Study on the Microflora Structure in a Litopenaeus vannamei–Sinonovacula constricta Tandem-Culture Model Based on High-Throughput Sequencing under Different Culture Densities
Abstract
1. Introduction
2. Materials and Methods
2.1. Basic Information of the Shrimp and Razor-Clam Tandem-Culture Tank
2.2. Experiment Design
2.3. Daily Feeding and Management
2.4. Measurement of Water-Quality Indicators in Culture Tanks
2.5. Sample Collection and DNA Extraction
2.6. PCR Amplification and High-Throughput Sequencing of 16S rDNA Sequences
2.7. Data Processing and Analysis
3. Results
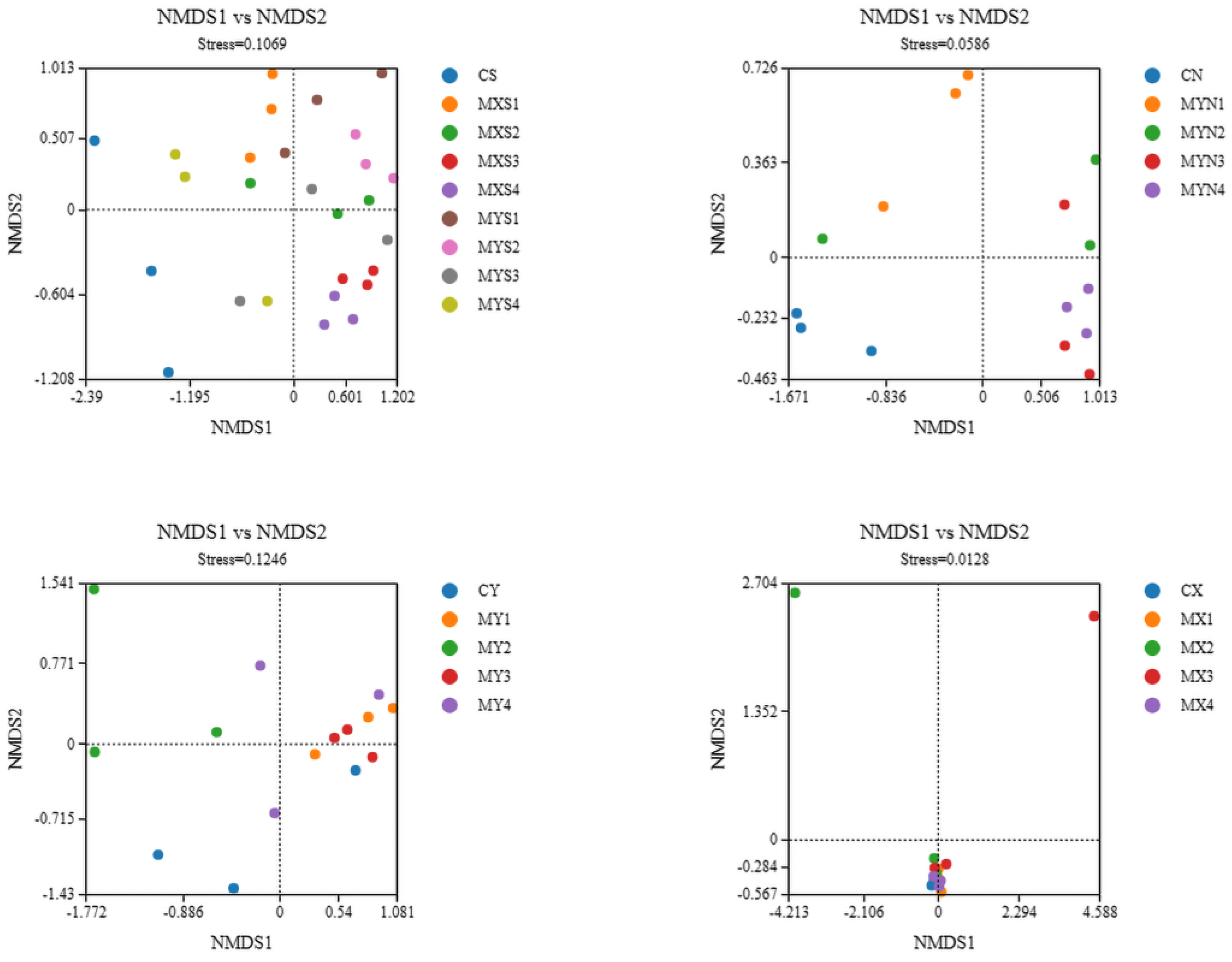
3.1. Structural Compositions and Differences of Bacterial Communities of Each Sample in Different Density Groups
3.1.1. Bacterial Community Structure of Each Sample Based on Phylum Level
3.1.2. Structural Compositions of Bacterial Communities for Each Sample Based on Genus Level
3.2. α Diversity Index
3.3. Correlation of Each Sample Flora with Water Environment Factors
4. Discussion
4.1. Bacterial Community Composition in Different Culture-Density Groups in the Tandem-Culture Model
4.2. Changes in α-Diversity Indexes of Bacterial Communities in Different Culture-Density Groups in the Tandem-Culture Model
4.3. Correlation between Bacterial Composition and Environmental Factors in Shrimp and Razor-Clam Culture Tanks of Different Density Groups in the Tandem-Culture Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Liu, L.; Zheng, B.; Zhu, Y.; Wang, X. Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar. Pollut. Bull. 2013, 72, 181–187. [Google Scholar] [CrossRef]
- Fan, L.; Chen, J.; Meng, S.; Song, C.; Qiu, L.; Hu, G.; Xu, P. Characterization of microbial communities in intensive GIFT tilapia (Oreochromis niloticus) pond systems during the peak period of breeding. Aquac. Res. 2017, 48, 459–472. [Google Scholar] [CrossRef]
- Michaud, L.; Giudice, A.L.; Troussellier, M.; Smedile, F.; Bruni, V.; Blancheton, J.P. Phylogenetic characterization of the heterotrophic bacterial communities inhabiting a marine recirculating aquaculture system. J. Appl. Microbiol. 2010, 107, 1935–1946. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Tao, P.; Tan, J.; Mu, H.; Peng, L.; Yang, D.; Tong, S.; Chen, L. Identification of Bacterial Community Composition in Freshwater Aquaculture System Farming of Litopenaeus vannamei Reveals Distinct Temperature-Driven Patterns. Int. J. Mol. Sci. 2014, 15, 13663–13680. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, J.; Zhang, C.; Qin, J.; Wang, Y. Bacterial composition, abundance and diversity in fish polyculture and mussel-fish integrated cultured ponds in China. Aquac. Res. 2017, 48, 3950–3961. [Google Scholar]
