analysis for Bray–Curtis distance; (B) PCoA analysis based on weighted UniFrac; (C) Similarity tree for all samples.
3.3. Taxonomic Composition
A total of 37 phyla were identified, and significant differences were observed in the relative abundance of intestinal microbes between the Y1, Y2 and Y3 groups (p < 0.05). Regarding the relative abundance of Y1 group individuals, there were three core phyla with a mean relative abundance greater than 1%, namely, Firmicutes (79.24%), Proteobacteria (17.09%) and Actinobacteriota (2.01%), which accounted for 98.33% of the total abundance. In the Y2 group, five core phyla were identified: Firmicutes (47.84%), Proteobacteria (44.22%), Actinobacteriota (1.83%), Acidobacteriota (1.66%) and Bacteroidetes (1.24%), accounting for 96.79% of the total abundance. Among the core phyla in the Y3 group, except for Acidobacterota, which was not identified, all core phyla were the same as those in the Y2 group, and their mean relative abundances were similar.
Among the intestinal microbes, Firmicutes, Proteobacteria and Actinobacteriota demonstrated a significant differential abundance between the Y1, Y2 and Y3 groups, especially Firmicutes, which showed a dramatic decrease in the abundance of community change with growth in age from one month old to two months old (as shown in Table 2). In addition, we assigned genera with a mean relative abundance of over 1% as core genera. Eight core genera were detected in the Y1 group, namely, Exiguobacterium (63.62%), Shewanella (7.19%), Lactococcus (5.33%), Streptococcus (1.88%), Acinetobacter (1.68%), Aeromonas (1.54%), Pseudomonas (1.38%), and Bacillus (1.25%) (Figure 3). Five core genera were detected in the Y2 group, namely, Lactococcus (28.51%), Candidatus Hepatoplasma (13.08%), Enterobacter (10.51%), Aeromonas (8.12%), and Streptococcus (1.58%). Eight core genera were detected in the Y3 group, namely, Lactococcus (30.55%), Aeromonas (17.97%), Acinetobacter (11.73%), Enterobacter (3.08%), Streptococcus (2.28%), Exiguobacterium (1.72%), Lactobacillus (1.65%), and Candidatus Hepatoplasma (1.16%).

Table 2.
Relative abundance of the core phyla in the microbial community in each group.
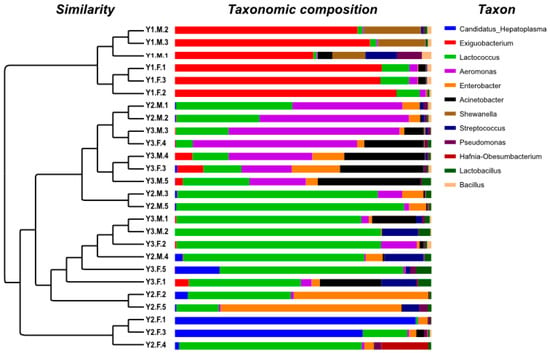
Figure 3.
Microbial community bar plot with cluster tree. The left of the figure shows the similarity in cluster analysis results based on community composition in each sample, and the right shows an abundance of community structure at all core genera levels of each sample.
3.4. Variance Analysis and Function Prediction
The results of the LEfSe and Wilcoxon analysis are shown in Figure 4A–C. The bar charts show microbial communities at the genera level, with significant differences between pairs in the three groups. A total of six genera and significant differences in abundance between pairs in the three groups were detected via LEfSe analysis, and all come from the core genera of the three groups. Among the six genera, four were identified both in pairs of Y1 vs. Y2 groups and Y1 vs. Y3 groups, but only three were detected between the Y1 and Y3 groups. According to the results of LEfSe analysis, the genera of Shewanella and Exiguobacterium were highly enriched in group Y1, Candidatus Hepatoplasma and Enterobacter shared highly increased specificity in group Y2, and Acinetobacter showed highly enriched specificity in group Y3. In the Wilcoxon analysis results, the genera of Shewanella were significantly more enriched in the Y1 group than in the Y2 group. The genera of Enterbacter and Candidatus Hepatoplasma were more enriched in the Y2 group than in the Y1 group, with p < 0.01 within the Wilcoxon analysis.
