An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri)
Abstract
1. Introduction
2. Materials and Methods
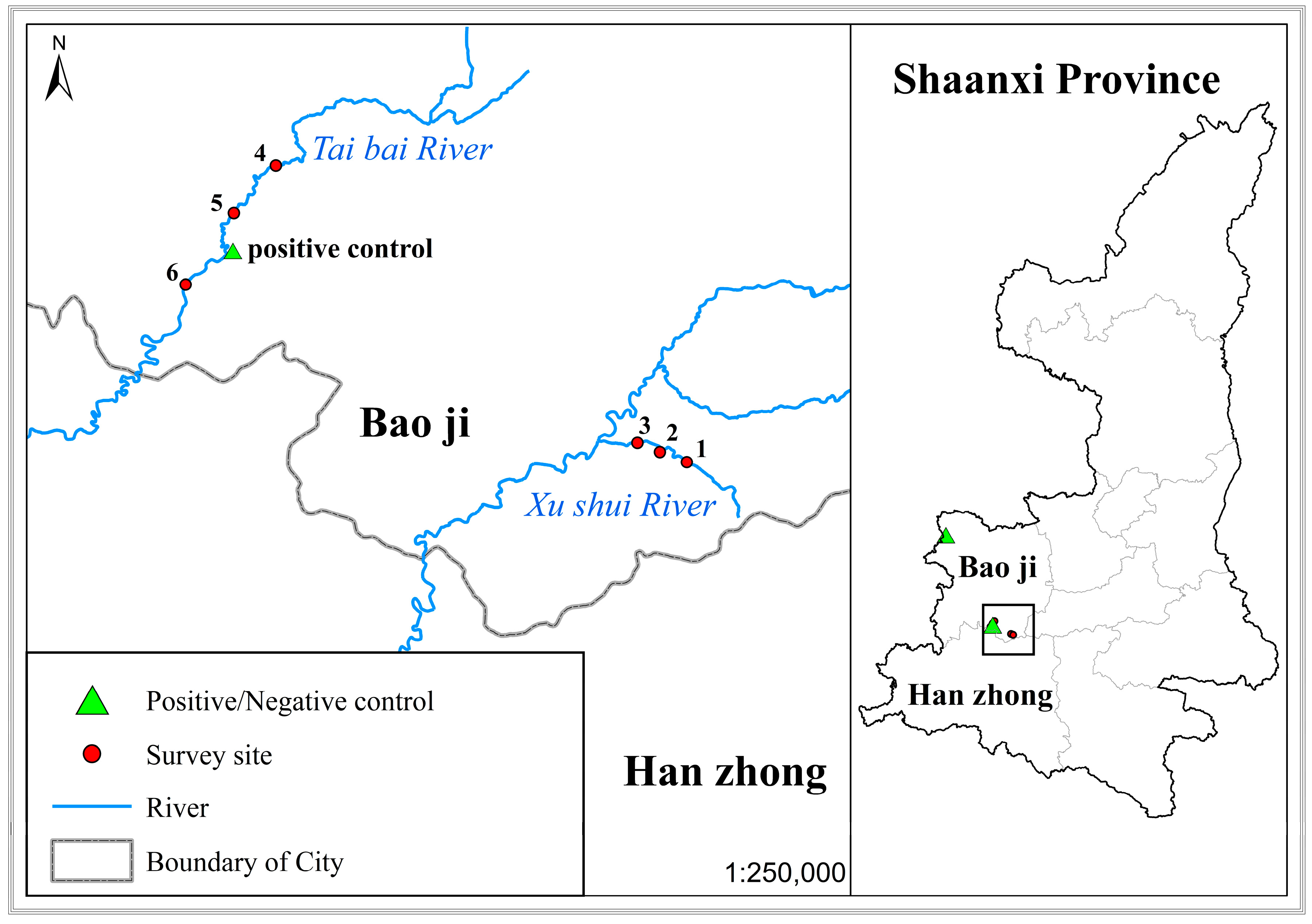
2.1. eDNA Sampling
2.2. Tissue DNA Collection and Extraction
2.3. eDNA Extraction
2.4. Primer Design and Screening
2.5. Optimization of PCR Condition and Verification
3. Results
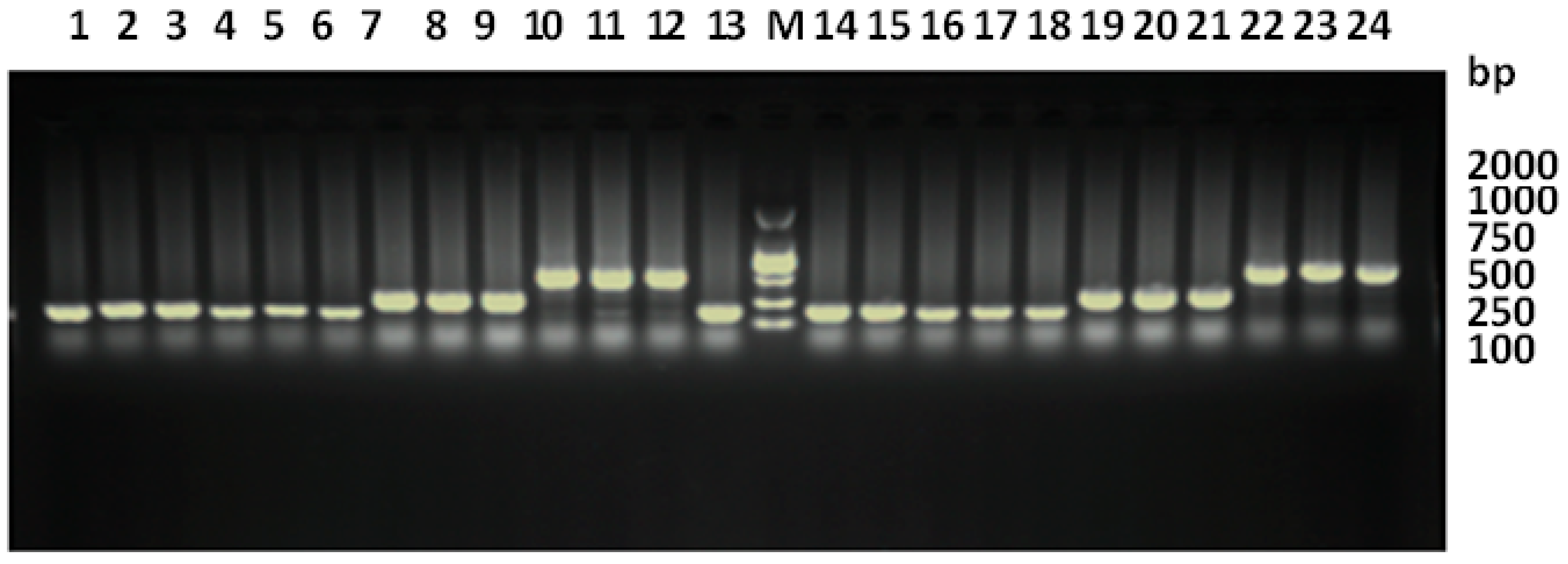
3.1. Primer Testing and PCR Amplification Optimization
3.2. Comparison of eDNA Isolation Methods from Water Samples
3.3. Selection of Sample Volume
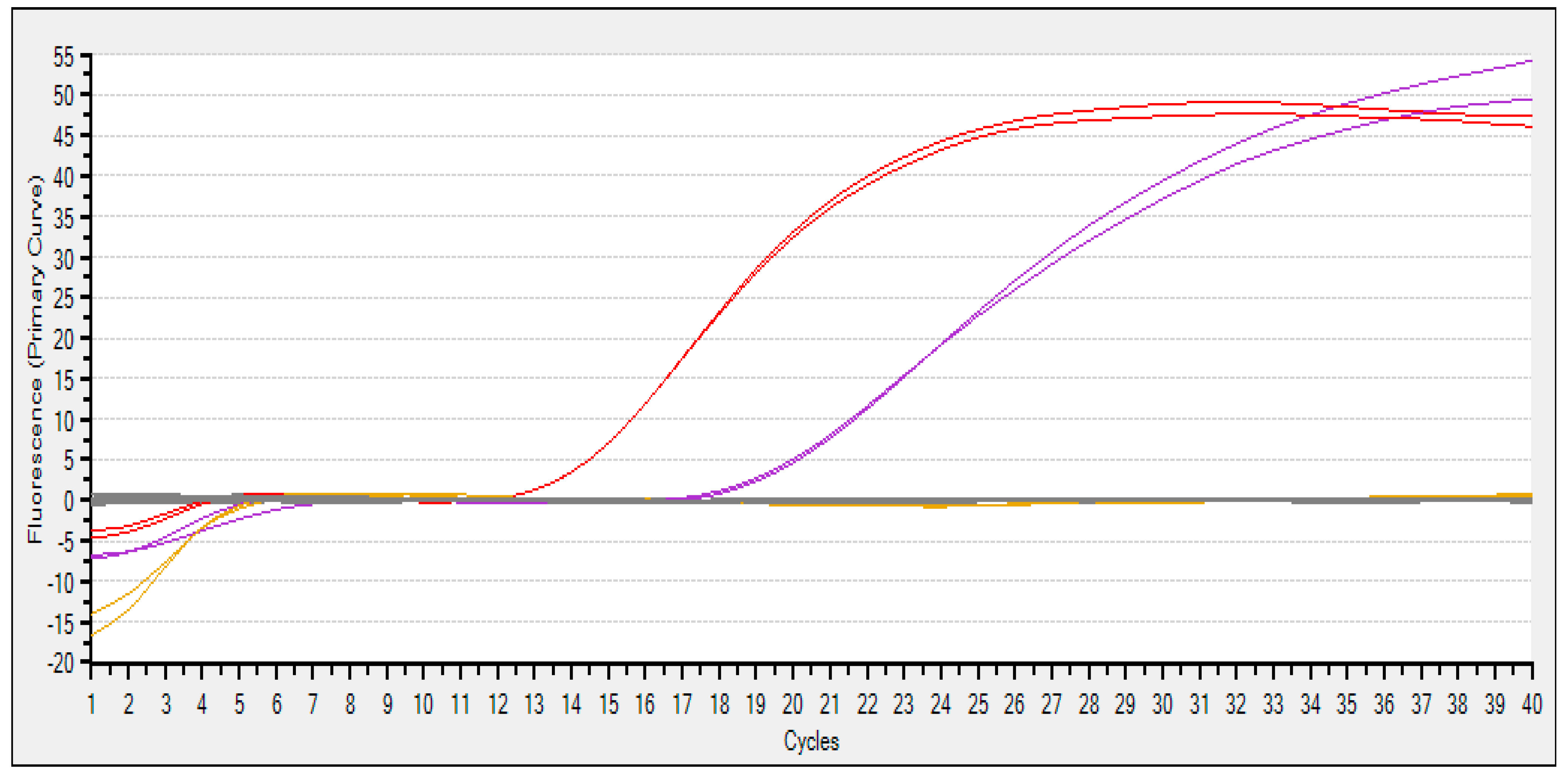
3.4. Quantitative PCR (qPCR) Analysis Using TaqMan Probes
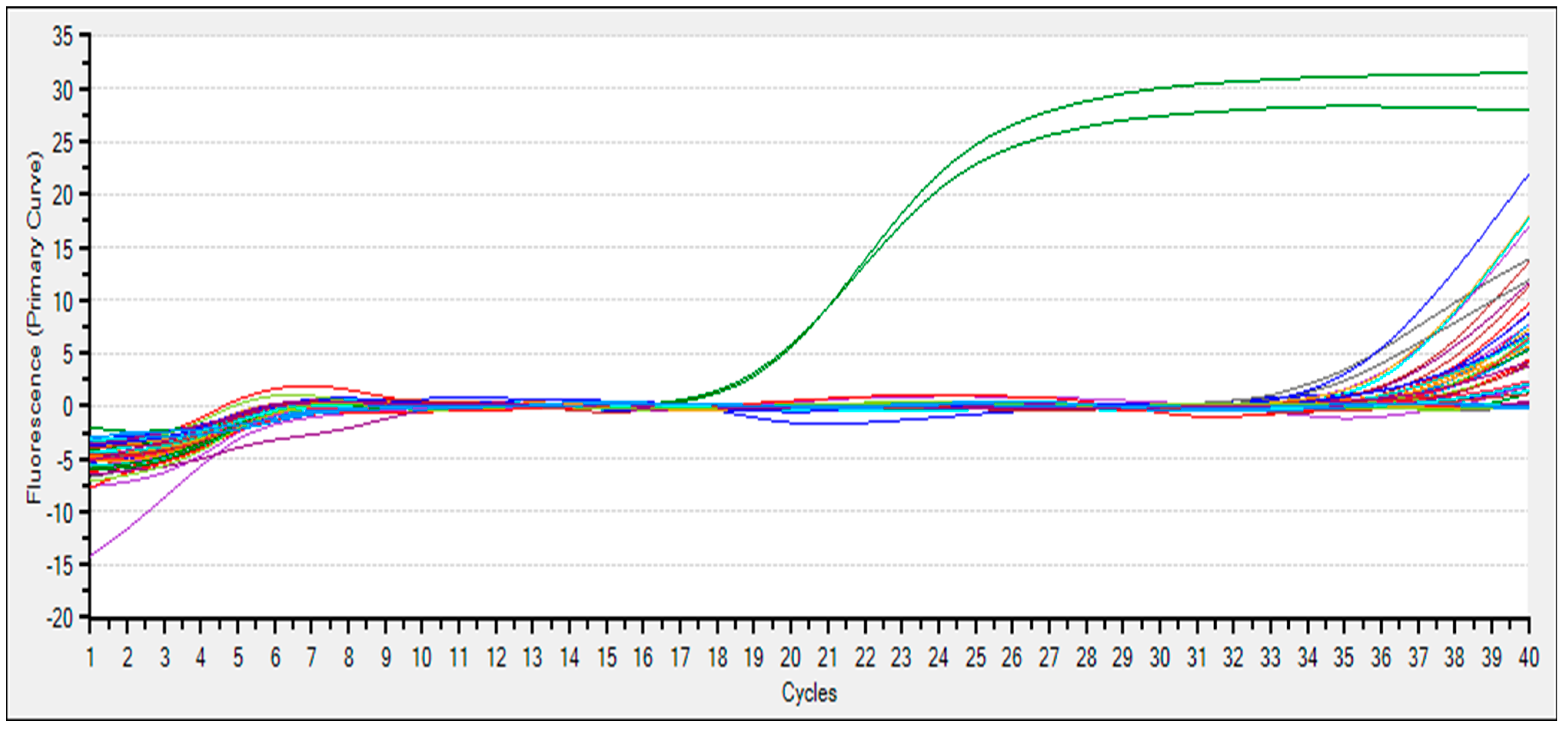
3.5. eDNA Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Snyder, C.D.; Young, J.A. Identification of management thresholds of urban development in support of aquatic biodiversity conservation. Ecol. Indic. 2020, 112, 106124. [Google Scholar] [CrossRef]
- Roper, B.B.; Capurso, J.M.; Paroz, Y.; Young, M.K. Conservation of aquatic biodiversity in the context of multiple-use management on National Forest System lands. Fisheries 2018, 43, 396–405. [Google Scholar] [CrossRef]
- Leng, X.Q.; Du, H.; Xiong, W.; Cheng, P.L.; Luo, J.; Wu, J.M. Successful ultrasonography-assisted artificial reproduction of critically endangered Sichuan taimen (Hucho bleekeri). Fishes 2023, 8, 152. [Google Scholar] [CrossRef]
- Eiler, A.; Lofgren, A.; Hjerne, O.; Norden, S.; Saetre, P. Environmental DNA (eDNA) detects the pool frog (Pelophylax lessonae) at times when traditional monitoring methods are insensitive. Sci. Rep. 2018, 8, 5452. [Google Scholar] [CrossRef] [PubMed]
- Schmelzle, M.C.; Kinziger, A.P. Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol. Ecol. Resour. 2016, 16, 895–908. [Google Scholar] [CrossRef]