- Wang, X.; Xie, X.; Jin, S.; Zhu, J.; Zhao, Q.; Zhou, S. Seasonal changes of the bacterial community structure of Sinonovacula constricta and its aquaculture ponds in high-throughput sequencing. J. Appl. Ecol. 2019, 30, 4267–4276. [Google Scholar]
- Guan, X.; Wang, B.; Jiang, J.; Tian, J.; Dong, Y.; Chen, Z.; Gao, S.; Wang, X.; Sun, B.; Duan, P.; et al. Comparative study on microbial communities in sediments of mixed culture ponds of Penaeus vannamei in China. Mar. Sci. Bull. 2020, 39, 730–739. [Google Scholar]
- Fabrini, C.; Gustavo, B.G.; Olav, V.; Schveitzer, R. Management of biofloc concentrations as an ecological strategy for microbial control in intensive shrimp culture. Aquaculture 2021, 543, 736969. [Google Scholar]
- Radhey, S. The phylogeny of proteobacteria: Relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 2000, 24, 367–402. [Google Scholar]
- Li, Q.; Zhang, Y.; Juck, D.; Fortin, N.; Greer, C.W.; Tang, Q. Phylogenetic analysis of bacterial communities in the shrimp and sea cucumber aquaculture environment in northern China by culturing and PCR–DGGE. Aquacult. Int. 2010, 18, 977–990. [Google Scholar] [CrossRef]
- Betina, L.; Yang, W.; Zheng, Y.; Nicholaus, R.; Zhu, J.; Zhu, J. Bioturbation by the razor clam (Sinonovacula constricta) on the microbial community and enzymatic activities in the sediment of an ecological aquaculture wastewater treatment system. Sci. Total Environ. 2018, 643, 1098–1107. [Google Scholar]
- Liu, C.; Jie, R.; Jiang, Z.; Zhang, M.; Wang, J. Development of a Blocking Primer to Inhibit the PCR Amplification of the 18S rDNA Sequences of Litopenaeus vannamei and Its Efficacy in Crassostrea hongkongensis. Front. Microbiol. 2019, 10, 830. [Google Scholar] [CrossRef]
- Mukherjee, A.; Rodiles, A.; Merrifield, D.L.; Chandra, G.; Ghosh, K. Exploring intestinal microbiome composition in three Indian major carps under polyculture system: A high-throughput sequencing based approach. Aquaculture 2020, 524, 735206. [Google Scholar] [CrossRef]
- Zhao, Y.; Guo, H.; Zhang, D. Effects of different farming modes on intestinal microbial community of Litopenaeus vannamei. J. Fish. China 2021, 45, 221–234. [Google Scholar]
- Adrien, F.; Yanath, B.; Francois, K.; Drider, D.; Dhulster, P.; Firdaous, L. Recovery of nisin from culture supernatants of Lactococcus lactis by ultrafiltration: Flux properties and separation efficiency. Food Bioprod. Process. 2022, 136, 196–210. [Google Scholar]
