Abstract
Oxygen level is an important environmental factor affecting the circadian rhythm. However, little is known about the molecular mechanism by which clock genes regulate the circadian rhythm in fish under hypoxia. To explore changes in the transcription and expression of clock genes and related molecular regulatory mechanisms in pearl gentian grouper under hypoxia, liver transcriptome data were analyzed after exposure to acute hypoxic stress (dissolved oxygen 0.5 mg/L) for 1, 3, 6, and 9 h. miR-210 and m0044-5p inhibited the expression of period3 (per3) and casein kinase 1 delta b (csnk1db) in the core loop of the circadian clock, respectively. The nuclear receptor subfamily 1 group d member 1 (nr1d1) and RAR-related orphan receptor b (rorb) genes in the auxiliary loop were jointly up-regulated by three miRNAs (miR-144-3p/5p, miR-361-5p, and miR-133) and the transcription factor nuclear receptor subfamily 1 group d member 2 (Nr1d2). The pearl gentian grouper maintains the stability of circadian clock systems and normal physiological metabolism under hypoxic stress by regulating the transcriptional expression of these genes via miRNAs and transcription factors to improve hypoxic tolerance. These findings provide important basic data for future research on hypoxic tolerance in pearl gentian grouper and provide new insights into the interaction between hypoxia and the circadian rhythm in fish.
Keywords:
pearl gentian grouper; acute hypoxia; circadian rhythm; clock genes; transcriptional expression Key Contribution:
The pearl gentian grouper maintains physiological metabolism and enhances tolerance to hypoxia by regulating the transcriptional expression of circadian clock genes via miRNAs and transcription factors.
1. Introduction
The circadian rhythm is an endogenous oscillation of biological processes in organisms that follows an approximately 24 h cycle for synchronization with changes in the external environment [,]. Circadian rhythm regulates the physiological behaviors and metabolic reactions of diverse organisms []. The circadian clock is the material basis of the circadian rhythm and is composed of the input pathway, core oscillator, and output pathway []. The core oscillator consists of clock genes and their proteins, forming a complex core loop (also known as the transcription–translation feedback loop) [,]. The basic helix–loop–helix ARNT-like (Bmal)/Clock circadian regulator (Clock) heterodimer binds to the E-box element of the gene promoter, activating the clock genes and clock-controlled gene transcription (forward regulation) []. Subsequently, the cryptochrome circadian regulator (Cry)/Period (Per) heterodimer inhibits the transcriptional activity of the Bmal/Clock heterodimer, thereby inhibiting cryptochrome (cry) and period (per) transcription (negative regulation) [,]. In addition, an auxiliary feedback loop composed of the nuclear receptor subfamily 1 group d (Nr1d) and RAR-related orphan receptor (Ror) proteins maintains the stability of the core loop []. The Nr1d and Ror proteins competitively bind to specific sequences of DNA, termed ROR response elements (ROREs), inhibiting and promoting the expression of the clock gene basic helix–loop–helix ARNT-like 1 (bmal1), respectively []. Expression levels of approximately 10% to 30% of genes in animals and plants are regulated by the circadian clock system, participating in growth, development, reproduction, metabolism, and environmental adaptation [,]. Thus, stabilization of the circadian clock system is beneficial for the response and adaptation to periodic environmental changes [].
Similar to other animals, fish show obvious circadian rhythms in behaviors, growth and development, and physiological metabolism []. Changes in environmental signals could disrupt the circadian clock system and alter the circadian rhythm in fish, leading to disruptions in behaviors and physiological processes [,]. Numerous studies have shown that, as the zeitgebers of the circadian clock system, light and temperature conditions could reset the clock and affect rhythms in fish [,]. Oxygen levels exhibit periodicity in the natural environment and affect the circadian clock system []. On the one hand, oxygen level fluctuations could cause changes in the amplitude of the circadian rhythm cycle and metabolism in fish []. Under hypoxia, the rate of hypoxia-inducible factor-1α (Hif-1α) binding to the clock gene period1 (per1) increased nearly four times in zebrafish, Danio rerio, significantly inhibiting the expression of per1, leading to a change in the circadian rhythm amplitude []. Both hypoxia and diclofenac exposure inhibited the transcription of clock circadian regulator (clock) and per1 and destroyed the circadian rhythm in the three-spined stickleback Gasterosteus aculeatus, thus affecting the transcription and activity rhythm of enzymes related to energy metabolism []. The metabolic rate of lake sturgeon Acipenser fulvescens exhibited a circadian rhythm, and its standard metabolic rate was not affected by hypoxia, but the maximum metabolic rate reduced as the oxygen level decreased []. These findings indicated that its metabolic rhythm was regulated by both external oxygen changes and the internal circadian rhythm []. On the other hand, circadian rhythm disorders would reduce the changes in physiological processes in fish under hypoxia, including changes in angiogenesis, red blood cell apoptosis, and oxygen transport capacity, ultimately leading to a decline in survival []. Sustained hypoxia could easily cause tissue damage in fish, while diel-cycling hypoxia could improve the ability of fish to tolerate hypoxia [,]. Collectively, several studies have explored the impacts of oxygen levels on the fish clock system, characterizing the complex interactions between the two. However, little is known about the molecular mechanism by which clock genes regulate the circadian rhythm in fish under hypoxia.
The pearl gentian grouper is a hybrid fish from interspecific hybridization between Epinephelus lanceolatus (♂) and Epinephelus fuscoguttatus (♀), also known as the dragon tiger grouper []. It has various beneficial characteristics, including strong disease resistance, a fast growth rate, high-quality meat, and rich nutrients []. This grouper is an economically important farmed fish in the coastal countries of Asia, particularly on the southeastern coast of China (e.g., Fujian, Guangdong, and Hainan provinces) []. Recently, with the promotion of high-density and intensive farming, a high stocking density, poor water exchange, and high oxygen consumption by the respiration of plankton and sediment microorganisms could easily lead to hypoxia in aquaculture waters []. Although this grouper has strong tolerance to hypoxia, its tolerance threshold is 0.24–0.70 mg/L [,]. However, hypoxia has negative effects on feeding and swimming behavior, growth and development, and physiological metabolism, and even causes a large number of deaths in fish []. Furthermore, hypoxia significantly alters energy metabolism, oxidative stress, and apoptosis pathways in this grouper [,]. As the circadian rhythm responds to environmental changes synchronously through the regulation of the circadian clock system, it plays an important role in maintaining behavioral activities, physiological metabolism, and environmental adaptation in fish [,]. Therefore, hypoxia would cause circadian rhythm cycle disturbances in fish, thus affecting the rhythm of their behavioral activities and physiological metabolism. In a transcriptomic analysis of the response to cold stress in the pearl gentian grouper, we found that nuclear receptor subfamily 1 group d member 2 (nr1d2) and protein phosphatase 1 catalytic subunit gamma (ppp1cc-a) were up-regulated via ssa-miR-25-3-5p and ccr-miR-489, respectively, and contributed to the regulation of the circadian rhythm core loop to prevent cold shock and enhance cold tolerance []. However, the effect of hypoxia on the circadian rhythm regulation of this grouper has not been explored.