- Snyder, D.E. Invited overview: Conclusions from a review of electrofishing and its harmful effects on fish. Rev. Fish Biol. Fish. 2003, 13, 445–453. [Google Scholar] [CrossRef]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef]
- Lydolph, M.C.; Jacobsen, J.; Arctander, P.; Gilbert, M.T.; Gilichinsky, D.A.; Hansen, A.J.; Willerslev, E.; Lange, L. Beringian paleoecology inferred from permafrost-preserved fungal DNA. Appl. Environ. Microbiol. 2005, 71, 1012–1017. [Google Scholar] [CrossRef]
- Valiere, N.; Taberlet, P. Urine collected in the field as a source of DNA for species and individual identification. Mol. Ecol. 2000, 9, 2150–2152. [Google Scholar] [CrossRef]
- Ma, H.Y.; Zhang, H.; Deng, J.; Zhao, H.; Kong, F.; Jiang, W.; Zhang, H.X.; Dong, X.G.; Wang, Q.J. Detection the eDNA of Batrachuperus taibaiensis from the Zhouzhi Heihe River using a nested PCR method and DNA barcoding. Animals 2022, 12, 1105. [Google Scholar] [CrossRef]
- Lam, I.P.Y.; Sung, Y.H.; Fong, J.J. Using eDNA techniques to find the endangered big-headed turtle (Platysternon megacephalum). PLoS ONE 2022, 17, e0262015. [Google Scholar] [CrossRef]
- Uchii, K.; Wakimura, K.; Kikko, T.; Yonekura, R.; Kawaguchi, R.; Komada, H.; Yamanaka, H.; Kenzaka, T.; Tani, K. Environmental DNA monitoring method of the commercially important and endangered fish Gnathopogon caerulescens. Limnology 2022, 23, 49–56. [Google Scholar] [CrossRef]
- Yu, D.; Shen, Z.; Chang, T.; Li, S.; Liu, H. Using environmental DNA methods to improve detectability in an endangered sturgeon (Acipenser sinensis) monitoring program. BMC Ecol. Evol. 2021, 21, 216. [Google Scholar] [CrossRef]