- Allison, S.D.; Martiny, J. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Ramakrishna, B.S. Role of the gut microbiota in human nutrition and metabolism. J. Gastroen. Hepatol. 2013, 28, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Dai, W.; Zhu, J.; Liu, K.; Dong, C.; Qiu, Q. The Underlying Ecological Processes of Gut Microbiota Among Cohabitating Retarded, Overgrown and Normal Shrimp. Microb. Ecol. 2017, 73, 988–999. [Google Scholar] [CrossRef]
- Bischoff, V.; Zucker, F.; Moraru, C. Marine Bacteriophages. Encycl. Virol. (Fourth Ed.) 2021, 4, 322–341. [Google Scholar]
- Asmani, K.; Petton, B.; Grand, J.L.; Mounier, J.; Robert, R.; Nicolas, J.L. Establishment of microbiota in larval culture of Pacific oyster, Crassostrea gigas. Aquaculture 2016, 464, 434–444. [Google Scholar] [CrossRef]
- Krüger, K.; Chafee, M.; Francis, T.B.; del Rio, T.G.; Becher, D.; Schweder, T.; Amann, R.I.; Teeling, H. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019, 13, 2800–2816. [Google Scholar] [CrossRef]
- Meng, D.; Li, J.; Liu, T.; Liu, Y.; Yan, M.; Hu, J.; Li, X.; Liu, X.; Liang, Y.; Liu, H.; et al. Effects of redox potential on soil cadmium solubility: Insight into microbial community. J. Environ. Sci. 2019, 75, 224–232. [Google Scholar] [CrossRef]
- Guo, C.; Zhang, X.; Luan, S.; Zhou, H.; Liu, L.; Qu, Y. Diversity and structure of soil bacterial community in intertidal zone of Daliao River estuary, Northeast China. Mar. Pollut. Bull. 2021, 163, 111965. [Google Scholar] [CrossRef] [PubMed]
- MußMann, M.; Pjevac, P.; Krüger, K.; Dyksma, S. Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 2017, 11, 1276–1281. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, K.; Renuka; Arun, K. New-Generation Probiotics. In Microbiome and Metabolome in Diagnosis, Therapy, and other Strategic Applications; Faintuch, J., Faintuch, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 417–424. [Google Scholar]
- Hester, E.R.; Harpenslager, S.F.; van Diggelen, J.M.H.; Lamers, L.L.; Jetten, M.S.M.; Luke, C.; Lucker, S.; Welte, C.U. Linking nitrogen load to the structure and function of wetland soil and rhizosphere microbial communities. mSystems 2018, 3, e00214-17. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, Y.; Liu, X.; Yuan, L.; Liu, X.; Wang, L.; Sun, H. Effects of endogenous potential probiotic Lactobacillus rhamnosus M2-4 on intestinal microflora and metabonomics in juvenile sea cucumber Apostichopus japonicus. Aquaculture 2022, 555, 738247. [Google Scholar] [CrossRef]
- Kong, N.; Han, S.; Fu, Q.; Yu, Z.; Wang, L.; Song, L. Impact of ocean acidification on the intestinal microflora of the Pacific oyster Crassostrea gigas. Aquaculture 2022, 546, 737365. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, Z.; Li, X.; Long, Z.; Pei, Y. Comprehensive analysis of the migration and transformation of nutrients between sediment and overlying water in complex habitat systems. Sci. Total Environ. 2022, 852, 158433. [Google Scholar] [CrossRef]
