Effects of LPS, Poly (I:C) and Edwardsiella tarda on the Expression Patterns of IL-17 Family Members and Their Receptors in Spotted Sea Bass (Lateolabrax maculatus)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Cloning and Identification of IL-17 Ligand and Receptor Genes
2.3. Sequence Analysis
2.4. Tissue Expression Analysis of the IL-17 Ligand and Receptor Genes
2.5. Expression Analysis of the IL-17 Ligand and Receptor Genes after LPS/Poly (I:C)/Edwardsiella tarda Infection
2.6. Statistical Analysis
3. Results and Discussion
3.1. Identification and Sequence Analysis of the IL-17 Ligand and Receptor Genes
3.2. Tissue Distribution of the IL-17 Ligands and Receptor
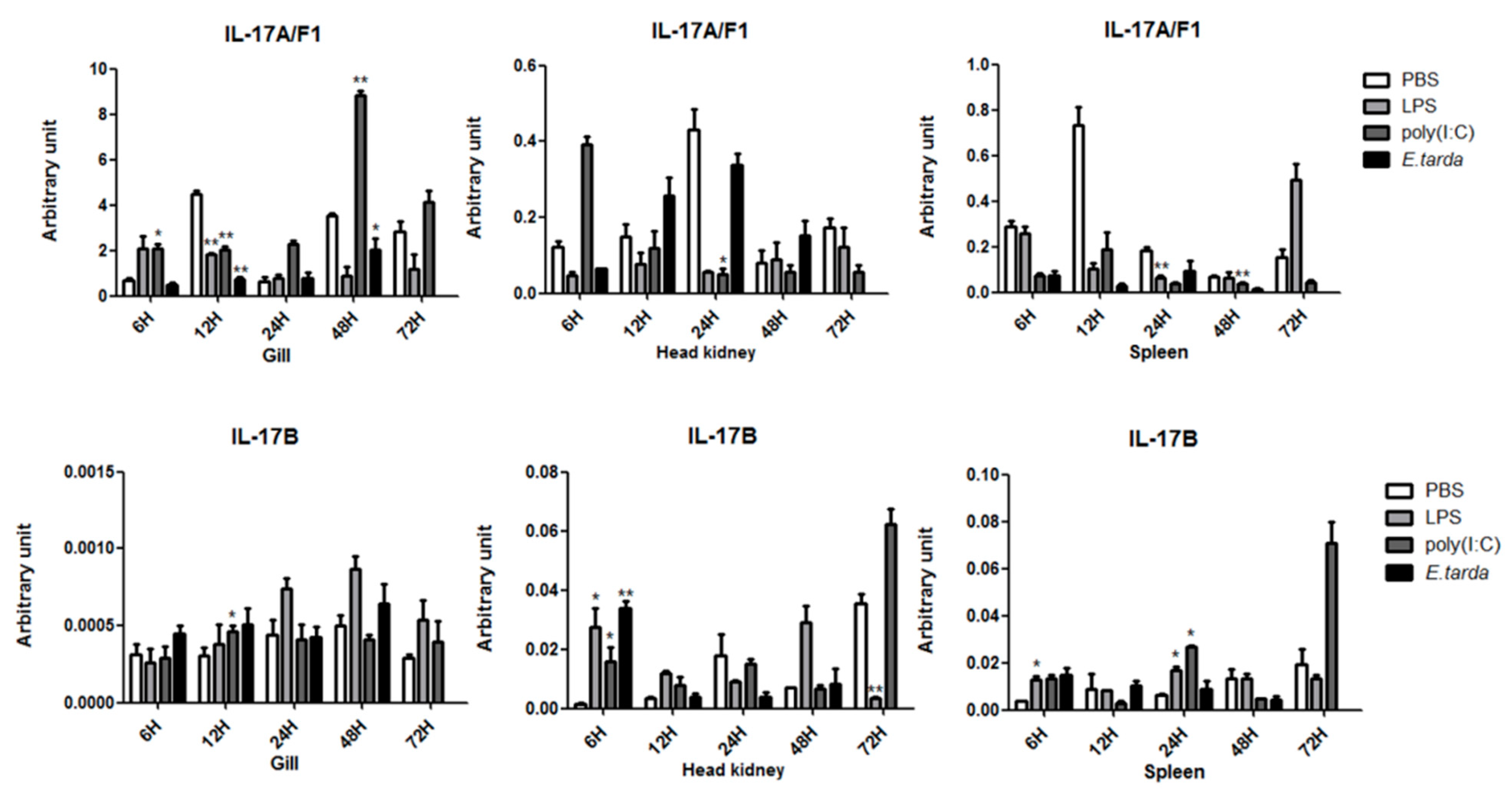
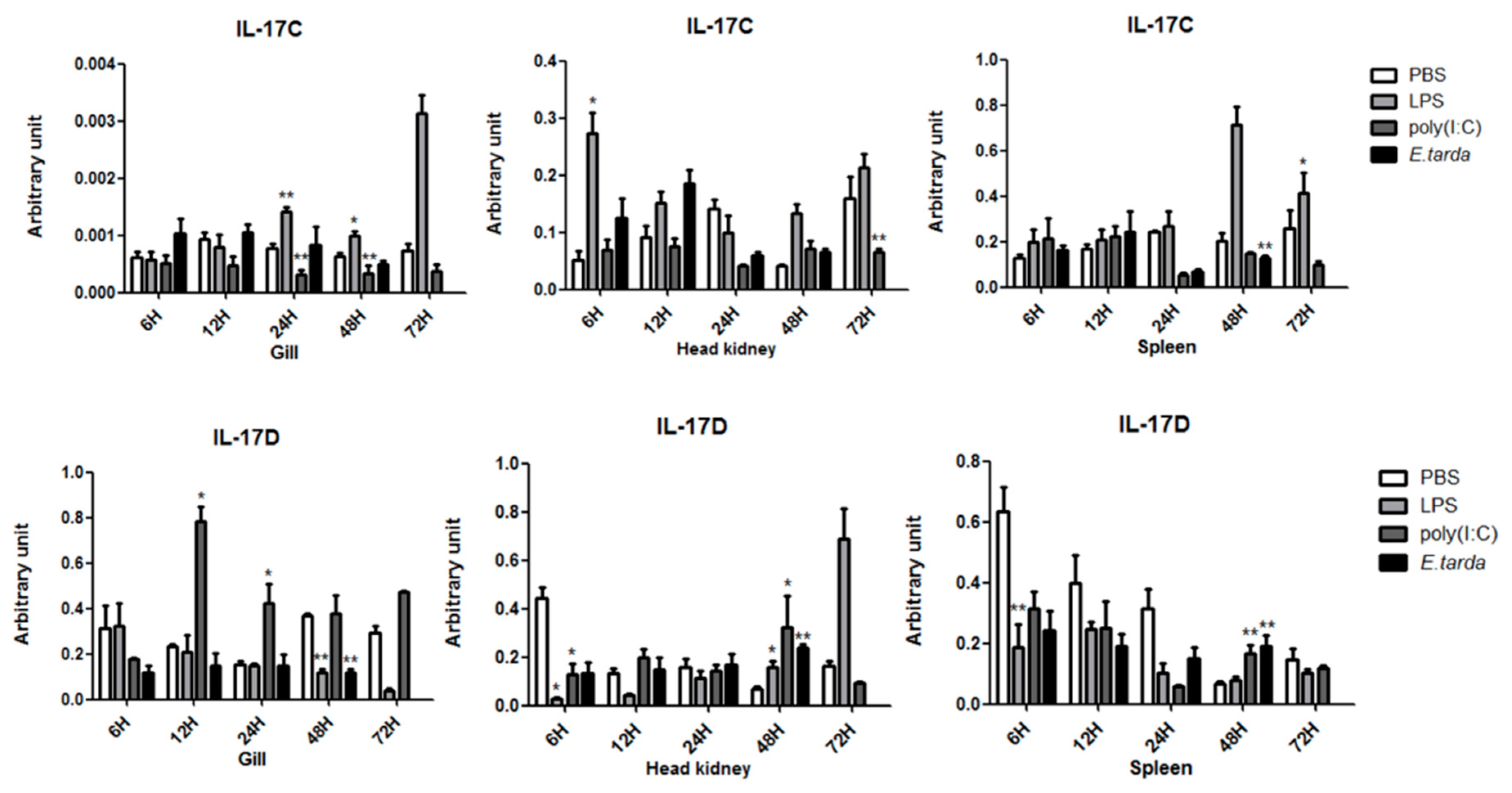
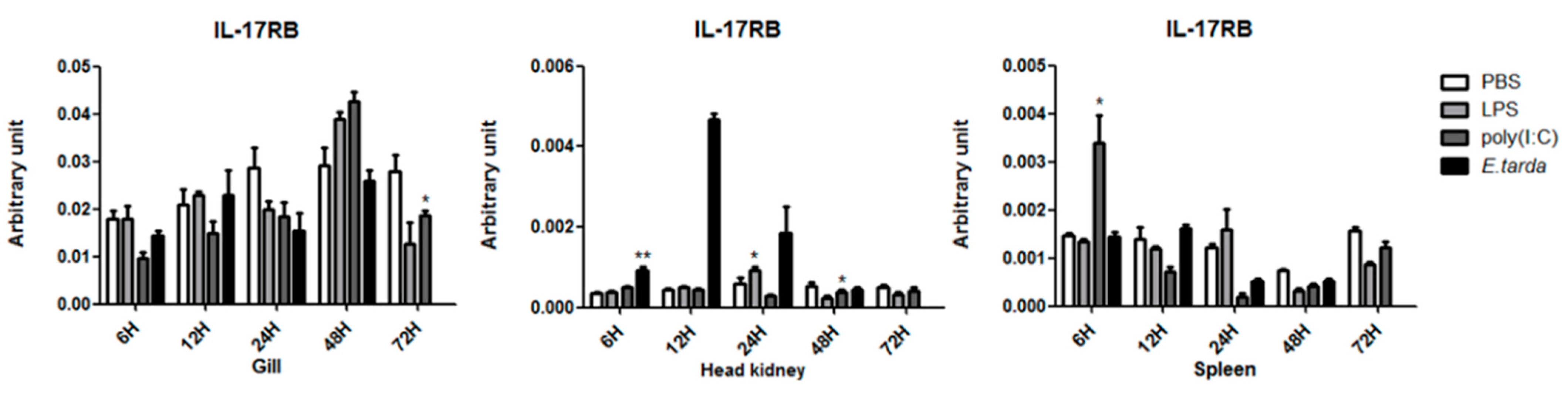
3.3. The mRNA Expression Status of the IL-17 Ligand and Receptor Genes after Injection with LPS/Poly (I:C)/E. tarda
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holdsworth, S.R.; Gan, P.Y. Cytokines: Names and Numbers You Should Care About. Clin. J. Am. Soc. Nephrol. 2015, 10, 2243–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Sun, Y.N.; Su, X.R.; Li, T.W.; Xu, T.J. Characterization of six IL-17 family genes in miiuy croaker and evolution analysis of vertebrate IL-17 family. Fish Shellfish Immun. 2016, 49, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenberger, P.; Kolls, J.K. Interleukin 17: An example for gene therapy as a tool to study cytokine mediated regulation of hematopoiesis. J. Cell Biochem. 2002, 85, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Z.; Chen, J.; Huang, A.; Stinson, J.; Heldens, S.; Foster, J.; Dowd, P.; Gurney, A.L.; Wood, W.I. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc. Natl. Acad. Sci. USA 2000, 97, 773–778. [Google Scholar] [CrossRef]
- Sanders, A.J.; Guo, X.; Mason, M.D.; Jiang, W.G. IL-17B Can Impact on Endothelial Cellular Traits Linked to Tumour Angiogenesis. J. Oncol. 2010, 2010, 817375. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Carrozzi, V.; Sambandam, A.; Luis, E.; Lin, Z.G.; Jeet, S.; Lesch, J.; Hackney, J.; Kim, J.; Zhou, M.J.; Lai, J.; et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat. Immunol. 2011, 12, 1159–1166. [Google Scholar] [CrossRef]
- Song, X.Y.; Zhu, S.; Shi, P.Q.; Liu, Y.; Shi, Y.F.; Levin, S.D.; Qian, Y.C. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat. Immunol. 2011, 12, 1151–1158. [Google Scholar] [CrossRef]
