Development of a Multiplex Polymerase Chain Reaction Method for Rapid and Accurate Identification of Girella punctata and G. leonina (Teleostei: Girellidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. PCR, Sequencing, Primer Design, and Data Analysis
2.3. Species-Specific Primer Design
2.4. Multiplex Species-Specific (MSS) PCR
2.5. Specificity and Sensitivity of MSS-PCR
3. Results and Discussion
3.1. Species Identification Based on mtDNA COI
3.2. Sequencing of COI Gene and Genetic Diversity Analysis
3.3. Multiplex Species-Specific Primer
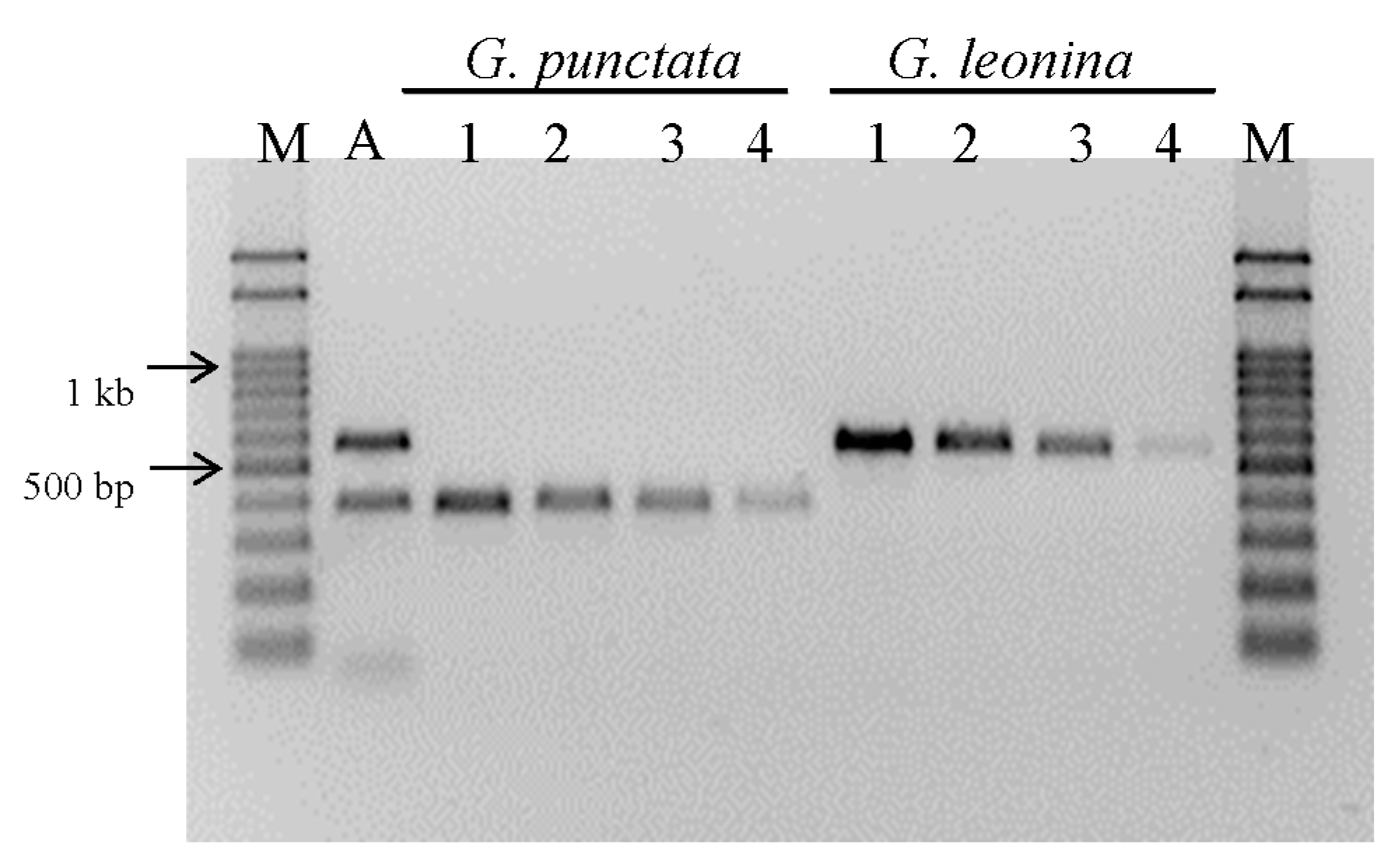
3.4. PCR Specificity and Sensitivity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2023. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 2 August 2023).