- Sheng, P.; Chen, Q.; Han, Q.; Lu, J.; Zhang, B.; Jiang, M.; Peng, R.; Jiang, X. Bacterial community in gut, shell sediment, and surrounding water of Hyriopsis cumingii fed three different microalgal diets. Aquaculture 2021, 540, 736701. [Google Scholar] [CrossRef]
- Zheng, Z.; Lin, B.; Wang, Y.; Liao, M.; Wang, L.; Rong, X. Analysis of the structural characteristics of digestive tract flora of Cynoglossus semilaevis cultured in ponds in high-throughput sequencing. J. Aquat. Biol. 2015, 39, 38–45. [Google Scholar]
- Lu, J.; Li, S.; He, X.; Tang, R.; Li, D. An in-pond tank culture system for high-intensive fish production: Effect of stocking density on growth of grass carp (Ctenopharyngodon idella Valenciennes, 1844) and blunt snout bream (Megalobrama amblycephala Yih, 1955). Aquaculture 2022, 549, 737808. [Google Scholar] [CrossRef]
- Aurobinda, U.; Himanshu, S.S.; Basanta, K.D.; Ramteke, M.H.; Kumar, V.; Krishna, G.; Mohanty, B.P.; Chadha, N.K.; Das, A.K. Stocking density matters in open water cage culture: Influence on growth, digestive enzymes, haemato-immuno and stress responses of Puntius sarana (Ham, 1822). Aquaculture 2022, 547, 737445. [Google Scholar]
- Zhou, L.; Qu, Y.; Qin, J.; Chen, L.; Han, F.; Li, E. Deep insight into bacterial community characterization and relationship in the pond water, sediment and the gut of shrimp (Penaeus japonicus). Aquaculture 2020, 539, 736658. [Google Scholar] [CrossRef]
- Alexia, O.; Regina, E.G.; Eduardo, Q.G.; Cristina, E.F.; Roberto, H.H.; Alberto, P.R. Digestive microbiota of shrimp Penaeus vannamei and oyster Crassostrea gigas co-cultured in integrated multi-trophic aquaculture system. Aquaculture 2020, 521, 735059. [Google Scholar]
- Fernandes, G.L.; Shenoy, B.D.; Damare, S.R. Diversity of bacterial community in the oxygen minimum zones of Arabian Sea and Bay of Bengal as deduced by illumina sequencing. Front. Microbiol. 2022, 10, 3153. [Google Scholar] [CrossRef]








| Cultured Organisms | Culture Tank Groups | |||
|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | Group 4 | |
| Stocking Density of Pacific White Shrimp (ind/m2) | 40 | 60 | 80 | 100 |
| Actual Number of Shrimp Raised (ind) | 8 | 12 | 16 | 20 |
| Stocking Density of Razor Clams (ind/m2) | 200 | 300 | 400 | 500 |
| Actual Number of Razor Clams Raised (ind) | 10 | 15 | 20 | 25 |
| Indicator | CS | MXS1 | MXS2 | MXS3 | MXS4 | MYS1 | MYS2 | MYS3 | MYS4 |
|---|---|---|---|---|---|---|---|---|---|