- Peng, T.; Chanthaphavong, R.S.; Sun, S.J.; Trigilio, J.A.; Phasouk, K.; Jin, L.; Layton, E.D.; Li, A.Z.; Correnti, C.E.; De van der Schueren, W.; et al. Keratinocytes produce IL-17c to protect peripheral nervous systems during human HSV-2 reactivation. J. Exp. Med. 2017, 214, 2315–2329. [Google Scholar] [CrossRef]
- Starnes, T.; Broxmeyer, H.E.; Robertson, M.J.; Hromas, R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J. Immunol. 2002, 169, 642–646. [Google Scholar] [CrossRef] [Green Version]
- Saddawi-Konefka, R.; Seelige, R.; Gross, E.T.E.; Levy, E.; Searles, S.C.; Washington, A.; Santosa, E.K.; Liu, B.C.; O’Sullivan, T.E.; Harismendy, O.; et al. Nrf2 Induces IL-17D to Mediate Tumor and Virus Surveillance. Cell Rep. 2016, 16, 2348–2358. [Google Scholar] [CrossRef] [Green Version]
- Fort, M.M.; Cheung, J.; Yen, D.; Li, J.; Zurawski, S.M.; Lo, S.; Menon, S.; Clifford, T.; Hunte, B.; Lesley, R.; et al. IL-25 induces IL-4 IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity 2001, 15, 985–995. [Google Scholar] [CrossRef] [Green Version]
- Hurst, S.D.; Muchamuel, T.; Gorman, D.M.; Gilbert, J.M.; Clifford, T.; Kwan, S.; Menon, S.; Seymour, B.; Jackson, C.; Kung, T.T.; et al. New IL-17 family members promote Th1 or Th2 responses in the lung: In vivo function of the novel cytokine IL-25. J. Immunol. 2002, 169, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Wright, J.F.; Guo, Y.; Quazi, A.; Luxenberg, D.P.; Bennett, F.; Ross, J.F.; Qiu, Y.; Whitters, M.J.; Tomkinson, K.N.; Dunussi-Joannopoulos, K.; et al. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J. Biol. Chem. 2007, 282, 13447–13455. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.H.; Dong, C. A novel heterodimeric cytokine consisting of IL-17 and IL-17F regulates inflammatory responses. Cell Res. 2007, 17, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Fernandez, C.; Chaves-Pozo, E.; Cuesta, A. Identification and Regulation of Interleukin-17 (IL-17) Family Ligands in the Teleost Fish European Sea Bass. Int. J. Mol. Sci. 2020, 21, 2439. [Google Scholar] [CrossRef] [Green Version]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Monin, L.; Gaffen, S.L. Interleukin 17 Family Cytokines: Signaling Mechanisms, Biological Activities, and Therapeutic Implications. Cold Spring Harb. Perspect. Biol. 2018, 10, a028522. [Google Scholar] [CrossRef]
- Ashfaq, H.; Soliman, H.; Saleh, M.; El-Matbouli, M. CD4: A vital player in the teleost fish immune system. Vet. Res. 2019, 50, 1. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Ai, C.; Mu, Y.; Ao, J.; Chen, X. Molecular characterization and evolution analysis of five interleukin-17 receptor genes in large yellow croaker Larimichthys crocea. Fish Shellfish Immun. 2016, 58, 332–339. [Google Scholar] [CrossRef]