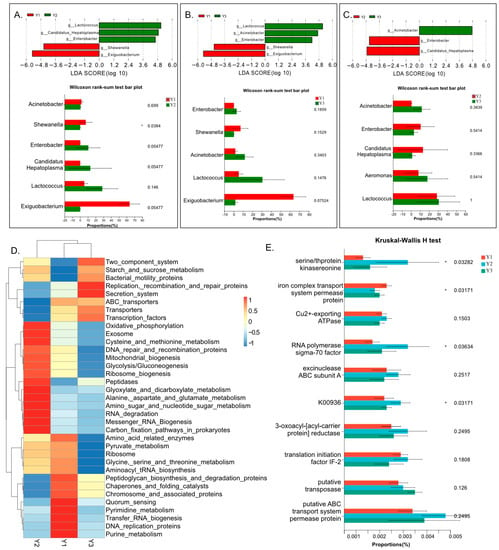
Figure 4.
Results of LEfSe and function prediction analysis showing differences in the intestinal microbiota in the three groups. (A–C) Bar charts showing the significantly different abundance of genera between three groups (LDA > 4); (D) Heatmap of functional clustering of the most enriched gut microbiomes across different groups; (E) Results of a Kruskal–Wallis test for the different functions of the communities in the different groups. The * indicate that the function of Kruskall-Wallis H test p-value is less than 0.05.
The result of the function predicted using PICRUSt2 indicated microbial communities at genera levels, with the most enriched function in the three groups (shown in Figure 4D). The microbial communities in the Y1 group were highly enriched in KEGG pathways such as pyrimidine and purine metabolism, transfer RNA biogenesis, DNA replication proteins, etc. The microbial communities in the Y2 group were highly enriched in KEGG pathways such as cysteine and methionine metabolism, amino sugar and nucleotide sugar metabolism, etc. The microbial communities in the Y3 group were highly enriched on KEGG pathways such as the secretion system, replication recombination and repair proteins, etc. According to the Kruskal–Wallis test, serine/thprotein kinasereonine and RNA polymerase had significantly higher functions in the Y2 group than in the other two groups, and the iron complex transport system had a significantly higher function in the Y1 group than in the other two groups.
4. Discussion
The composition of the intestinal microbial community may vary across developmental stages, and also reflects the health status, digestion, and absorption capacity of the host animal. For example, a shift in diet from yolk or redworms as a juvenile to shrimp or crab as an adult comes with a change in the composition of the intestinal microbial community in the Chinese giant salamander (Andrias davidianus) [22]. A previous study found that Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria were the dominant phyla in zoeae, and varied throughout the larval development of freshwater prawns (Macrobrachium rosenbergii) [8]. In our study, only the three phyla Firmicutes, Proteobacteria and Actinobacteria were dominant in one-month-old prawns, and the relative abundance of Firmicutes decreased, while that of Proteobacteria increased with the growth of freshwater prawns under pond culture conditions. This result indicated that diet and digestive and absorption capacity are very different at one month of age in Macrobrachium rosenbergii.
The intestinal microbial composition can reflect diet composition and digestive and absorption capacity, and then affect the growth of shrimp and crab hosts [23,24]. The intestinal microbiota plays a role in promoting health and growth in fishes, and it has also been found that the bacterial community can improve the metabolic capacity of carbohydrates and amino acids to enhance growth and survival [25]. In our study, an increasing trend of community richness and diversity of the intestinal microbiota with age was found, but there were no significant changes in diversity after two months of age in Macrobrachium rosenbergii. Previous studies have shown that more diverse gut communities exert greater protective effects on the host [26,27]. The diversity of the intestinal microbiota was lower in the one-month-old freshwater prawn group than in the two- and three-month-old groups. Therefore, prawns before one month of age might be more vulnerable to the homeostatic imbalance of the intestinal microbiota than those after two months of age.