| DO | 7.96 ± 0.27 a | 7.39 ± 0.09 b | 7.27 ± 0.06 b | 7.17 ± 0.08 b | 7.11 ± 0.10 b | 6.55 ± 0.07 c | 6.44 ± 0.07 cd | 6.32 ± 0.11 cd | 6.23 ± 0.12 d |
| pH | 7.97 ± 0.01 c | 7.99 ± 0.06 bc | 8.03 ± 0.01 abc | 8.05 ± 0.01 ab | 8.02 ± 0.02 abc | 8.05 ± 0.04 ab | 8.03 ± 0.01 abc | 8.08 ± 0.05 a | 8.02 ± 0.02 abc |
| COD | 0.64 ± 0.01 f | 5.75 ± 0.15 de | 5.53 ± 0.02 e | 6.10 ± 0.11 bc | 6.40 ± 0.26 a | 5.84 ± 0.08 cd | 5.69 ± 0.13 de | 5.84 ± 0.11 cd | 6.25 ± 0.11 ab |
| TN | 1.10 ± 0.00 f | 8.71 ± 0.63 e | 10.44 ± 0.69 cd | 12.16 ± 0.57 b | 14.91 ± 0.47 a | 9.50 ± 1.04 de | 11.33 ± 0.45 bc | 12.64 ± 0.51 b | 14.01 ± 0.49 a |
| NO3−-N | 0.51 ± 0.00 e | 3.24 ± 0.23 cd | 3.94 ± 0.83 bc | 4.37 ± 0.48 b | 6.06 ± 0.71 a | 2.71 ± 0.44 d | 2.67 ± 0.30 d | 3.08 ± 0.23 cd | 4.13 ± 0.57 bc |
| NO2−-N | 0.01 ± 0.00 d | 0.11 ± 0.02 c | 0.17 ± 0.01 b | 0.24 ± 0.02 a | 0.24 ± 0.02 a | 0.10 ± 0.01 c | 0.17 ± 0.02 b | 0.21 ± 0.00 a | 0.23 ± 0.02 a |
| NH4+-N | 0.002 ± 0.00 e | 0.11 ± 0.09 cd | 0.14 ± 0.02 bcd | 0.18 ± 0.00 abc | 0.23 ± 0.03 ab | 0.08 ± 0.05 de | 0.14 ± 0.03 bcd | 0.16 ± 0.05 abcd | 0.25 ± 0.03 a |
| TP | 0.25 ± 0.00 e | 1.72 ± 0.13 d | 2.14 ± 0.17 c | 2.58 ± 0.12 ab | 2.33 ± 0.23 bc | 1.45 ± 0.04 d | 2.09 ± 0.26 c | 2.79 ± 0.19 a | 2.44 ± 0.18 abc |
| PO43−-P | 0.13 ± 0.00 c | 0.17 ± 0.01 c | 0.24 ± 0.04 b | 0.31 ± 0.02 a | 0.25 ± 0.03 b | 0.17 ± 0.03 c | 0.24 ± 0.03 b | 0.31 ± 0.02 a | 0.26 ± 0.03 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Xu, J.; Xu, S.; Bao, G.; Wang, D. Study on the Microflora Structure in a Litopenaeus vannamei–Sinonovacula constricta Tandem-Culture Model Based on High-Throughput Sequencing under Different Culture Densities. Fishes 2023, 8, 287. https://doi.org/10.3390/fishes8060287
Zhao C, Xu J, Xu S, Bao G, Wang D. Study on the Microflora Structure in a Litopenaeus vannamei–Sinonovacula constricta Tandem-Culture Model Based on High-Throughput Sequencing under Different Culture Densities. Fishes. 2023; 8(6):287. https://doi.org/10.3390/fishes8060287
Chicago/Turabian StyleZhao, Chunpu, Jilin Xu, Shanliang Xu, Gege Bao, and Danli Wang. 2023. "Study on the Microflora Structure in a Litopenaeus vannamei–Sinonovacula constricta Tandem-Culture Model Based on High-Throughput Sequencing under Different Culture Densities" Fishes 8, no. 6: 287. https://doi.org/10.3390/fishes8060287
APA StyleZhao, C., Xu, J., Xu, S., Bao, G., & Wang, D. (2023). Study on the Microflora Structure in a Litopenaeus vannamei–Sinonovacula constricta Tandem-Culture Model Based on High-Throughput Sequencing under Different Culture Densities. Fishes, 8(6), 287. https://doi.org/10.3390/fishes8060287