- Yao, Z.B.; Fanslow, W.C.; Seldin, M.F.; Rousseau, A.M.; Painter, S.L.; Comeau, M.R.; Cohen, J.I.; Spriggs, M.K. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity 1995, 3, 811–821. [Google Scholar] [CrossRef] [Green Version]
- Doyle, M.S.; Collins, E.S.; FitzGerald, O.M.; Pennington, S.R. New insight into the functions of the interleukin-17 receptor adaptor protein Act1 in psoriatic arthritis. Arthritis Res. Ther. 2012, 14, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novatchkova, M.; Leibbrandt, A.; Werzowa, J.; Neubuser, A.; Eisenhaber, F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem. Sci. 2003, 28, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 2009, 9, 556–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, J.F.; Bennett, F.; Li, B.L.; Brooks, J.; Luxenberg, D.P.; Whitters, M.J.; Tomkinson, K.N.; Fitz, L.J.; Wolfman, N.M.; Collins, M.; et al. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J. Immunol. 2008, 181, 2799–2805. [Google Scholar] [CrossRef] [Green Version]
- Korenaga, H.; Kono, T.; Sakai, M. Isolation of seven IL-17 family genes from the Japanese pufferfish Takifugu rubripes. Fish Shellfish Immun. 2010, 28, 809–818. [Google Scholar] [CrossRef]
- Chang, S.H.; Reynolds, J.M.; Pappu, B.P.; Chen, G.; Martinez, G.J.; Dong, C. Interleukin-17C Promotes Th17 Cell Responses and Autoimmune Disease via Interleukin-17 Receptor E. Immunity 2011, 35, 611–621. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Ho, W.H.; Maruoka, M.; Corpuz, R.T.; Baldwin, D.T.; Foster, J.S.; Goddard, A.D.; Yansura, D.G.; Vandlen, R.L.; Wood, W.I.; et al. IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J. Biol. Chem. 2001, 276, 1660–1664. [Google Scholar] [CrossRef] [Green Version]
- Rong, Z.L.; Wang, A.A.; Li, Z.Y.; Ren, Y.M.; Cheng, L.; Li, Y.H.; Wang, Y.Y.; Ren, F.L.; Zhang, X.N.; Hu, J.; et al. IL-17RD (Sef or IL-17RLM) interacts with IL-17 receptor and mediates IL-17 signaling. Cell Res. 2009, 19, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Song, X.Y.; Qian, Y.C. The activation and regulation of IL-17 receptor mediated signaling. Cytokine 2013, 62, 175–182. [Google Scholar] [CrossRef]
- Gunimaladevi, I.; Savan, R.; Sakai, M. Identification, cloning and characterization of interleukin-17 and its family from zebrafish. Fish Shellfish Immun. 2006, 21, 393–403. [Google Scholar] [CrossRef]
- Kono, T.; Korenaga, H.; Sakai, M. Genomics of fish IL-17 ligand and receptors: A review. Fish Shellfish Immun. 2011, 31, 635–643. [Google Scholar] [CrossRef]