- Kumar, G.; Eble, J.E.; Gaither, M.R. A practical guide to sample preservation and pre-PCR processing of aquatic environmental DNA. Mol. Ecol. Resour. 2020, 20, 29–39. [Google Scholar] [CrossRef]
- Boyd, S.H.; Niemiller, K.D.K.; Dooley, K.E.; Nix, J.; Niemiller, M.L. Using environmental DNA methods to survey for rare groundwater fauna: Detection of an endangered endemic cave crayfish in northern Alabama. PLoS ONE 2020, 15, e0242741. [Google Scholar] [CrossRef]
- Hinlo, R.; Gleeson, D.; Lintermans, M.; Furlan, E. Methods to maximise recovery of environmental DNA from water samples. PLoS ONE 2017, 12, e0179251. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Deiner, K.; Walser, J.C.; Machler, E.; Altermatt, F. Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol. Conserv. 2015, 183, 53–63. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Li, P.L.; Wong, J.K. Advantages of using the QIAshredder instead of restriction digestion to prepare DNA for droplet digital PCR. Biotechniques 2014, 56, 194–196. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular detection of vertebrates in stream water: A demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 2011, 6, e0022746. [Google Scholar] [CrossRef]
- Zhang, Y.; Luan, P.; Ren, G.; Hu, G.; Yin, J. Estimating the inbreeding level and genetic relatedness in an isolated population of critically endangered Sichuan taimen (Hucho bleekeri) using genome-wide SNP markers. Ecol. Evol. 2020, 10, 1390–1400. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.H.; Wang, D.Q.; Wu, J.M.; Wang, C.Y.; Wei, Q.W. Conservation genetics assessment and phylogenetic relationships of critically endangered Hucho bleekeri in China. J. Appl. Ichthyol. 2016, 32, 343–349. [Google Scholar] [CrossRef]