Diversity differences in the intestinal microbiota have been reported in a number of shrimp species, such as Indian white shrimp (Penaeus indicus), Pacific white shrimp (Litopenaeus vannamei), red claw crayfish (Cherax quadricarinatus) and red swamp crayfish (Procambarus clarkii) [28,29,30]. The intestinal microbiota of Indian white shrimp differed among different stages and showed decreasing diversity with age. The farmed Pacific white shrimp microbiota composition was less diverse than that in the wild environment. In crayfish, the diversity of microbiota composition differed among species. In our study, there were significant differences in the microbiota composition diversity between different stages, and Exiguobacterium, Lactococcus and Aeromonas were the three most significantly differentially abundant genera between stages. Exiguobacterium is one main microbial component in the gut that has been frequently isolated from various habitats, including seawater, marine sediment, marine algae [31], soil [32], and freshwater [33] and has unique characteristics of salt and alkali resistance and environmental stress resistance [34]. In addition, Exiguobacterium has the highest ion transport function based on genome sequencing [35], which is consistent with the result of function analysis in our study. The identification of Exiguobacterium is important evidence for the adaptation of Macrobrachium rosenbergii to seawater at the early stage of development after hatching, and indicates that the intestinal microbiota’s composition plays an important role in Macrobrachium rosenbergii’s adaptation to different environments.
Stage-specific, sex-specific, and species-specific microbiota compositions in growth responses have been reported in vertebrates and invertebrates, such as Cetobacterium in the giant salamander (Andrias davidianus) [36], Rhodobacterales in threespine stickleback (Gasterosteus aculeatus) [37] and Vibrio coralliilyticus in the red claw crayfish (Cherax quadricarinatus) [30]. Candidatus Hepatoplasma was detected only in the two- and three-month-old prawn groups. Candidatus hepatoplasma is a genus of mycoplasma in the family Mycoplasmataceae. It has been reported in the digestive tract of the Norway lobster Nephrops norvegicus [38], showed higher abundance under low-quality food supply in most isopods, and maintained a higher survival rate [39]. Candidatus Hepatoplasma has been found in the gut of juvenile spiny lobster P. ornatus, and plays a symbiotic role in nourishment [40]. In our study, Candidatus Hepatoplasma showed stage-specific differences, with the highest abundance in two-month-old prawns, a stage with higher growth rate and nutrition demand [41], which suggests that it has a strong nourishment function in Macrobrachium rosenbergii. This finding regarding their relationship and mechanism needs to be further assessed.
Lactococcus and Acinetobacter were genera whose relative abundance dramatically increased with age. A previous study indicated that Lactococcus can provide effective preventive and certain reparative functions against high-protein meal-induced adverse effects [42]. Lactococcus has been used as a dietary probiotic for aquatic feed, and its benefits on growth, immunity, and digestive enzyme activity in shrimp culture have been reported by several authors [43,44]. Lactococcus demonstrated a positive relationship with weight-at-age in our study. The results suggested that Lactococcus plays a role as a growth promoter in Macrobrachium rosenbergii, as well as in Litopenaeus vannamei [45]. Acinetobacter has also been found to play a vital role in producing a wide range of secondary metabolites for improving growth function for host animals [46]. Moreover, when growth-related functions, such as serine/thprotein kinasereonine and RNA polymerase, were enhanced, early-stage growth in animals was improved [47,48]. In our study, Acinetobacter showed a significant increase with age, suggesting that the growth metabolism gradually increases before the age of three months. Based on the results, we suggest that dietary supplementation of the intestinal microbe Lactococcus and Acinetobacter might also promote the growth performance and digestive enzyme activity of Macrobrachium rosenbergii in the developmental stage before sexual maturation.
Shewanella and Enterobacter were other genera whose relative abundance dramatically changed with age. Recent reports have shown that Shewanella is the dominant prokaryotic genus, is closely related to the phage communities in the shrimp intestine, and affects shrimp health through direct or indirect phage–prokaryote interactions [49]. In addition, the effects of Shewanella on the immune system have been proven by feeding experiments. Previous studies have found that Shewanella can protect Litopenaeus vannamei from bacterial infection to improve survival [50]. In our study, Shewanella was found to be significantly enriched in one-month-old juvenile shrimp with enhanced immune-related metabolites such as glycine serine and threonine metabolism [51]. These results were consistent with the relative abundance of Shewanella in Litopenaeus vannamei [49]. However, its function at different life-stages of Macrobrachium rosenbergii still needs to be assessed. Enterobacter also showed a dramatically changed microbial composition. Interestingly, it exhibited a similar trend of change as peptidase in three developmental stages in Macrobrachium rosenbergii. Peptidase, as an important intestinal digestive enzyme, not only promotes digestion and metabolism [52] but also has the function of producing immune functional peptides [53]. Enterobacter has been reported to inhibit Flavobacterium psychrophilum, which can cause bacterial cold-water disease in rainbow trout Oncorhynchus mykiss [54]. In our study, Enterobacter and peptidase had a higher abundance in the two-month group. These results indicate that in the development from one month old to two months old, intestinal microbial composition significantly changes to adapt to the transformation in salinity of the environment and enhances the digestive and adaptive ability to meet growth needs at different life stages of Macrobrachium rosenbergii.