- Wang, T.H.; Martin, S.A.M.; Secombes, C.J. Two interleukin-17C-like genes exist in rainbow trout Oncorhynchus mykiss that are differentially expressed and modulated. Dev. Comp. Immunol. 2010, 34, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.M.; Wang, T.H.; Holland, J.W.; Zou, J.; Secombes, C.J. Cloning and Characterization of Rainbow Trout Interleukin-17A/F2 (IL-17A/F2) and IL-17 Receptor A: Expression during Infection and Bioactivity of Recombinant IL-17A/F2. Infect. Immun. 2013, 81, 340–353. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.H.; Jiang, Y.S.; Wang, A.; Husain, M.; Xu, Q.Q.; Secombes, C.J. Identification of the salmonid IL-17A/F1a/b, IL-17A/F2b, IL-17A/F3 and IL-17N genes and analysis of their expression following in vitro stimulation and infection. Immunogenetics 2015, 67, 395–412. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Q.; Li, C.; Thongda, W.; Luo, Y.P.; Beck, B.; Peatman, E. Characterization and mucosal responses of interleukin 17 family ligand and receptor genes in channel catfish Ictalurus punctatus. Fish Shellfish Immun. 2014, 38, 47–55. [Google Scholar] [CrossRef]
- Dong, C.J.; Kong, S.N.; Zheng, X.H.; Zhang, J.F.; Nie, G.X.; Li, X.J.; Xu, P. Genome-wide identification of interleukin-17 (IL17) in common carp (Cyprinus carpio) and its expression following Aeromonas hydrophila infection. Gene 2019, 686, 68–75. [Google Scholar] [CrossRef]
- Li, H.X.; Yu, J.H.; Li, J.L.; Tang, Y.K.; Yu, F.; Zhou, J.; Yu, W.J. Cloning and characterization of two duplicated interleukin-17A/F2 genes in common carp (Cyprinus carpio L.): Transcripts expression and bioactivity of recombinant IL-17A/F2. Fish Shellfish Immun. 2016, 51, 303–312. [Google Scholar] [CrossRef]
- Li, H.X.; Zhang, L.; Li, J.L.; Yu, F.; Wang, M.Y.; Wang, Q.; Wu, Y.S.; Zhang, Q.Y.; Tang, Y.K.; Yu, J.H. Identification, expression and pro-inflammatory effect of interleukin-17 N in common carp (Cyprinus carpio L.). Fish Shellfish Immun. 2021, 111, 6–15. [Google Scholar] [CrossRef]
- Ding, Y.; Ao, J.Q.; Ai, C.X.; Chen, X.H. Molecular and functional identification of three interleukin-17A/F (IL-17A/F) homologues in large yellow croaker (Larimichthys crocea). Dev. Comp. Immunol. 2016, 55, 221–232. [Google Scholar] [CrossRef]
- Ding, Y.; Ao, J.Q.; Chen, X.H. Comparative study of interleukin-17C (IL-17C) and IL-17D in large yellow croaker Larimichthys crocea reveals their similar but differential functional activity. Dev. Comp. Immunol. 2017, 76, 34–44. [Google Scholar] [CrossRef]