Based on the results of previous studies, we speculate that hypoxic conditions may also affect the transcriptional regulation of circadian clock genes in pearl gentian grouper, playing an important role in maintaining circadian rhythm homeostasis and normal physiological metabolism and improving its tolerance to hypoxia. This means that changes in oxygen levels are closely related to circadian rhythms. Thus, we combined previous mRNA-Seq and newly obtained miRNA-Seq data for pearl gentian grouper under acute hypoxia to obtain differentially expressed genes (DEGs), microRNAs, and transcription factors (TFs) related to circadian rhythm in this study. Using bioinformatics analyses, we further explored the transcriptional regulatory changes of the circadian clock system in the grouper under acute hypoxia. Our study provides important basic data and a reference for improving tolerance to hypoxia in this grouper under high-density and intensive farming. Furthermore, these results improve our understanding of the molecular regulatory mechanism of the circadian rhythm in fish adaptation to the hypoxic environment.
2. Materials and Methods
2.1. Acute Hypoxic Stress and Sampling
The experimental samples in this study were derived from our previous analysis of the transcriptome under hypoxic stress []. Briefly, 200 juvenile pearl gentian groupers (average standard length: 10.7 ± 0.58 cm; average standard weight: 42.8 ± 6.99 g) were temporarily cultured for 2 weeks at a water temperature of 25.0 ± 1 °C, salinity of 24.0 ± 1 ‰, and DO of 5.5 ± 0.5 mg/L. Thereafter, the water DO content in three rearing tanks was rapidly reduced from 5.5 mg/L to 0.5 mg/L within 1 h. After 0.5 mg/L DO for 1 h, 3 h, 6 h, and 9 h, three fish were randomly selected from each tank from which to take liver tissue samples; these treatment groups were named Hy1, Hy3, Hy6, and Hy9. Liver samples were taken before hypoxic stress (i.e., at 5.5 mg/L DO), named Hy0 (control group). Samples were treated with liquid nitrogen and stored at −80 °C until sequencing and RT-qPCR validation experiments.
2.2. MiRNA Sequencing and Prediction of miRNA Target Genes
High-quality RNAs from 15 liver tissues analyzed by previous transcriptome sequencing (with three duplicates per group) were sent to Genedenovo Biotechnology Co., Ltd. (Guangzhou, China) for miRNA sequencing. Differentially expressed miRNAs (DE miRNAs) were screened with p < 0.05 and |log2 (Fold Change)| > 1 as criteria. Three methods, RNAhybrid v2.1.2 + SVM_light v6.01 [,], miRanda v3.3a [], and TargetScan v7.0 [], were used to predict potential target genes of DE miRNAs. Based on our previously detected (DEGs), miRNA-mRNA regulatory pairs with negative correlations were obtained.
2.3. KEGG Enrichment, Pathway Network, and STEM Analyses
Based on an integration analysis of mRNA-Seq and miRNA-Seq, a KEGG enrichment analysis of genes in miRNA-mRNA regulatory pairs was performed using the OmicShare tools (https://www.omicshare.com/tools, accessed on 22 February 2023), and significant KEGG pathways related to hypoxia were selected (p < 0.05). Interactions among these pathways were evaluated, and hub pathways were determined by a pathway network analysis []. The pathway network was visualized and edited using Cytoscape v3.8.2 (Cytoscape Consortium, San Diego, CA, USA) [].
A trend analysis of genes in miRNA-mRNA pairs was carried out using Short Time- series Expression Miner (STEM) v1.3.13 (Carnegie Mellon University, Pittsburgh, PA, USA) [] to obtain gene expression profiles related to the duration of hypoxic stress. The main parameters for the STEM analysis were as follows: module significance was set to p < 0.05, the number of modules was 20, and the minimum fold change was 2.
2.4. Construction of a PPI Network and miRNA-TF-mRNA Regulatory Network Based on Target Genes
Amino acid sequences of the target genes were submitted to STRING v11.5 (https://cn.string-db.org, accessed on 27 February 2023), and a protein–protein interaction (PPI) network was constructed with zebrafish as the reference species []. The analysis parameters were set with a confidence level of 0.9 and an FDR stringency of 0.05%.
The DEGs were aligned to the AnimalTFDB [] using Blastp to identify potential TFs, predict target genes, and obtain TF-mRNA regulatory pairs. Based on the regulatory relationships between miRNAs, TF, and DEGs, a miRNA-TF-mRNA regulatory network was constructed using Cytoscape [].
2.5. RT-qPCR Validation Experiment
Expression levels of key genes and miRNAs in 15 liver samples from one control group and four treatment groups were verified by real-time fluorescent quantitative PCR (RT-qPCR). Specific primers for genes and miRNAs were designed using Primer Premier 5 (PREMIER Biosoft International, Palo Alto, CA, USA) (Table 1). After the total RNA was extracted from each sample with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), RNA quality and concentration were evaluated via 1% agarose gel electrophoresis (the extracted RNA showed in Figures S1 and S2) and NanoDrop2000 (Thermo Scientific, Waltham, MA, USA). The reverse transcriptions were performed using the One-Step gDNA Removal Kit (TransGen Biotech, Beijing, China) and miRNA First Strand cDNA Synthesis Kit (Sangon Biotech, Shanghai, China) to obtain cDNA templates for genes and miRNAs, respectively. PCR systems for genes and miRNAs were configured using the PerfectStart® Green qPCR SuperMix Kit (TransGen Biotech, Beijing, China) and the miRNA Fluorescence Quantitative PCR Kit (Sangon Biotech, Shanghai, China), respectively. The selected reference gene for RT-qPCR was 18S rRNA []. Triplicate experiments were conducted for each sample, and a negative reaction without a template was set as the control. PCR was performed using the LightCycler96 (Roche, Mannheim, Germany) under the following reaction conditions: preincubation at 95 °C for 30 s, and 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The melting curve adopted instrument default values of 95 °C for 10 s, 65 °C for 60 s, and 97 °C for 1 s to determine primer dimer and non-specific amplification. Relative expression levels of genes and miRNAs were calculated using the comparative CT method (2−ΔΔCT), and line charts were drawn using GraphPad Prism v8.0.2 (GraphPad Software, San Diego, CA, USA).

Table 1.
Primer sequences of key genes and miRNAs.