5. Conclusions
Microbial community richness and diversity increased with growth, which might be one of the reasons that the prawn maintained a fast growth rate before sexual maturation in Macrobranchium rosenbergii. Diet, digestive and absorption capacities are very different, and the microbiota composition diversity also changed from one to three months of age. The results indicate that the intestinal microbiota’s composition might play an essential role in Macrobrachium rosenbergii adaptation to different environments. Shewanella and Enterobacter dramatically changed in the one-month group and two-month group, which may relate to the changing salinity of the environment, and enhanced the resistance and digestive ability of Macrobrachium rosenbergii to meet growth needs in different life stages.
Author Contributions
Conceptualization, J.R.; methodology, validation, investigation, writing—review and editing, H.X. and J.R.; resource, X.X.; funding acquisition, J.R.; supervision and project administration, B.L. and D.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Zhejiang Provincial Natural Science Foundation, grant number LY21C190001 and the APC was funded by Zhejiang Provincial Natural Science Foundation.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by Committee of Laboratory Animal Experimentation at Zhejiang Academy of Agricultural Sciences (No. 2023ZAASLA35).
Data Availability Statement
All raw supported data can be found in the NCBI (National Center for Biotechnology Information), with the accession code PRJNA1007864.
Acknowledgments
We would like to express gratitude to Dongmin Zhu at Yonggang Aquatic Seedlings Co., Ltd., Ningbo, Zhejiang province, for his kind help in collecting samples.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hameed, A.S.; Yoganandhan, K.; Sri Widada, J.; Bonami, J.R. Experimental transmission and tissue tropism of Macrobrachium rosenbergii nodavirus (MrNV) and its associated extra small virus (XSV). Dis. Aquat. Org. 2004, 62, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Hossain, F.E.; Ramim, A.M.; Rahman, M.S.; Faruque, A.; Naser, M.N.; Karim, M.M. Identification of bacterial agents causing mortality in postlarvae of giant freshwater prawn (Macrobrachium rosenbergii) in south-west coastal districts of bangladesh. Aquac. Res. 2017, 48, 3545–3555. [Google Scholar] [CrossRef]
- Bruyn, M.D.; Nugroho, E.; Hossain, M.M.; Wilson, J.C.; Mather, P.B. Phylogeographic evidence for the existence of an ancient biogeographic barrier: The isthmus of Kra seaway. Heredity 2005, 94, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H.; Lee, A.; Santin, E. Microbiome and pathogen interaction with the immune system. Poult. Sci. 2020, 99, 1906–1913. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Holt, C.C.; Bass, D.; Stentiford, G.D.; van der Giezen, M. Understanding the role of the shrimp gut microbiome in health and disease. J. Invertebr. Pathol. 2021, 186, 107387. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, B.; Wang, N.; Yang, J.; Zhou, Q.; Sun, C.; Zhao, Y. Low fish meal diet supplemented with probiotics ameliorates intestinal barrier and immunological function of Macrobrachium rosenbergii via the targeted modulation of gut microbes and derived secondary metabolites. Front. Immunol. 2022, 13, 1074399. [Google Scholar] [CrossRef]
- Ma, R.; Wang, Y.; Zhao, S.; Yin, M.; Fang, W. The composition of the microbial community associated with Macrobrachium rosenbergii zoeae varies throughout larval development. Fish Dis. 2020, 43, 413–421. [Google Scholar] [CrossRef]
- Liu, B.; Song, C.; Gao, Q.; Liu, B.; Zhou, Q.; Sun, C.; Zhang, H.; Liu, M.; Tadese, D.A. Maternal and environmental microbes dominate offspring microbial colonization in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Total Environ. 2021, 790, 148062. [Google Scholar] [CrossRef]