- Loeza-Quintana, T.; Crookes, S.; Li, P.Y.; Reid, D.P.; Smith, M.; Hanner, R.H. Environmental DNA detection of endangered and invasive species in Kejimkujik National Park and historic site. Genome 2021, 64, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Lor, Y.; Schreier, T.M.; Waller, D.L.; Merkes, C.M. Using environmental DNA (eDNA) to detect the endangered Spectaclecase Mussel (Margaritifera monodonta). Freshw. Sci. 2020, 39, 837–847. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Standards for methods utilizing environmental DNA for detection of fish species. Genes 2020, 11, 296. [Google Scholar] [CrossRef]
- Turner, C.R.; Barnes, M.A.; Xu, C.C.Y.; Jones, S.E.; Jerde, C.L.; Lodge, D.M. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 2014, 5, 676–684. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Belle, C.C.; Stoeckle, B.C.; Geist, J. Taxonomic and geographical representation of freshwater environmental DNA research in aquatic conservation. Aquat. Conserv. 2019, 29, 1996–2009. [Google Scholar] [CrossRef]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.L.; Buckley, T.R.; Cruickshank, R.; Dopheide, A.; Handley, K.M.; Hermans, S.; et al. Methods for the extraction, storage, amplification and sequencing of DNA from environmental samples. N. Z. J. Ecol. 2018, 42, 10. [Google Scholar] [CrossRef]
- Rajendhran, J.; Gunasekaran, P. Strategies for accessing soil metagenome for desired applications. Biotechnol. Adv. 2008, 26, 576–590. [Google Scholar] [CrossRef]
- Tringe, S.G.; Rubin, E.M. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005, 6, 805–814. [Google Scholar] [CrossRef]