3. Results
3.1. KEGG Enrichment and Pathway Network Analyses
After data filtering, mRNA and miRNA sequencing yielded clean reads ranging from 35,152,534 to 66,514,474 and 8,691,172 to 26,262,181, respectively. In total, 2383 DEGs and 91 DE miRNAs were screened from mRNA and miRNA sequencing data, respectively. By combining the mRNA-Seq and miRNA-Seq datasets, a total of 562 miRNA-mRNA regulatory pairs were obtained, including 72 DE miRNAs and 358 DEGs. Based on a KEGG enrichment analysis of 358 DEGs, the circadian rhythm pathway was the most significant pathway except for cancer-related pathways among the top 20 pathways (Figure 1). During the whole hypoxic stress process, we did not observe disease symptoms in the experimental fish. The four genes period3 (per3), casein kinase 1, delta b (csnk1db), nuclear receptor subfamily 1, group d, member 1 (nr1d1), and RAR-related orphan receptor b (rorb) were significantly enriched in the circadian rhythm pathway, and these were likely to play an important role in the acute hypoxic response in pearl gentian grouper. Further pathway network analysis revealed that the circadian rhythm pathway (ko04710) was connected with other pathways by directly acting on ubiquitin-mediated proteolysis (ko04120) and the MAPK signaling pathway (ko04010), thereby regulating the acute hypoxia response in pearl gentian grouper (Figure 2). Moreover, ko04710 was associated with other circadian-related pathways, including circadian rhythm-fly (ko04711), circadian entrainment (ko04713), and African trypanosomiasis (ko05143), although these three pathways were not connected to other pathways.
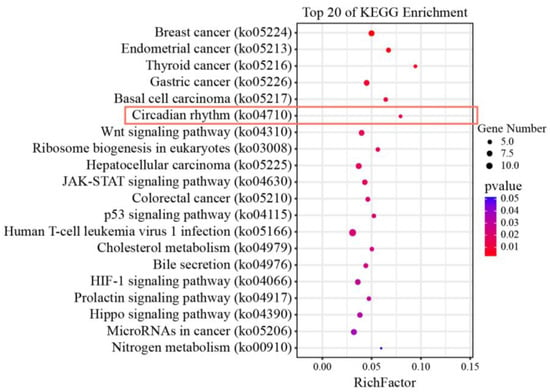
Figure 1.
KEGG enrichment analysis of 358 target genes.
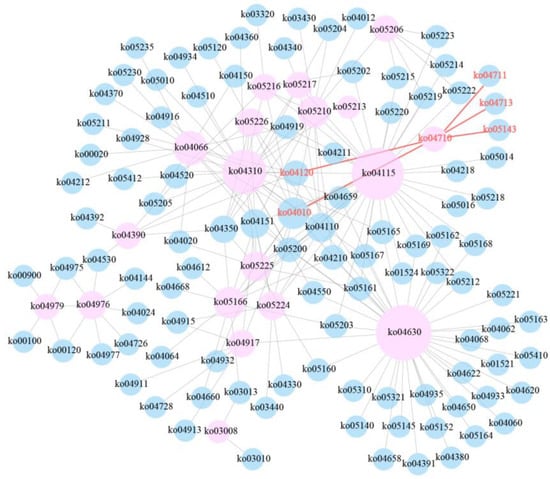
Figure 2.
Pathway interaction network. The nodes represent pathways (pink represent significantly enriched pathways, blue represent non-significant pathways), the size of each node represents its connectivity (where a larger node indicates a more important pathway in the network), and the lines represent the interactions between pathways.
3.2. Expression Trends of Target Genes
In the STEM analysis, 358 target genes (from 562 miRNA–mRNA regulatory pairs in the integration analysis) were clustered into 20 gene expression patterns (Figure 3). Three expression profiles were significant (p < 0.05) and contained 63.41% of the target genes. The expression levels of 122 genes in Profile 0 decreased consistently during acute hypoxic stress, whereas the expression levels of 31 genes in Profile 19 continued to increase. In Profile 18, the expression levels of 74 genes increased in the early stage of hypoxia (0–3 h) and then decreased slightly (6–9 h), with an overall increasing trend. The expression patterns of the four circadian rhythm genes belonged to Profile 0 (per3 and csnk1db), Profile 19 (nr1d1), and Profile 17 (rorb). Obviously, the diversity of gene expression patterns indicates that a complex regulatory mechanism was involved in the response to hypoxia in pearl gentian grouper.
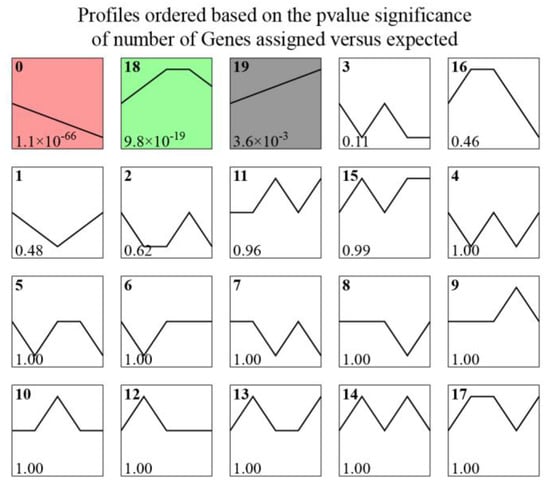
Figure 3.
Twenty gene expression profiles involving 358 target genes. The number at the bottom left represent p-value, colors represent significant profiles (p < 0.05), white represent non-significant profiles.
3.3. PPI Network and miRNA-TF-mRNA Regulatory Network of Circadian Rhythm Genes
A PPI network analysis of four circadian rhythm genes (per3, csnk1db, nr1d1, and rorb) was performed using zebrafish as the reference species. As shown in Figure 4, the PPI network was composed of 24 proteins with direct or indirect interactions, mainly related to the circadian rhythm. Among these, 4, 1, and 13 proteins showed direct interactions with Period3 (Per3), Casein kinase 1 delta b (Csnk1db), and Nuclear receptor subfamily 1 group d member 1 (Nr1d1), respectively. However, there were no interactions between RAR-related orphan receptor b (Rorb) and other proteins in the PPI network. This may be explained by the limited research on the functional role of Rorb in fish.
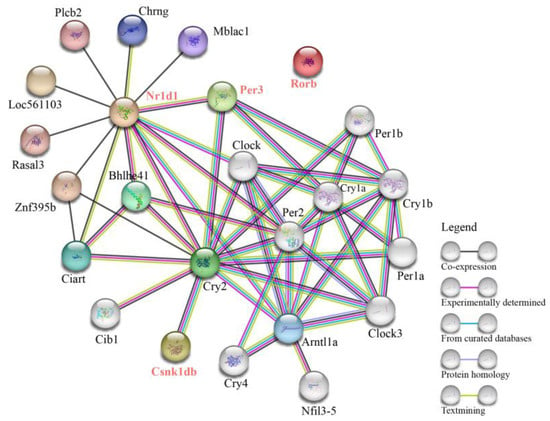
Figure 4.
Protein–protein interaction network of the four circadian rhythm genes with zebrafish as the reference species.
The TFs and their target genes formed 33 TF-mRNA regulatory pairs, including 28 TF and 24 DEGs. Combined with 562 miRNA-mRNA pairs, we detected seven regulatory pairs related to the circadian clock (Figure 5). The miRNA-TF-mRNA regulatory network included six DE miRNAs, one TF, and four DEGs. The per3 and csnk1db genes were regulated by miR-210 and m0044-5p, respectively. The nr1d1 gene was co-regulated by miR-144-3p, miR-144-5p, and miR-361-5p. rorb was co-regulated by miR-133 and the TF Nuclear receptor subfamily 1group d member 2 (Nr1d2).