- Du, L.Y.; Qin, L.; Wang, X.Y.; Zhang, A.Y.; Wei, H.; Zhou, H. Characterization of grass carp (Ctenopharyngodon idella) IL-17D: Molecular cloning, functional implication and signal transduction. Dev. Comp. Immunol. 2014, 42, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Du, L.Y.; Feng, S.Y.; Yin, L.C.; Wang, X.Y.; Zhang, A.Y.; Yang, K.; Zhou, H. Identification and functional characterization of grass carp IL-17A/F1: An evaluation of the immunoregulatory role of teleost IL-17A/F1. Dev. Comp. Immunol. 2015, 51, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Song, R.X.; Wang, X.W.; Hu, H.X.; Zhang, Z.B. Peritoneal bacterial infection repressed the expression of IL17D in Siberia sturgeon a chondrostean fish in the early immune response. Fish Shellfish Immun. 2017, 64, 39–48. [Google Scholar] [CrossRef]
- Chi, H.; Sun, L. Comparative study of four interleukin 17 cytokines of tongue sole Cynoglossus semilaevis: Genomic structure, expression pattern, and promoter activity. Fish Shellfish Immun. 2015, 47, 321–330. [Google Scholar] [CrossRef]
- Costa, M.M.; Pereiro, P.; Wang, T.; Secombes, C.J.; Figueras, A.; Novoa, B. Characterization and gene expression analysis of the two main Th17 cytokines (IL-17A/F and IL-22) in turbot, Scophthalmus maximus. Dev. Comp. Immunol. 2012, 38, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.B.; Tian, Y.; Wen, H.S.; Liu, Y.; Sun, Y.L.; Yanglang, A.; Li, Y. Effects of Vibrio harveyi infection on serum biochemical parameters and expression profiles of interleukin-17 (IL-17)/interleukin-17 receptor (IL-17R) genes in spotted sea bass. Dev. Comp. Immunol. 2020, 110, 103731. [Google Scholar] [CrossRef]
- Okamura, Y.; Morimoto, N.; Sawada, S.; Kono, T.; Hikima, J.; Sakai, M. Molecular characterization and expression of two interleukin-17 receptor A genes on different chromosomes in Japanese medaka, Oryzias latipes. Comp. Biochem. Phys. B 2020, 240, 110386. [Google Scholar] [CrossRef]
- Harada, N.; Okamura, Y.; Kono, T.; Sakai, M.; Hikima, J. Identification of two interleukin 17 receptor C (IL-17RC) genes and their binding activities to three IL-17A/F ligands in the Japanese medaka, Oryzias latipes. Dev. Comp. Immunol. 2021, 124, 104179. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Cao, M.; Xue, T.; Yu, H.H.; Yang, T.Z.; Yan, X.; Li, C. Characterization of IL-17/IL-17R gene family in Sebastes schlegelii and their expression profiles under Aeromonas salmonicida and Edwardsiella piscicida infections. Aquaculture 2022, 551, 737901. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, X.X.; Zhang, G.R.; Ji, W.; Shi, Z.C.; Ma, X.F.; Wei, K.J. Molecular characterization, expression and function analysis of interleukin-17B, C and D genes in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2022, 552, 737962. [Google Scholar] [CrossRef]