- Cornejo-Granados, F.; Gallardo-Becerra, L.; Leonardo-Reza, M.; Ochoa-Romo, J.P.; Ochoa-Leyva, A. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. Peer J. 2018, 6, e5382. [Google Scholar] [CrossRef]
- Huang, Z.; Li, X.; Wang, L.; Shao, Z. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus Vannamei. Aquac. Res. 2016, 47, 1737–1746. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, M.; Liu, J.; Qiao, Y.; Wang, L.; Li, Z.; Zhang, X.-H.; Yu, M. Bacterial community associated with healthy and diseased Pacific white shrimp (Litopenaeus vannamei) larvae and rearing water across different growth stages. Front. Microbiol. 2017, 8, 1362. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Huang, Z.; Zeng, S.; Liu, J.; Wei, D.; Deng, X.; Weng, S.; He, Z.; He, J. Environmental factors shape water microbial community structure and function in shrimp cultural enclosure ecosystems. Front. Microbiol. 2017, 8, 2359. [Google Scholar] [CrossRef] [PubMed]
- Hammer, T.J.; McMillan, W.O.; Fierer, N. Metamorphosis of a butterfly-associated bacterial community. PLoS ONE 2014, 9, e86995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, X. Core gut microbiota of shrimp sunction as a regulator to maintain immune homeostasis in response to WSSV infection. Microbiol. Spectr. 2022, 10, e0246521. [Google Scholar]
- Muhammad, A.; Fang, Y.; Hou, Y.; Shi, Z. The gut entomotype of red palm weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae) and their rffect on host nutrition metabolism. Front. Microbiol. 2017, 8, 2291. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Ng, K.M.; Correia, M.B.; Cabral, V.; Shi, H.; Sonnenburg, J.L.; Huang, K.C.; Xavier, K.B. Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat. Microbiol. 2020, 5, 630–641. [Google Scholar] [CrossRef]
- Manfioletti, G.; Schneider, C. A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res. 1988, 16, 2873–2884. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Li, M.; Shao, D.; Zhou, J.; Gu, J.; Qin, J.; Chen, W.; Wei, W. Signatures within esophageal microbiota with progression of esophageal squamous cell carcinoma. Chin. J. Cancer Res. 2020, 32, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gaughan, S.; Chang, Q.; Chen, H.; Lu, G.; Wang, X.; Xu, L.; Zhu, L.; Jiang, J. Age-related changes in the gut microbiota of the Chinese giant salamander (Andrias davidianus). Microbiologyopen 2019, 8, e00778. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Fan, W.; Huang, M.; Liu, M. Effects of dietary Lactobacillus delbrueckii on growth performance, body composition, digestive and absorptive capacity, and gene expression of common carp (Cyprinus carpio Huanghe var). Aquac. Nutr. 2019, 25, 166–175. [Google Scholar] [CrossRef]
- Castex, M.; Leclercq, E.; Lemaire, P.; Chim, L. Dietary probiotic Pediococcus acidilactici MA18/5M improves the growth, feed performance and antioxidant status of penaeid shrimp Litopenaeus stylirostris: A Growth-Ration-Size Approach. Animals 2021, 11, 3451. [Google Scholar] [CrossRef]
- Amenyogbe, E.; Luo, J.; Fu, W.J.; Abarike, E.D.; Wang, Z.L.; Huang, J.S.; Ayisi, C.L.; Chen, G. Effects of autochthonous strains mixture on gut microbiota and metabolic profile in cobia (Rachycentron canadum). Sci. Rep. 2022, 12, 17410. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Najnine, F.; Han, H.; Wu, B.; Cai, J. BALOs Improved gut microbiota health in postlarval shrimp (Litopenaeus vannamei) after being subjected to salinity reduction treatment. Front. Microbiol. 2020, 11, 1296. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, L.; Liang, Y.; Li, P.; Zhang, T.; Meng, F.; Liu, B.; Zhang, H.; Fu, W.; Wang, W.; et al. Effects of dietary yeast culture on health status in digestive tract of juvenile Pacific white shrimp Litopenaeus Vannamei. Fish Shellfish Immunol. Rep. 2022, 3, 100065. [Google Scholar]