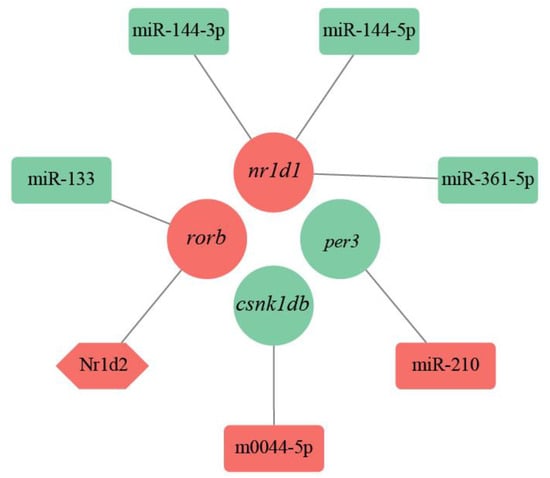
Figure 5.
miRNA-TF-mRNA regulatory network for the circadian rhythm.
3.4. RT-qPCR Validation of Key Genes and miRNAs
Relative expression levels of four circadian rhythm genes (per3, csnk1db, nr1d1, and rorb) and four transcriptional regulators (miR-210, m0044-5p, miR-361-5p, and TF Nr1d2) in all groups were quantitatively verified by RT-qPCR. As illustrated in Figure 6, the expression levels of nr1d1, rorb, miR-210, and m0044-5p were up-regulated, while the expression levels of per3, csnk1db, and miR-361-5p were down-regulated overall. The nr1d2 expression fluctuated. RT-qPCR expression levels of genes and miRNAs were consistent with the relative expression trends obtained by sequencing, supporting the reliability and accuracy of mRNA-Seq and miRNA-Seq results.
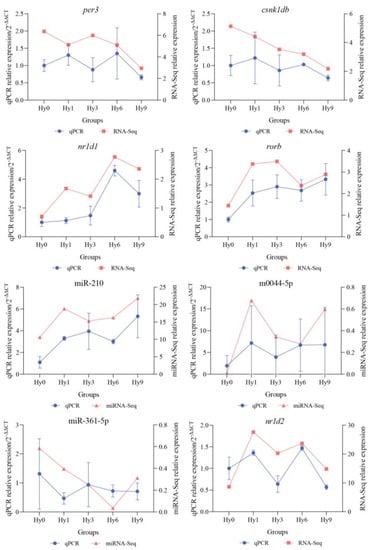
Figure 6.
Comparison of relative expression levels obtained by RT-qPCR validation and sequencing.
4. Discussion
Among the 358 DEGs detected in this study, the four genes per3, csnk1db, rorb, and nr1d1 were significantly enriched in the circadian rhythm pathway. Moreover, regulatory relationships between these four target genes and seven regulatory factors (miR-210, m0044-5p, miR-133, miR-144-3p, miR-144-5p, miR-361-5p, and TF Nr1d2) were determined. By further exploring the functions and pathways related to these molecules, we confirmed that they play crucial roles in regulating the circadian rhythm cycle and circadian clock system of pearl gentian grouper exposed to acute hypoxia.
4.1. Expression Changes of Core Clock Genes and Stability of the Circadian Cycle under Hypoxia
Period genes (including per1, per2, and per3 isoforms) are negative regulatory elements of the circadian clock [,]. Zheng et al. [] demonstrated that the double knockout of the per1 and per2 genes leads to the loss of circadian rhythm in mice. That is, these two genes play a direct and important role in the regulation of the animal circadian rhythm []. The precise regulatory roles of per1 and per2 in the circadian rhythm of fish under hypoxia and cold stress have been confirmed by several studies [,,]. However, the functional role of per3 in the animal circadian rhythm is still controversial. Shearman et al. [] found that a per3 deficiency does not directly cause the loss of the circadian rhythm in mice, suggesting that the gene is not necessary for the circadian rhythm. Furthermore, Bae et al. [] reported that the changes in the circadian rhythm in mice with per1/per3 and per2/per3 double deficiencies were similar to those of mice with per1 or per2 deficiency alone. In contrast, Yagita et al. [] confirmed that the Per3 protein encoded by per3 promotes nuclear translocation and stability of Per1 and Per2 in mammals. In a circadian rhythm transcriptome study of the gilthead sea bream Sparus aurata, Yúfera et al. [] found that the expression peaks of per3 appeared before those of per1 and per2, indicating that per3 functions in the regulation of per1 and per2. Recently, per3 expression in various tissues of the darkbarbel catfish Pelteobagrus vachellii was observed to show obvious circadian rhythms []. In the liver of pearl gentian grouper under hypoxia, the up-regulation of miR-210 inhibited the expression of per3 (Figure 7). MiR-210, which is a hypoxia-induced miRNA with an essential regulatory role in the animal cell cycle, DNA damage repair, angiogenesis, stem cell differentiation, mitochondrial metabolism, and cancer treatment []. It is important for cells to maintain normal metabolic function in hypoxic environments []. Similarly, Nagel et al. [] reported that miR-192 and miR-194 could inhibit the transcriptional expression of the period gene family, which may shorten the circadian rhythm period in animals. Hung et al. [] found that cold tolerance in zebrafish larvae could be improved by regulating the expression of per2 via dre-miR-29b. Therefore, we infer that hypoxia induces the up-regulation of miR-210 expression in pearl gentian grouper to inhibit the expression of per3, resulting in decreased period protein levels and Cry/Per heterodimer activity, thereby weakening the inhibitory effect on the Bmal/Clock heterodimer [,]. This change ultimately shortens the length of its circadian rhythm cycle, which is conducive to enhancing tolerance to hypoxia in pearl gentian grouper. To our knowledge, this study reveals, for the first time, the functional role of per3 in enhancing hypoxia tolerance in fish by regulating the circadian rhythm.
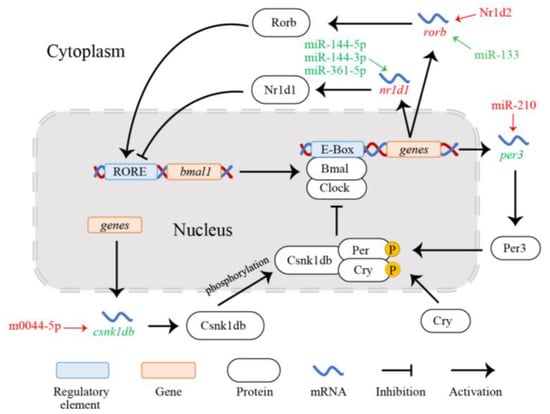
Figure 7.
Regulatory network for the circadian rhythm in pearl gentian grouper under hypoxia.