- Evans, N.T.; Lamberti, G.A. Freshwater fisheries assessment using environmental DNA: A primer on the method, its potential, and shortcomings as a conservation tool. Fish. Res. 2018, 197, 60–66. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA-An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. The detection of aquatic animal species using environmental DNA-a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Yang, J.H.; Zhang, X.W.; Xie, Y.W.; Song, C.; Zhang, Y.; Yu, H.X.; Burton, G.A. Zooplankton community profiling in a eutrophic freshwater ecosystem-lake Tai basin by DNA metabarcoding. Sci. Rep. 2017, 7, 1773. [Google Scholar] [CrossRef]
- Sigut, M.; Kostovcik, M.; Sigutova, H.; Hulcr, J.; Drozd, P.; Hrcek, J. Performance of DNA metabarcoding, standard barcoding, and morphological approach in the identification of host-parasitoid interactions. PLoS ONE 2017, 12, e0187803. [Google Scholar] [CrossRef]
- Abad, D.; Albaina, A.; Aguirre, M.; Laza-Martinez, A.; Uriarte, I.; Iriarte, A.; Villate, F.; Estonba, A. Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Mar. Biol. 2016, 163, 149. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar] [CrossRef]
- Li, C.H.; Ling, L.X.; Tan, J.; Lin, X.L.; Wang, H.; Sun, B.J.; Li, Z. Challenge, breakthrough and future perspectives of environmental DNA technology in monitoring aquatic organisms. J. Shanghai Ocean. Univ. 2023, 32, 564–574. (In Chinese) [Google Scholar] [CrossRef]
- Pedersen, M.W.; Overballe-Petersen, S.; Ermini, L.; Sarkissian, C.D.; Haile, J.; Hellstrom, M.; Spens, J.; Thomsen, P.F.; Bohmann, K.; Cappellini, E.; et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. B 2015, 370, 20130383. [Google Scholar] [CrossRef]
- Knapp, M.; Clarke, A.C.; Horsburgh, K.A.; Matisoo-Smith, E.A. Setting the stage-building and working in an ancient DNA laboratory. Ann. Anat. 2012, 194, 3–6. [Google Scholar] [CrossRef] [PubMed]