- Sun, Z.S.; Qin, Y.T.; Liu, D.J.; Wang, B.J.; Jia, Z.; Wang, J.Y.; Gao, Q.; Zou, J.; Pang, Y. The evolution and functional characterization of CXC chemokines and receptors in lamprey. Dev. Comp. Immunol. 2021, 116, 103905. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Oliver, T.; Schmidt, B.; Nathan, D.; Clemens, R.; Maskell, D. Using reconfigurable hardware to accelerate multiple sequence alignment with ClustalW. Bioinformatics 2005, 21, 3431–3432. [Google Scholar] [CrossRef] [PubMed]
- Atteson, K. The performance of neighbor-joining methods of phylogenetic reconstruction. Algorithmica 1999, 25, 251–278. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Yi, L.; Nie, P.; Yu, H.B.; Xie, H.X. Regulation of Type III Secretion of Translocon and Effector Proteins by the EsaB/EsaL/EsaM Complex in Edwardsiella tarda. Infect. Immun. 2017, 85, e00322-1. [Google Scholar] [CrossRef] [Green Version]
- Pressley, M.E.; Phelan, P.E.; Witten, P.E.; Mellon, M.T.; Kim, C.H. Pathogenesis and inflammatory response to Edwardsiella tarda infection in the zebrafish. Dev. Comp. Immunol. 2005, 29, 501–513. [Google Scholar] [CrossRef]
- Huang, X.D.; Zhang, H.; He, M.X. Comparative and Evolutionary Analysis of the Interleukin 17 Gene Family in Invertebrates. PLoS ONE 2015, 10, e0132802. [Google Scholar] [CrossRef] [PubMed]
- Swain, P.; Nayak, S.K.; Nanda, P.K.; Dash, S. Biological effects of bacterial lipopolysaccharide (endotoxin) in fish: A review. Fish Shellfish Immun. 2008, 25, 191–201. [Google Scholar] [CrossRef]
- Okuda, J.; Arikawa, Y.; Takeuchi, Y.; Mahmoud, M.M.; Suzaki, E.; Kataoka, K.; Suzuki, T.; Okinaka, Y.; Nakai, T. Intracellular replication of Edwardsiella tarda in murine macrophage is dependent on the type III secretion system and induces an up-regulation of anti-apoptotic NF-kappa B target genes protecting the macrophage from staurosporine-induced apoptosis. Microb. Pathog. 2006, 41, 226–240. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Yuan, S.Y.; Sun, Z.S.; Lei, L.N.; Wan, S.; Wang, J.Y.; Zou, J.; Gao, Q. Gene identification and functional analysis of peptidoglycan recognition protein from the spotted sea bass (Lateolabrax maculatus). Fish Shellfish Immun. 2020, 106, 1014–1024. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, K.W.; Jung, H.S.; Kim, J.; Yun, A.; Cho, S.H.; Kwon, M. Effects of dietary inclusion of yacon, ginger and blueberry on growth, body composition and challenge test of juvenile rockfish (Sebastes schlegeli) against Edwardsiella tarda. Aquacult. Nutr. 2018, 24, 1048–1055. [Google Scholar] [CrossRef]