We also observed that the up-regulation of the novel miRNA m0044-5p inhibited the expression of the csnk1db gene (Figure 7). The CK1δ kinase encoded by csnk1db belongs to the casein kinase family, which directly promotes the degradation of PER or changes its location in cells via phosphorylation [,]. Phosphorylation modification is an important regulatory mechanism for the circadian clock, affecting the stability, activity, and interactions of circadian clock proteins []. When the activity of CK1δ kinase is inhibited or deactivated, the animal circadian cycle is prolonged []. Its overexpression would shorten the circadian cycle []. Moreover, CK1 kinase cooperates with protein phosphatase 1 (PP1) to jointly regulate the activity and stability of a variety of circadian clock proteins and participate in regulating the circadian cycle [,]. Smadja Storz et al. [] found that the rhythmic expression of per and arylalkylamine N-acetyltransferase 2 (aanat2) in zebrafish disappears with the inhibition of CK1δ kinase activity, with effects on the behavioral rhythm. Miao et al. [] reported that ccr-miR-489 in the liver of pearl gentian grouper shortens the circadian cycle by regulating the transcriptional expression of ppp1cc-a and participating in the phosphorylation of PER under cold stress. The csnk1db gene regulates the activity and stability of circadian clock elements and determines the length of the circadian cycle by the autophosphorylation of CK1 kinase or in collaboration with other genes []. Therefore, the down-regulation of csnk1db means that the phosphorylation of the CK1 kinase was inhibited by hypoxia in pearl gentian grouper, thereby reducing the phosphorylation and degradation of the Cry/Per heterodimer. This could lead to increased Bmal/Clock heterodimer activity [,], thus prolonging the length of the circadian cycle in this grouper.
To sum up, miR-210 and m0044-5p inhibited the transcription of per3 and csnk1db, respectively, and shortened and extended the circadian cycle, maintaining the stability of the circadian cycle of pearl gentian grouper under hypoxia, which is conducive to physiological and biochemical reactions and survival. This regulatory relationship between oxygen and the circadian clock is similar to the temperature compensation mechanism of the circadian rhythm []. That is, a change in the oxygen content does not have a significant impact on the length of the circadian cycle in animals within a certain level and duration of hypoxia. Circadian clock genes are capable of self-regulation to maintain the stability of the circadian cycle under environmental change.
4.2. Regulation of the Auxiliary Loop and Stability of Circadian Clock Systems
The auxiliary loop of the circadian clock is mainly mediated by the Nr1d and Ror proteins, which regulate the transcriptional activity of the clock gene bmal1 in the core loop by competitive binding []. In this study, we found that the down-regulation of three miRNAs (i.e., miR-144-3p, miR-144-5p, and miR-361-5p) collectively promotes the expression of the nr1d1 gene (Figure 7). The nr1d1 gene is a regulatory gene in the circadian clock system; it is expressed in various tissues and exhibits periodic changes []. The Nr1d1 protein encoded by nr1d1 inhibits bmal1 gene expression by binding to the promoter RORE element, thus participating in the circadian rhythm and lipid metabolism to maintain the normal metabolic function of animals [,]. Previous studies have shown that nr1d1 deficiency could disrupt the circadian rhythm, affect the expression of core clock genes and downstream genes, and influence glucose and lipid metabolism in animals [,]. The knockout of nr1d1 altered the expression of circadian rhythm genes and metabolism in the liver of mice []. The nr1d1 gene also plays a pivotal role in the circadian clock system and movement rhythm in various fishes, such as zebrafish [], Atlantic cod Gadus morhua [], Chinese perch Siniperca chuatsi [], and the goldfish Carassius auratus []. Three regulators (miR-144-3p, miR-144-5p, and miR-361-5p) of the nr1d1 gene participate in the regulation of the circadian rhythm via clock genes or related signaling pathways [,]. Therefore, the above three miRNAs co-regulated nr1d1, thereby inhibiting the transcriptional expression of bmal1 and facilitating circadian rhythm stability and normal physiological metabolism under hypoxic conditions in pearl gentian grouper. In the previous cold stress transcriptome analysis, we detected increased nr1d2 gene expression via ssa-miR-25-3-5p, and this gene participated in the maintenance of the circadian rhythm balance and lipid homeostasis to enhance cold tolerance []. As a homologue of nr1d1, nr1d2 is also considered to be involved in the regulation of the circadian rhythm, energy metabolism, cellular autophagy, and other biological processes [,]. The nr1d2 gene was not significantly enriched in the circadian rhythm pathway, in our study. However, the results of the previous cold stress analysis suggest that nr1d1 and nr1d2 may both contribute to the regulation of circadian rhythms under stress and may have complementary roles [].
Furthermore, we observed that the up-regulation of rorb in the auxiliary loop is jointly regulated by the TF Nr1d2 and miR-133 (Figure 7). The rorb gene is a member of the ror gene family and encodes the retinoid-related orphan receptor involved in the auxiliary loop and maintaining the stability of the core loop by binding to the RORE element of bmal1 and activating gene expression [,]. The RORE element of rorb binds to the Nr1d protein, and its transcriptional expression is regulated by Nr1d []. Masana et al. [] reported that mice lacking Rorb showed abnormal circadian behavior, confirming that rorb plays a critical role in animal circadian regulation. Thus, we believe that the TF Nr1d2 up-regulated the expression of rorb, which is beneficial for the stable regulation of the circadian clock system in the pearl gentian grouper under hypoxia. In addition, miR-133, which regulates rorb, is involved in the regulation of embryonic and vascular development, muscle regeneration, cardiac rhythm, and energy metabolism in fish []. It has been demonstrated that miR-133-3p participates in the regulation of glucose and lipid metabolism in the largemouth bass Micropterus salmoides during acute hypoxia []. Although the function of miR-133 in the circadian clock system has not been established, our results show that rorb is a potential target gene for miR-133, and the down-regulation of miR-133 significantly increases the expression of rorb. This suggests that miR-133 plays a key role in the regulation of the circadian rhythm in pearl gentian grouper under hypoxia. Based on the above analysis, we conclude that rorb activates the transcriptional expression of bmal1 under the co-regulation of the TF Nr1d2 and miR-133 to maintain the normal circadian rhythm of pearl gentian grouper exposed to acute hypoxia.
In summary, various miRNAs, TFs, and target genes mediate the transcriptional expression of the core gene bmal1 to stabilize the core loop of the circadian clock, thereby preventing disruptions of the circadian rhythm caused by hypoxia in the pearl gentian grouper. This transcriptional regulation would be conducive to the survival of this grouper under hypoxia and the maintenance of normal physiological and metabolic activities.