| Names | Sequence | Annealing Temperature (°C) | Production Size (bp) |
|---|---|---|---|
| PNL | F 5′-TAAGAACCGACCAACG-3′ | 58.0 | 814 |
| R 5′-GTGCCAAATGTCATAAAG-3′ | |||
| PN1 | F 5′-AAAGCCGAATGTAATGC-3′ | 58.5 | 325 |
| R 5′-AGGATCGTTCAGCGTAG-3′ | |||
| PN2 | F 5′-AAGAACCGACCAACGA-3′ | 59.0 | 193 |
| R 5′-TGGGTAACGGGCAATA-3′ | |||
| P1 | F 5′-AAAACACGGATAACCACC-3′ | 59.0 | 223 |
| R 5′- ACCAAATGCCAGGAATAA-3′ | |||
| Ptaq | F 5′-CCCCTTCATAATTAAAGTATACATTA-3′ | 60.8 | 82 |
| R 5′-GGTCTGATGACAGTGTTG-3′ | 61.5 | ||
| P (FAM) TAACACACTTTATGACATTTGGCACCG (Eclipse) | 69.3 |
| Well | Template Type | Target Name | Sample Name | Ct(CP) |
|---|---|---|---|---|
| C1 | Tissue | JQ675732.1 | B-1 | 19.48 |
| C2 | Tissue | JQ675732.1 | B-1 | 19.66 |
| C3 | Tissue | HM804473.1 | H-2 | 14.09 |
| C4 | Tissue | HM804473.1 | H-2 | 14.09 |
| C5 | BC | HM804473.1 | H2O | -- |
| C6 | BC | HM804473.1 | H2O | -- |
| Well | Sample Type | Sample Name | Ct(CP) | Ct(SDM) |
|---|---|---|---|---|
| B3 | UNKN | XJG1-1 | -- | -- |
| C3 | UNKN | XJG1-1 | -- | -- |
| B4 | UNKN | XJG1-2 | 38.01 | 37.07 |
| C4 | UNKN | XJG1-2 | 37.92 | 36.84 |
| B5 | UNKN | XJG1-3 | 38.02 | 36.77 |
| C5 | UNKN | XJG1-3 | 37.91 | 36.78 |
| B6 | UNKN | XJG2-1 | 39.09 | -- |
| C6 | UNKN | XJG2-1 | -- | -- |
| B7 | UNKN | XJG2-2 | 38.53 | -- |
| C7 | UNKN | XJG2-2 | 38.38 | 37.63 |
| B8 | UNKN | XJG2-3 | 36.97 | 36.14 |
| C8 | UNKN | XJG2-3 | 38.64 | 36.85 |
| B9 | UNKN | XJG3-1 | 37.4 | 37.11 |
| C9 | UNKN | XJG3-1 | 38.87 | 37.31 |
| B10 | UNKN | XJG3-2 | -- | -- |
| C10 | UNKN | XJG3-2 | -- | -- |
| D3 | UNKN | XJG3-3 | 37.8 | 37.5 |
| E3 | UNKN | XJG3-3 | 38.87 | 37.81 |
| D4 | UNKN | TBH2-1 | 33.38 | 33.53 |
| E4 | UNKN | TBH2-1 | 32.66 | 33.1 |