- Zeng, S.; Khoruamkid, S.; Kongpakdee, W.; Wei, D.; Yu, L.; Wang, H.; Deng, Z.; Weng, S.; Huang, Z.; He, J.; et al. Dissimilarity of microbial diversity of pond water, shrimp intestine and sediment in aquamimicry system. AMB Express 2020, 10, 180. [Google Scholar] [CrossRef]
- Patil, P.K.; Vinay, T.N.; Ghate, S.D.; Baskaran, V.; Avunje, S. 16 S rRNA gene diversity and gut microbial composition of the Indian white shrimp (Penaeus indicus). Antonie Van Leeuwenhoek 2021, 114, 2019–2031. [Google Scholar] [CrossRef]
- Chen, H.; Liu, F.; Ouyang, M.; Zhou, H.; Lou, B. Differences in intestinal microbial composition between red claw crayfish (Cherax quadricarinatus) and red swamp crayfish (Procambarus clarkii) cultured in pond. Fishes 2022, 7, 241. [Google Scholar] [CrossRef]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An overview of a versatile genus with potential in industry and agriculture. Crit. Rev. Biotechnol. 2018, 38, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.K.; Raichand, R.; Kaur, I.; Kaur, C.; Pareek, S.; Mayilraj, S. Exiguobacterium himgiriensis sp. nov. a novel member of the genus Exiguobacterium, isolated from the Indian Himalayas. Antonie Van Leeuwenhoek 2013, 103, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Raichand, R.; Pareek, S.; Singh, N.K.; Mayilraj, S. Exiguobacterium aquaticum sp. nov., a member of the genus Exiguobacterium. Int. J. Syst. Evol. Microbiol. 2012, 62, 2150–2155. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, Z.; Li, Y.; Li, X.; Guan, Z.; Zheng, J. Comparative genomics of Exiguobacterium reveals what makes a cosmopolitan bacterium. Msystems 2021, 6, e0038321. [Google Scholar] [CrossRef] [PubMed]
- Cong, M.; Jiang, Q.; Xu, X.; Huang, L.; Su, Y.; Yan, Q. The complete genome sequence of Exiguobacterium arabatum W-01 reveals potential probiotic functions. MicrobiologyOpen 2017, 6, e496. [Google Scholar] [CrossRef]
- Cai, M.; Deng, H.; Sun, H.; Si, W.; Li, X.; Hu, J.; Huang, M.; Fan, W. Changes of intestinal microbiota in the giant salamander (Andrias davidianus) during growth based on high-throughput sequencing. Front. Microbiol. 2023, 14, 1052824. [Google Scholar] [CrossRef]
- Ling, F.; Steinel, N.; Weber, J.; Ma, L.; Smith, C.; Correa, D.; Zhu, B.; Bolnick, D.; Wang, G. The gut microbiota response to helminth infection depends on host sex and genotype. ISME J. 2020, 14, 1141–1153. [Google Scholar] [CrossRef]
- Meziti, A.; Mente, E.; Kormas, K.A. Gut bacteria associated with different diets in reared Nephrops norvegicus. Syst. Appl. Microbiol. 2012, 35, 473–482. [Google Scholar] [CrossRef]
- Fraune, S.; Zimmer, M. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 2008, 10, 2497–2504. [Google Scholar] [CrossRef]
- Ooi, M.C.; Goulden, E.F.; Smith, G.G.; Nowak, B.F.; Bridle, A.R. Developmental and gut-related changes to microbiomes of the cultured juvenile spiny lobster Panulirus ornatus. FEMS Microbiol. Ecol. 2017, 93, 1–10. [Google Scholar] [CrossRef]
- Sui, J.; Luan, S.; Yang, G.; Xia, Z.; Luo, K.; Tang, Q.; Lu, X.; Meng, X.; Kong, J. Genetic parameters and selection response for the harvest body weight of the giant freshwater prawn (Macrobrachium rosenbergii) in a breeding program in China. PLoS ONE 2019, 14, e0218379. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.L.; Liu, Z.Y.; Jin, Y.M.; Liu, Z.X.; Zhang, B.Y.; Yuan, Z.H.; Ye, J.D.; Sun, Y.Z. Preventive and reparative functions of host-associated probiotics against soybean meal induced growth, immune suppression and gut injury in Japanese seabass (Lateolabraxjaponicus). Fish Shellfish Immunol. 2022, 128, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Amoah, K.; Huang, Q.C.; Tan, B.P.; Zhang, S.; Chi, S.Y.; Yang, Q.H.; Liu, H.Y.; Dong, X.H. Dietary supplementation of probiotic Bacillus coagulans ATCC 7050, improves the growth performance, intestinal morphology, microflora, immune response, and disease confrontation of Pacific white shrimp, Litopenaeus Vannamei. Fish Shellfish Immunol. 2019, 87, 796–808. [Google Scholar] [CrossRef]