5. Conclusions
Based on analyses of mRNA and miRNA data, we detected a close correlation between changes in the circadian rhythm of pearl gentian grouper and acute hypoxia and obtained insight into the molecular regulatory mechanism underlying the circadian rhythm under hypoxia. Clock-related genes (per3, csnk1db, nr1d1, and rorb) maintained the circadian rhythm cycle and the overall stability of the circadian clock system via multiple miRNAs and TF Nr1d2. This self-regulation of the circadian rhythm by the grouper with a certain intensity and duration of hypoxia contributes to the maintenance of normal physiological metabolism and enhanced hypoxia tolerance. These findings provide an important reference for improving hypoxic tolerance in pearl gentian grouper under high-density and intensive farming. Notably, the role of the per3 gene and miR-133 in the regulation of the circadian rhythm in the fish response to hypoxia was identified for the first time in this study. Collectively, these results improve our understanding of the regulation of fish circadian rhythm under environmental stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes8070358/s1, Figure S1: RNA extractions and migration-1; Figure S2: RNA extractions and migration-2.
Author Contributions
Conceptualization, R.-X.W., J.Z. and S.-F.N.; methodology, R.-X.W., S.-F.N. and Y.-S.L.; validation, R.-X.W. and Y.-S.L.; formal analysis, Y.-S.L.; investigation, Y.-S.L., J.Z. and Z.-B.L.; data curation, R.-X.W. and Y.-S.L.; writing—original draft preparation, R.-X.W. and Y.-S.L.; writing—review and editing, R.-X.W., J.Z. and Y.-S.L.; visualization, R.-X.W. and Y.-S.L.; supervision, R.-X.W., J.Z., S.-F.N. and B.-G.T.; project administration, R.-X.W., J.Z. and B.-G.T.; funding acquisition, J.Z., R.-X.W. and B.-G.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), China (No. ZJW-2019-06); the Science and Technology Planning Project of Guangdong Province, China (No. 2017A030303077).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board (Ethics Committee) of the Animal Experimental Ethics Committee of Guangdong Ocean University (approval number: 0301-2020).
Data Availability Statement
mRNA-Seq and miRNA-Seq raw reads data have been uploaded to NCBI database (accession: PRJNA801908 and PRJNA967800).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dunlap, J.C. Molecular bases for circadian clocks. Cell 1999, 96, 271–290. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Panda, S.; Kay, S.A. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 2001, 17, 215–253. [Google Scholar] [CrossRef] [PubMed]
- Philpott, J.M.; Torgrimson, M.R.; Harold, R.L.; Partch, C.L. Biochemical mechanisms of period control within the mammalian circadian clock. Semin. Cell Dev. Biol. 2022, 126, 71–78. [Google Scholar] [CrossRef]
- Morris, A.R.; Stanton, D.L.; Roman, D.; Liu, A.C. Systems level understanding of circadian integration with cell physiology. J. Mol. Biol. 2020, 432, 3547–3564. [Google Scholar] [CrossRef]
- Rey, G.; Reddy, A.B. Connecting cellular metabolism to circadian clocks. Trends Cell Biol. 2013, 23, 234–241. [Google Scholar] [CrossRef]
- Amaral, I.P.G.; Johnston, I.A. Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R193–R206. [Google Scholar] [CrossRef]
- Solt, L.A.; Kojetin, D.J.; Burris, T.P. The REV-ERBs and RORs: Molecular links between circadian rhythms and lipid homeostasis. Future Med. Chem. 2011, 3, 623–638. [Google Scholar] [CrossRef]
- Covington, M.; Maloof, J.; Straume, M.; Kay, S.; Harmer, S. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef]
- Krittika, S.; Yadav, P. Circadian clocks: An overview on its adaptive significance. Biol. Rhythm Res. 2020, 51, 1109–1132. [Google Scholar] [CrossRef]
- Reebs, S. Plasticity of diel and circadian rhythms in fishes. Rev. Fish Biol. Fish. 2002, 12, 349–371. [Google Scholar] [CrossRef]
- Costa, L.S.; Serrano, I.; Sánchez-Vázquez, F.J.; López-Olmeda, J.F. Circadian rhythms of clock gene expression in nile tilapia (Oreochromis niloticus) central and peripheral tissues: Influence of different lighting and feeding conditions. J. Comp. Physiol. B 2016, 186, 775–785. [Google Scholar] [CrossRef]
- Prokkola, J.M.; Nikinmaa, M. Circadian rhythms and environmental disturbances—Underexplored interactions. J. Exp. Biol. 2018, 221, jeb179267. [Google Scholar] [CrossRef] [PubMed]
- López-Olmeda, J.F.; Madrid, J.A.; Sánchez-Vázquez, F.J. Light and temperature cycles as zeitgebers of zebrafish (Danio rerio) circadian activity rhythms. Chronobiol. Int. 2006, 23, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Mortola, J.P. Hypoxia and circadian patterns. Respir. Physiol. Neurobiol. 2007, 158, 274–279. [Google Scholar] [CrossRef]
- Sandbichler, A.M.; Jansen, B.; Peer, B.A.; Paulitsch, M.; Pelster, B.; Egg, M. Metabolic plasticity enables circadian adaptation to acute hypoxia in zebrafish cells. Cell. Physiol. Biochem. 2018, 46, 1159–1174. [Google Scholar] [CrossRef] [PubMed]
- Egg, M.; Köblitz, L.; Hirayama, J.; Schwerte, T.; Folterbauer, C.; Kurz, A.; Fiechtner, B.; Möst, M.; Salvenmoser, W.; Sassone-Corsi, P.; et al. Linking oxygen to time: The bidirectional interaction between the hypoxic signaling pathway and the circadian clock. Chronobiol. Int. 2013, 30, 510–529. [Google Scholar] [CrossRef] [PubMed]
- Prokkola, J.M.; Nikinmaa, M.; Lubiana, P.; Kanerva, M.; McCairns, R.J.S.; Götting, M. Hypoxia and the pharmaceutical diclofenac influence the circadian responses of three-spined stickleback. Aquat. Toxicol. 2015, 158, 116–124. [Google Scholar] [CrossRef]
- Svendsen, J.C.; Genz, J.; Anderson, W.G.; Stol, J.A.; Watkinson, D.A.; Enders, E.C. Evidence of circadian rhythm, oxygen regulation capacity, metabolic repeatability and positive correlations between forced and spontaneous maximal metabolic rates in lake sturgeon Acipenser fulvescens. PLoS ONE 2014, 9, e94693. [Google Scholar] [CrossRef]
- Pelster, B.; Egg, M. Multiplicity of hypoxia-inducible transcription factors and their connection to the circadian clock in the zebrafish. Physiol. Biochem. Zool. 2015, 88, 146–157. [Google Scholar] [CrossRef]