| D5 | UNKN | TBH2-2 | 33.08 | 33.16 |
| E5 | UNKN | TBH2-2 | 32.97 | 33.28 |
| D6 | UNKN | TBH2-3 | 37.03 | 36.87 |
| E6 | UNKN | TBH2-3 | 36.72 | 36.78 |
| D9 | UNKN | SJG4-1 | 37.66 | 38.18 |
| E9 | UNKN | SJG4-1 | 39.5 | -- |
| F2 | UNKN | SJG4-2 | 38.03 | 37.53 |
| G2 | UNKN | SJG4-2 | 35.64 | 35.81 |
| F3 | UNKN | SJG4-3 | 38.54 | 38.19 |
| G3 | UNKN | SJG4-3 | -- | -- |
| D10 | UNKN | TBH4-1 | 35.64 | 35.95 |
| E10 | UNKN | TBH4-1 | 36.39 | 36.62 |
| F3 | UNKN | TBH4-2 | 35.22 | 35.27 |
| G3 | UNKN | TBH4-2 | 35.85 | 35.81 |
| F4 | UNKN | TBH4-3 | 35.96 | 36.35 |
| G4 | UNKN | TBH4-3 | 37.35 | 37.83 |
| F5 | NTC | negative | 34.29 | 34.66 |
| G5 | NTC | negative | 35.37 | 35.28 |
| G8 | BC | H2O | -- | -- |
| G9 | BC | H2O | -- | -- |
| G10 | PTC | positive | 21.48 | 21.27 |
| G11 | PTC | positive | 21.66 | 21.34 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, J.; Zhang, H.; Wang, Q.; Kong, F.; Zhao, H.; Zhang, L.; Jiang, W. An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri). Fishes 2023, 8, 339. https://doi.org/10.3390/fishes8070339
Deng J, Zhang H, Wang Q, Kong F, Zhao H, Zhang L, Jiang W. An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri). Fishes. 2023; 8(7):339. https://doi.org/10.3390/fishes8070339
Chicago/Turabian StyleDeng, Jie, Hongxing Zhang, Qijun Wang, Fei Kong, Hu Zhao, Lu Zhang, and Wei Jiang. 2023. "An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri)" Fishes 8, no. 7: 339. https://doi.org/10.3390/fishes8070339
APA StyleDeng, J., Zhang, H., Wang, Q., Kong, F., Zhao, H., Zhang, L., & Jiang, W. (2023). An Optimized Environmental DNA Method to Improve Detectability of the Endangered Sichuan Taimen (Hucho bleekeri). Fishes, 8(7), 339. https://doi.org/10.3390/fishes8070339