- Lim, S.G.; Jeong, M.H.; Lee, T.H.; Gil, H.W.; Park, I.S. Comparison of Morphological Characteristics between Smallscale Blackfish, Girella leonine and Largescale Blackfish, G. punctata. J. Fish. Mar. Sci. Educ. 2016, 28, 1848–1857. [Google Scholar]
- Yagishita, N.; Nakabo, T. Evolutionary trend in feeding habits of Girella (Perciformes: Girellidae). Ichthyol. Res. 2003, 50, 358–366. [Google Scholar] [CrossRef]
- Yagishita, N.; Nakabo, T. Revision of the genus Girella (Girellidae) from East Asia. Ichthyol. Res. 2000, 47, 119–135. [Google Scholar] [CrossRef]
- Itoi, S.; Saito, T.; Shimojo, M.; Washio, S.; Sugita, H. Identification of Girella punctate and G. leonine by PCR-RFLP analysis. J. Mar. Sci. 2007, 64, 328–331. [Google Scholar]
- Okuno, R. Distribution of youngs of two reef fishes, Girella punctata Gray and G. melanichthys (Richardson), in Tanabe Bay and the relationship found between their schooling behaviors. Publ. Seto Mar. Biol. Lab. 1962, 10, 293–306. [Google Scholar] [CrossRef]
- Okuno, R. Observations and discussions on the social behaviors of marine fishes. Publ. Seto Mar. Biol. Lab. 1963, 11, 281–336. [Google Scholar] [CrossRef]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Lee, G.Y.; Suh, S.M.; Lee, Y.M.; Kim, H.Y. Multiplex PCR Assay for Simultaneous Identification of Five Types of Tuna (Katsuwonus pelamis, Thunnus alalonga, T. albacares, T. obesus and T. thynnus). Foods 2022, 11, 280. [Google Scholar] [CrossRef]
- Noh, E.S.; Lee, M.L.; Kim, E.M.; Park, J.Y.; Noh, J.K.; An, C.M.; Kang, J.H. Development of a Multiplex PCR Assay for Rapid Identification of Larimichthys polyactis, L. crocea, Atrobucca nibe, and Peseudotolithus elongates. J. Life Sci. 2017, 27, 746–753. [Google Scholar]
- Asensio Gil, L. PCR-based methods for fish and fishery products authentication. Trends Food Sci. Technol. 2007, 18, 558–566. [Google Scholar] [CrossRef]
- Axayacatl, R.O.; Juan, P.C.G. Molecular identification of dolphinfish species (genus Coryphaena) using multiplex haplotype-specific PCR of mitochondrial DNA. Ichthyol. Res. 2008, 55, 389–393. [Google Scholar]
- Dawnay, N.; Ogden, R.; Thorpe, R.S.; Pope, L.C.; Dawson, D.A.; McEwing, R. A forensic STR profiling system for the Eurasian badger: A framework for developing profiling systems for wildlife species. Forensic Sci. Int. Genet. 2008, 2, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Dong, C.M.; Lee, M.N.; Noh, J.K.; Noh, E.S.; Nam, B.H.; Kim, Y.O.; Jung, H.S. Development of multiplex species-specific PCR for the simultaneous identification of three closely related species in the genera Misgurnus and Paramisgurnus. Aquac. Rep. 2022, 24, 101–144. [Google Scholar] [CrossRef]
- Parvez, I.; Mahajebin, T.; Clarke, M.L.; Chhanda, M.S.; Sultana, S. Genetic variation of native and introduced climbing perch Anabas testudineus (Bloch, 1792) derived from mitochondrial DNA analyses. Ecol. Genet. Genom. 2020, 17, 100067. [Google Scholar] [CrossRef]
- Lee, Y.W.; Lee, S.H.; Xin, C.F.; Shin, J.H.; Shin, E.H. Development of a multiplex PCR system for the simultaneous detection of the shrimp species Fenneropenaeus chinensis, Litopenaeus vannamei, and Penaeus monodon. J. AOAC Int. 2017, 100, 104–108. [Google Scholar] [CrossRef]