- Williams, K.J.; Cassidy, A.A.; Verhille, C.E.; Lamarre, S.G.; MacCormack, T.J. Diel cycling hypoxia enhances hypoxia-tolerance in rainbow trout (Oncorhynchus mykiss): Evidence of physiological and metabolic plasticity. J. Exp. Biol. 2019, 222, jeb.206045. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Y.; Wang, Z.; Xu, K.; Xiao, X.; Mu, W. Comparison of effects in sustained and diel-cycling hypoxia on hypoxia tolerance, histology, physiology and expression of clock genes in high latitude fish Phoxinus lagowskii. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2021, 260, 111020. [Google Scholar] [CrossRef] [PubMed]
- Rahimnejad, S.; Bang, I.C.; Park, J.Y.; Sade, A.; Choi, J.; Lee, S.M. Effects of dietary protein and lipid levels on growth performance, feed utilization and body composition of juvenile hybrid grouper, Epinephelus fuscoguttatus × E. lanceolatus. Aquaculture 2015, 446, 283–289. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, Z.; Ding, N.; Xiong, W.; Zheng, G.; Lin, Q.; Zhang, G. Effects of temperature on the survival, feeding, and growth of pearl gentian grouper (female Epinephelus fuscoguttatus × male Epinephelus lanceolatus). Fish. Sci. 2018, 84, 399–404. [Google Scholar] [CrossRef]
- Ding, S.X.; Liu, Q.H.; Wu, H.H.; Qu, M. A review of research advances on the biology and artificial breeding of groupers. J. Fish. Sci. China 2018, 25, 737–752. [Google Scholar] [CrossRef]
- Ruan, W.; Ji, W.W.; Zheng, L.; Yue, D.D.; Fang, H. On hypoxia stress in fish and its nutritional regulation and response. Mar. Fish. 2020, 42, 751–761. [Google Scholar] [CrossRef]
- Chen, W.Q.; Wu, H.X.; Wu, L.; Ma, J.Z. Oxygen consumption rate and suffocation point of the juveniles for five species of mariculture fish. J. Mar. Sci. 2015, 33, 76–81. [Google Scholar] [CrossRef]
- Lin, G.W. The effect of water temperature, salinity and dissolved oxygen changes on survival for pearl gentian grouper (Epinephelus fuscoguttatus female × E. lanceolatus male). J. Aquac. 2020, 41, 29–32. [Google Scholar] [CrossRef]
- Abdel-Tawwab, M.; Monier, M.N.; Hoseinifar, S.H.; Faggio, C. Fish response to hypoxia stress: Growth, physiological, and immunological biomarkers. Fish Physiol. Biochem. 2019, 45, 997–1013. [Google Scholar] [CrossRef]
- Liang, Y.S.; Wu, R.X.; Niu, S.F.; Miao, B.B.; Liang, Z.B.; Zhai, Y. Liver transcriptome analysis reveals changes in energy metabolism, oxidative stress, and apoptosis in pearl gentian grouper exposed to acute hypoxia. Aquaculture 2022, 561, 738635. [Google Scholar] [CrossRef]
- Lu, Z.F.; Huang, H.; Huang, X.M.; Huang, W.Z. Effects of hypoxic stress on antioxidant and energy metabolism of hybrid grouper (Epinephelus fuscoguttatus female × Epinephelus lanceolatus male). J. Guangdong Ocean. Univ. 2022, 42, 13–19. [Google Scholar] [CrossRef]
- Miao, B.B.; Niu, S.F.; Wu, R.X.; Liang, Z.B.; Zhai, Y. The microRNAs-transcription factors-mRNA regulatory network plays an important role in resistance to cold stress in the pearl gentian grouper. Front. Mar. Sci. 2022, 8, 824533. [Google Scholar] [CrossRef]
- Krüger, J.; Rehmsmeier, M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Joachims, T. Support vector machines. In Learning to Classify Text Using Support Vector Madchines; Joachims, T., Ed.; Springer: Boston, MA, USA, 2002; pp. 35–44. [Google Scholar]
- Turner, D.A. Miranda: A non-strict functional language with polymorphic types. In Functional Programming Languages and Computer Architecture; Jouannaud, J.P., Ed.; Springer: Berlin/Heidelberg, Germany, 1985; pp. 1–16. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zheng, J.; Shen, N.; Wang, G.; Zhou, G.; Fang, Y.; Lin, J.; Zhao, J. Identification of pathways and genes associated with synovitis in osteoarthritis using bioinformatics analyses. Sci. Rep. 2018, 8, 10050. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Zhang, H.M.; Chen, H.; Liu, W.; Liu, H.; Gong, J.; Wang, H.; Guo, A.Y. AnimalTFDB: A comprehensive animal transcription factor database. Nucleic Acids Res. 2012, 40, D144–D149. [Google Scholar] [CrossRef]
- Wang, H. Comparative analysis of period genes in teleost fish genomes. J. Mol. Evol. 2008, 67, 29–40. [Google Scholar] [CrossRef]
- Im, J.S.; Jung, B.H.; Kim, S.E.; Lee, K.H.; Lee, J.K. Per3, a circadian gene, is required for chk2 activation in human cells. FEBS Lett. 2010, 584, 4731–4734. [Google Scholar] [CrossRef]
- Zheng, B.; Albrecht, U.; Kaasik, K.; Sage, M.; Lu, W.; Vaishnav, S.; Li, Q.; Sun, Z.S.; Eichele, G.; Bradley, A.; et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 2001, 105, 683–694. [Google Scholar] [CrossRef]
- Fahrenkrug, J.; Georg, B.; Hannibal, J.; Hindersson, P.; Gräs, S. Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary. Endocrinology 2006, 147, 3769–3776. [Google Scholar] [CrossRef] [PubMed]
- Hung, I.C.; Hsiao, Y.C.; Sun, H.S.; Chen, T.M.; Lee, S.J. MicroRNAs regulate gene plasticity during cold shock in zebrafish larvae. BMC Genom. 2016, 17, 922. [Google Scholar] [CrossRef] [PubMed]
- Shearman, L.P.; Jin, X.; Lee, C.; Reppert, S.M.; Weaver, D.R. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol. Cell. Biol. 2000, 20, 6269–6275. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M.; Weaver, D.R. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 2001, 30, 525–536. [Google Scholar] [CrossRef]
- Yagita, K.; Yamaguchi, S.; Tamanini, F.; van der Horst, G.T.; Hoeijmakers, J.H.; Yasui, A.; Loros, J.J.; Dunlap, J.C.; Okamura, H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000, 14, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Yúfera, M.; Perera, E.; Mata-Sotres, J.A.; Calduch-Giner, J.; Martínez-Rodríguez, G.; Pérez-Sánchez, J. The circadian transcriptome of marine fish (Sparus aurata) larvae reveals highly synchronized biological processes at the whole organism level. Sci. Rep. 2017, 7, 12943. [Google Scholar] [CrossRef]
- Qin, C.; Shao, T.; Liao, X.; He, Y.; Wang, J.; Hu, P. Diurnal expression of circadian clock genes period 1 and period 3 in Pelteobagrus vachellii. J. Oceanol. Limnol. 2021, 39, 652–660. [Google Scholar] [CrossRef]