- Ling, S.H.M.; Wang, X.H.; Xie, L.; Lim, T.M.; Leung, K.Y. Use of green fluorescent protein (GFP) to study the invasion pathways of Edwardsiella tarda in in vivo and in vitro fish models. Microbiology 2000, 146, 7–19. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, B.R.; Sahoo, P.K. Edwardsiellosis in fish: A brief review. J. Biosci. 2007, 32, 1331–1344. [Google Scholar] [CrossRef]
- Wu, J.Y.; Liu, G.B.; Sun, Y.C.; Wang, X.P.; Fang, H.; Jiang, H.; Guo, Z.M.; Dong, J.G. The role of regulator FucP in Edwardsiella tarda pathogenesis and the inflammatory cytokine response in tilapia. Fish Shellfish Immun. 2018, 80, 624–630. [Google Scholar] [CrossRef]
- Reddacliff, G.L.; Hornitzky, M.; Whittington, R.J. Edwardsiella tarda septicaemia in rainbow trout (Oncorhynchus mykiss). Aust. Vet. J. 1996, 73, 30. [Google Scholar] [CrossRef]
- Li, M.; Wang, Q.L.; Lu, Y.; Chen, S.L.; Li, Q.; Sha, Z.X. Expression profiles of NODs in channel catfish (Ictalurus punctatus) after infection with Edwardsiella tarda, Aeromonas hydrophila, Streptococcus iniae and channel catfish hemorrhage reovirus. Fish Shellfish Immun. 2012, 33, 1033–1041. [Google Scholar] [CrossRef]



| Gene | Primers | Sequence (5′ to 3′) | Application |
|---|---|---|---|
| IL-17A/F1 | F-long | GGTGACATCATCAGAGGAAGTCTATGC | PCR cloning |
| R-long | TGCTGGTGTTGGATGATGGGTC | PCR cloning | |
| F-short | TGACGGCGATCAGGATAAATGT | PCR cloning | |
| R-short | ACAGCGATGAGGCGGGACT | PCR cloning | |
| 5′R-long | AAAGCGTTGGGGTCAAGCTGCAG | 5′-RACE | |
| 5′R-short | GAGTGTCCTCGTCCTCCTTCTTGGTTC | 5′-RACE | |
| 3′F-long | CAGCATCAAAGTGCTCCAAATGACGGC | 3′-RACE | |
| 3′F-short | GTTCCTAGCAGACACATCAGGCCACT | 3′-RACE | |
| F | GGACACACAACATCAGCCATGATGAC | qRT-PCR | |
| R | AGCAGCACCTGGTGCATGAT | qRT-PCR | |
| IL-17B | F-PCR | ATGGAGCCGCGGGTCACACTCA | PCR cloning |
| R-PCR | AGGCCACTGGGAGCATAAGAACTT | PCR cloning | |
| 5′R-long | CCCGTCGATGATGCAGCCCTCAC | 5′-RACE | |
| 5′R-short | CGACGGTCTCGTGCCTCCAGTTG | 5′-RACE | |
| 3′F-long | CGTGATATCGCTTTTGCCGAGTGTGT | 3′-RACE | |
| 3′F-short | CAGGAAGTTCTTATGCTCCCAGTGGC | 3′-RACE | |
| F | TGGACACACAGAATAGACCGAGACG | qRT-PCR | |
| R | CAGGCCACTGGGAGCATAAGAACTT | qRT-PCR | |
| IL-17C | F-PCR | ATGATAGCAAGTAAAAGCAAAGCCGAACC | PCR cloning |
| R-PCR | GTACAGCCCACAGCTACTTCC | PCR cloning | |
| 5′R-long | CCGGTTCATCTTGCACGTCCACACG | 5′-RACE | |