- Won, S.; Hamidoghli, A.; Choi, W.; Bae, J.; Jang, W.J.; Lee, S.; Bai, S.C. Evaluation of potential probiotics Bacillus subtilis WB60, Pediococcus pentosaceus, and Lactococcus lactis on growth performance, immune response, gut histology and immune-related genes in whiteleg shrimp, Litopenaeus Vannamei. Microorg. 2020, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- Adel, M.; El-Sayed, A.M.; Yeganeh, S.; Dadar, M.; Giri, S.S. Effect of potential probiotic Lactococcus lactis Subsp. lactis on growth performance, intestinal microbiota, digestive enzyme activities, and disease resistance of Litopenaeus vannamei. Probiotics Antimicrob. Proteins 2017, 9, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Wen, M.; Shen, H.; Zhang, S.; Jiang, G.; Qiao, Y.; Cheng, J.; Cao, X.; Wan, X.; Sun, X. Intestinal microbiota differences in Litopenaeus vannamei shrimp between greenhouse and aquaponic rearing. Life 2023, 13, 525. [Google Scholar] [CrossRef] [PubMed]
- Elizarov, S.M.; Mironov, V.A.; Danilenko, V.N. Dynamics of serine/threonine protein kinase activity during the growth of the wild-type Streptomyces avermitilis strain and its chloramphenicol-resistant mutant. Microbiology 2000, 69, 281–286. [Google Scholar] [CrossRef]
- Richardson, L.A.; Reed, B.J.; Charette, J.M.; Freed, E.F.; Fredrickson, E.K.; Locke, M.N.; Baserga, S.J.; Gardner, R.G. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep. 2012, 2, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Zeng, S.; Zhou, R.; Hou, D.; Bao, S.; Zhang, L.; Hou, Q.; Li, X.; Weng, S.; He, J.; et al. Phage-prokaryote coexistence strategy mediates microbial community diversity in the intestine and sediment microhabitats of shrimp culture pond ecosystem. Front. Microbiol. 2022, 13, 1011342. [Google Scholar] [CrossRef] [PubMed]
- Interaminense, J.A.; Vogeley, J.L.; Gouveia, C.K.; Portela, R.S.; Oliveira, J.P.; Silva, S.M.B.C.; Coimbra, M.R.M.; Peixoto, S.M.; Soares, R.B.; Buarque, D.S.; et al. Effects of dietary Bacillus subtilis and Shewanella algae in expression profile of immune-related genes from hemolymph of Litopenaeus vannamei challenged with Vibrio parahaemolyticus. Fish Shellfish Immunol. 2019, 86, 253–259. [Google Scholar] [CrossRef]
- Natnan, M.E.; Low, C.F.; Chong, C.M.; Bunawan, H.; Baharum, S.N. Oleic acid as potential immunostimulant in metabolism pathways of hybrid grouper fingerlings (Epinephelus fuscoguttatus × Epinephelus lanceolatus) infected with Vibrio vulnificus. Sci. Rep. 2023, 13, 12830. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.J. Intestinal peptidases. Gut 1970, 11, 720–725. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tassanakajon, A.; Rimphanitchayakit, V.; Visetnan, S.; Amparyup, P.; Somboonwiwat, K.; Charoensapsri, W.; Tang, S. Shrimp humoral responses against pathogens: Antimicrobial peptides and melanization. Dev. Comp. Immunol. 2018, 80, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Schubiger, C.B.; Orfe, L.H.; Sudheesh, P.S.; Cain, K.D.; Shah, D.H.; Call, D.R. Entericidin is required for a probiotic treatment (Enterobacter sp. strain C6-6) to protect trout from cold-water disease challenge. Appl. Environ. Microbiol. 2015, 81, 658–665. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).