- Huang, X.; Le, Q.T.; Giaccia, A.J. MiR-210–Micromanager of the hypoxia pathway. Trends Mol. Med. 2010, 16, 230–237. [Google Scholar] [CrossRef]
- Nagel, R.; Clijsters, L.; Agami, R. The miRNA-192/194 cluster regulates the period gene family and the circadian clock. FEBS J. 2009, 276, 5447–5455. [Google Scholar] [CrossRef] [PubMed]
- Dilão, R.; Mota, B. The transcriptional regulation of PER protein in drosophila. J. Theor. Biol. 2019, 469, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Albornoz, A.; Yáñez, J.M.; Foerster, C.; Aguirre, C.; Pereiro, L.; Burzio, V.; Moraga, M.; Reyes, A.E.; Antonelli, M. The CK1 gene family: Expression patterning in zebrafish development. Biol. Res. 2007, 40, 251–266. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Yuan, B.; Xie, P.; Gu, Y.; Liu, Z.; Wang, T.; Li, Z.; Xu, Y.; Liu, Y. Decoupling PER phosphorylation, stability and rhythmic expression from circadian clock function by abolishing PER-CK1 interaction. Nat. Commun. 2022, 13, 3991. [Google Scholar] [CrossRef]
- Lee, C.; Etchegaray, J.P.; Cagampang, F.R.A.; Loudon, A.S.I.; Reppert, S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001, 107, 855–867. [Google Scholar] [CrossRef]
- Lee, H.; Chen, R.; Lee, Y.; Yoo, S.; Lee, C. Essential roles of CKIδ and CKIε in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 21359–21364. [Google Scholar] [CrossRef]
- Xu, P.; Ianes, C.; Gärtner, F.; Liu, C.; Burster, T.; Bakulev, V.; Rachidi, N.; Knippschild, U.; Bischof, J. Structure, regulation, and (patho-)physiological functions of the stress-induced protein kinase CK1 delta (CSNK1D). Gene 2019, 715, 144005. [Google Scholar] [CrossRef]
- Gallego, M.; Kang, H.; Virshup, D.M. Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 2006, 399, 169–175. [Google Scholar] [CrossRef]
- Lee, H.; Chen, R.; Kim, H.; Etchegaray, J.P.; Weaver, D.R.; Lee, C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. USA 2011, 108, 16451–16456. [Google Scholar] [CrossRef]
- Smadja Storz, S.; Tovin, A.; Mracek, P.; Alon, S.; Foulkes, N.S.; Gothilf, Y. Casein kinase 1δ activity: A key element in the zebrafish circadian timing system. PLoS ONE 2013, 8, e54189. [Google Scholar] [CrossRef]
- Song, H.; Pu, J.; Wang, L.; Wu, L.; Xiao, J.; Liu, Q.; Chen, J.; Zhang, M.; Liu, Y.; Ni, M.; et al. ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy 2015, 11, 1308–1325. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.J.; Logunova, L.; Maywood, E.S.; Gallego, M.; Lebiecki, J.; Brown, T.M.; Sládek, M.; Semikhodskii, A.S.; Glossop, N.R.J.; Piggins, H.D.; et al. Setting clock speed in mammals: The CK1ε tau mutation in mice accelerates the circadian pacemaker by selectively destabilizing PERIOD proteins. Neuron 2008, 58, 78–88. [Google Scholar] [CrossRef]
- Lahiri, K.; Vallone, D.; Gondi, S.B.; Santoriello, C.; Dickmeis, T.; Foulkes, N.S. Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol. 2005, 3, e351. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Boronat, M.; De Pedro, N.; Alonso-Gómez, Á.L.; Delgado, M.J.; Isorna, E. Nuclear receptors (PPARs, REV-ERBs, RORs) and clock gene rhythms in goldfish (Carassius auratus) are differently regulated in hypothalamus and liver. Front. Physiol. 2022, 13, 9037799. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Xia, H.B. Progresses on nuclear receptor Rev-erbs. Life Sci. Res. 2013, 17, 548–553. [Google Scholar] [CrossRef]
- Bugge, A.; Feng, D.; Everett, L.J.; Briggs, E.R.; Mullican, S.E.; Wang, F.; Jager, J.; Lazar, M.A. Rev-Erbα and Rev-Erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012, 26, 657–667. [Google Scholar] [CrossRef]
- Cho, H.; Zhao, X.; Hatori, M.; Yu, R.T.; Barish, G.D.; Lam, M.T.; Chong, L.W.; DiTacchio, L.; Atkins, A.R.; Glass, C.K.; et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012, 485, 123–127. [Google Scholar] [CrossRef]
- Huang, G.; Zhang, F.; Ye, Q.; Wang, H. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev-Erbα and indirectly via Cebpb/(C/Ebpβ) in zebrafish. Autophagy 2016, 12, 1292–1309. [Google Scholar] [CrossRef]
- Lazado, C.C.; Kumaratunga, H.P.S.; Nagasawa, K.; Babiak, I.; Giannetto, A.; Fernandes, J.M.O. Daily rhythmicity of clock gene transcripts in atlantic cod fast skeletal muscle. PLoS ONE 2014, 9, e99172. [Google Scholar] [CrossRef]
- Wu, P.; Li, Y.L.; Cheng, J.; Chen, L.; Zhu, X.; Feng, Z.G.; Zhang, J.S.; Chu, W.Y. Daily rhythmicity of clock gene transcript levels in fast and slow muscle fibers from Chinese perch (Siniperca chuatsi). BMC Genom. 2016, 17, 1008. [Google Scholar] [CrossRef]
- Figueredo, D.d.S.; Barbosa, M.R.; Gitaí, D.L.G.; de Andrade, T.G. Predicted microRNAs for mammalian circadian rhythms. J. Biol. Rhythms 2013, 28, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, K.; Chen, H.; Zhao, M.; Ji, G.; Zhang, Y.; Cao, H.; Kan, G.; Li, Y.; Qu, L. Functional annotation of extensively and divergently expressed miRNAs in suprachiasmatic nucleus of ClockΔ19 mutant mice. Biosci. Rep. 2018, 38, BSR20180233. [Google Scholar] [CrossRef] [PubMed]
- Flores, M.V.; Hall, C.; Jury, A.; Crosier, K.; Crosier, P. The zebrafish retinoid-related orphan receptor (Ror) gene family. Gene Expr. Patterns 2007, 7, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Ueda, H.R.; Hayashi, S.; Chen, W.; Sano, M.; Machida, M.; Shigeyoshi, Y.; Iino, M.; Hashimoto, S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005, 37, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Masana, M.I.; Sumaya, I.C.; Becker-Andre, M.; Dubocovich, M.L. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R2357–R2367. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Sharma, A.R.; Sharma, G.; Patra, B.C.; Nam, J.S.; Chakraborty, C.; Lee, S.S. The crucial role and regulations of miRNAs in zebrafish development. Protoplasma 2017, 254, 17–31. [Google Scholar] [CrossRef]
- Sun, J.L.; Zhao, L.L.; He, K.; Liu, Q.; Luo, J.; Zhang, D.M.; Liang, J.; Liao, L.; Yang, S. MiRNA-mRNA integration analysis reveals the regulatory roles of miRNAs in the metabolism of largemouth bass (Micropterus salmoides) livers during acute hypoxic stress. Aquaculture 2020, 526, 735362. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).