| 5′R-short | TGTGAACGAGGGGTTCGGCTTTGCT | 5′-RACE | |
| 3′F-long | CAGGATGCATCCTAATTCCGGACAAGTC | 3′-RACE | |
| 3′F-short | AAGAGGGAGCTCTGCAGTGATGGAAAG | 3′-RACE | |
| F | CTCATCTTTGGTCTTATCCTCG | qRT-PCR | |
| R | GCTCCTCAGCTTCCTCTCTACT | qRT-PCR | |
| IL-17D | F-long | TCCTGCTGCTGCTGCACCT | PCR cloning |
| R-long | CCTTTCTTGCCCGCAGAGACGA | PCR cloning | |
| F-short | TGCTGCTGGGCTGGTCGG | PCR cloning | |
| R-short | CGGCTTTGGGCTCTGCTCT | PCR cloning | |
| 5′R-long | CAGGCTCTGGTTGCTGTTGTGGC | 5′-RACE | |
| 5′R-short | CTTCCTGGTGGCCTTCTTCCGCA | 5′-RACE | |
| 3′F-long | CAGAGGCGTACTGCCTGTGT | 3′-RACE | |
| 3′F-short | CTGAGGAGGATGGGCTCCTGTGT | 3′-RACE | |
| F | CTACGCTCCGTCCGTCATC | qRT-PCR | |
| R | CGGCTTTGGGCTCTGCTCT | qRT-PCR | |
| IL-17RB | F-PCR | CCCTCCTATCAGTTCTCACCCT | PCR cloning |
| R-PCR | AGCTTCCCTTAACTGCGCTGC | PCR cloning | |
| 5′R-long | CCGCTGATGCTGTAGATGCCAGTGCTT | 5′-RACE | |
| 5′R-short | CACCAGCATCACTTTTAAGTCGGCCAGG | 5′-RACE | |
| 3′F-long | ATCCCAGCTGCAGCTCATGACCTTTACC | 3′-RACE | |
| 3′F-short | GCAGGACAAGAGGCCTCATGACCTGG | 3′-RACE | |
| F | GCCTGCCAGTCTTTTTGTCT | qRT-PCR | |
| R | TGCTCTGCTGATGTCCTTCT | qRT-PCR | |
| EF-1α | EF-1α F | ATCTCTGGATGGCACGGAGA | qRT-PCR |
| EF-1α R | CAGTGTGGTTCCGCTAGCATC | qRT-PCR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, S.; Sun, Z.; Zhang, C.; Pan, T.; Yuan, S.; Chen, Y.; Zou, J.; Gao, Q. Effects of LPS, Poly (I:C) and Edwardsiella tarda on the Expression Patterns of IL-17 Family Members and Their Receptors in Spotted Sea Bass (Lateolabrax maculatus). Fishes 2023, 8, 405. https://doi.org/10.3390/fishes8080405
Wan S, Sun Z, Zhang C, Pan T, Yuan S, Chen Y, Zou J, Gao Q. Effects of LPS, Poly (I:C) and Edwardsiella tarda on the Expression Patterns of IL-17 Family Members and Their Receptors in Spotted Sea Bass (Lateolabrax maculatus). Fishes. 2023; 8(8):405. https://doi.org/10.3390/fishes8080405
Chicago/Turabian StyleWan, Shuai, Zhaosheng Sun, Chang Zhang, Tingshuang Pan, Shuya Yuan, Yuxi Chen, Jun Zou, and Qian Gao. 2023. "Effects of LPS, Poly (I:C) and Edwardsiella tarda on the Expression Patterns of IL-17 Family Members and Their Receptors in Spotted Sea Bass (Lateolabrax maculatus)" Fishes 8, no. 8: 405. https://doi.org/10.3390/fishes8080405
APA StyleWan, S., Sun, Z., Zhang, C., Pan, T., Yuan, S., Chen, Y., Zou, J., & Gao, Q. (2023). Effects of LPS, Poly (I:C) and Edwardsiella tarda on the Expression Patterns of IL-17 Family Members and Their Receptors in Spotted Sea Bass (Lateolabrax maculatus). Fishes, 8(8), 405. https://doi.org/10.3390/fishes8080405