- Zuo, T.; Li, Z.; Lv, Y.; Duan, G.; Wang, C.; Tang, Q.; Xue, C. Rapid identification of sea cucumber species with multiplex-PCR. Food Control 2012, 26, 58–62. [Google Scholar] [CrossRef]
- Henegariu, O.; Heerema, N.A.; Dlouhy, S.R.; Vance, G.H.; Vogt, P.H. Multiplex PCR: Critical parameters and step-by-step protocol. Biotechniques 1997, 23, 504–511. [Google Scholar] [CrossRef] [PubMed]




| Oligo Name | Sequence (5′ → 3′) | Mer | Tm (°C) | GC (%) | Product Size |
|---|---|---|---|---|---|
| GR-207F | AGT AAT ACC AAT TAT GAT TGG A | 22 | 56 | 28 | 991 bp |
| GR-1181R | ATA GTG GGA ATC AGT GTA | 18 | 39 |
| Oligo Name | Sequence (5′ → 3′) | Target Species | Tm (°C) | Product Size (bp) |
|---|---|---|---|---|
| GP-825F | CTA CAT GGG TAT AGT TTG A | Girella punctata | 50 | 391 |
| GL-639F | AAT ACT TCT CAC AGA CCG A | Girella leonina | 579 | |
| GR-1181R | ATA GTG GGA ATC AGT GTA | - |
| Species | Polymorphic Sites | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Base Pair (bp) | 288 | 291 | 327 | 336 | 345 | 363 | 369 | 378 | 396 | 429 | 441 |
| Girella punctata | A | A | T | A | T | A | C | A | T | A | T |
| Girella leonina | G | G | C | G | C | T | A | G | C | G | C |
| 475 | 483 | 486 | 495 | 501 | 504 | 534 | 546 | 558 | 564 | 582 | |
| T | T | T | T | C | C | A | T | G | A | C | |
| C | A | C | C | T | T | G | C | A | G | A | |
| 583 | 594 | 600 | 621 | 630 | 633 | 636 | 639 | 678 | 696 | 717 | |
| C | T | T | C | T | G | T | G | T | G | T | |
| T | C | C | A | C | A | C | A | A | A | C | |
| 756 | 768 | 810 | 816 | 819 | 825 | 849 | 852 | 864 | 867 | 873 | |
| T | T | C | T | A | A | A | C | G | T | C | |
| C | C | T | C | G | G | T | T | A | C | T | |
| 966 | 975 | 978 | 984 | 996 | 1008 | 1017 | 1023 | 1035 | 1044 | 1047 | |
| T | G | T | T | T | G | A | C | T | T | A | |
| C | A | C | C | C | C | C | T | C | C | G | |
| 1065 | 1089 | 1107 | |||||||||
| A | G | T | |||||||||
| G | A | C | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.-M.; Lee, M.-N.; Dong, C.-M.; Noh, J.-K.; Noh, E.-S.; Kim, W.-J.; Nam, B.-H.; Kim, Y.-O. Development of a Multiplex Polymerase Chain Reaction Method for Rapid and Accurate Identification of Girella punctata and G. leonina (Teleostei: Girellidae). Fishes 2023, 8, 415. https://doi.org/10.3390/fishes8080415
Kim E-M, Lee M-N, Dong C-M, Noh J-K, Noh E-S, Kim W-J, Nam B-H, Kim Y-O. Development of a Multiplex Polymerase Chain Reaction Method for Rapid and Accurate Identification of Girella punctata and G. leonina (Teleostei: Girellidae). Fishes. 2023; 8(8):415. https://doi.org/10.3390/fishes8080415
Chicago/Turabian StyleKim, Eun-Mi, Mi-Nan Lee, Chun-Mae Dong, Jae-Koo Noh, Eun-Soo Noh, Woo-Jin Kim, Bo-Hye Nam, and Young-Ok Kim. 2023. "Development of a Multiplex Polymerase Chain Reaction Method for Rapid and Accurate Identification of Girella punctata and G. leonina (Teleostei: Girellidae)" Fishes 8, no. 8: 415. https://doi.org/10.3390/fishes8080415
APA StyleKim, E.-M., Lee, M.-N., Dong, C.-M., Noh, J.-K., Noh, E.-S., Kim, W.-J., Nam, B.-H., & Kim, Y.-O. (2023). Development of a Multiplex Polymerase Chain Reaction Method for Rapid and Accurate Identification of Girella punctata and G. leonina (Teleostei: Girellidae). Fishes, 8(8), 415. https://doi.org/10.3390/fishes8080415






