Diseased Fish Detection in the Underwater Environment Using an Improved YOLOV5 Network for Intensive Aquaculture
Abstract
1. Introduction
2. Materials and Methods
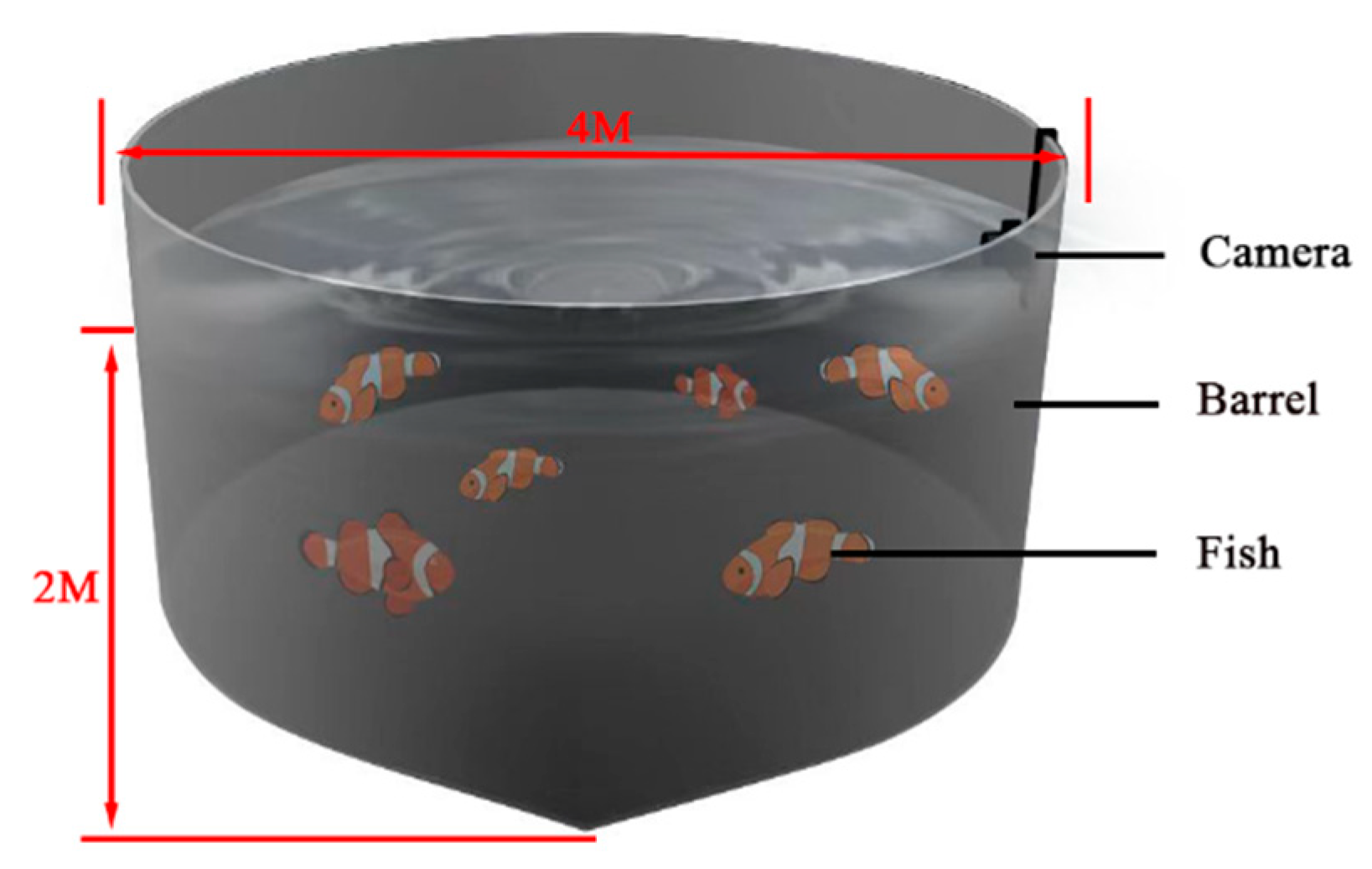
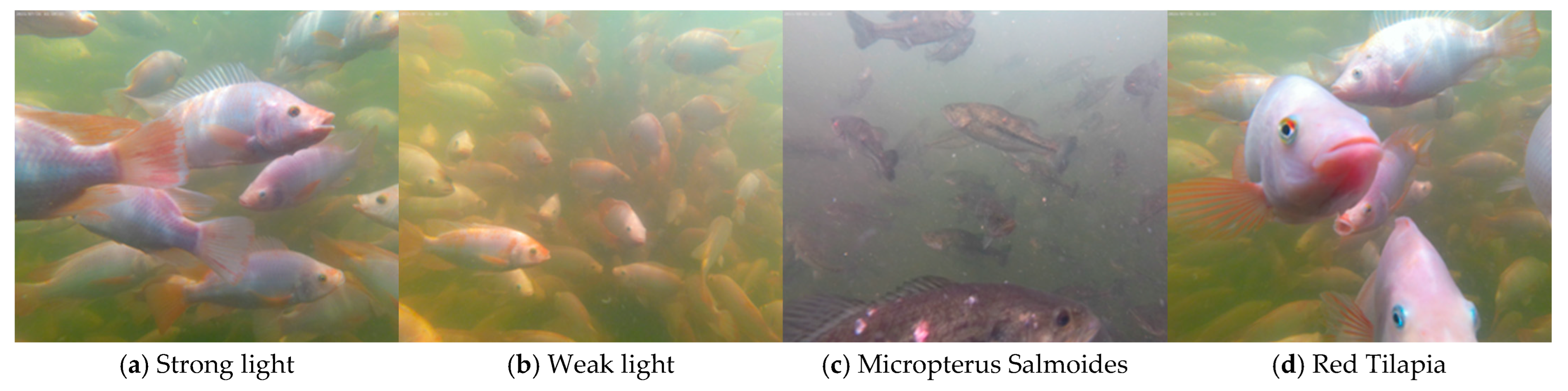
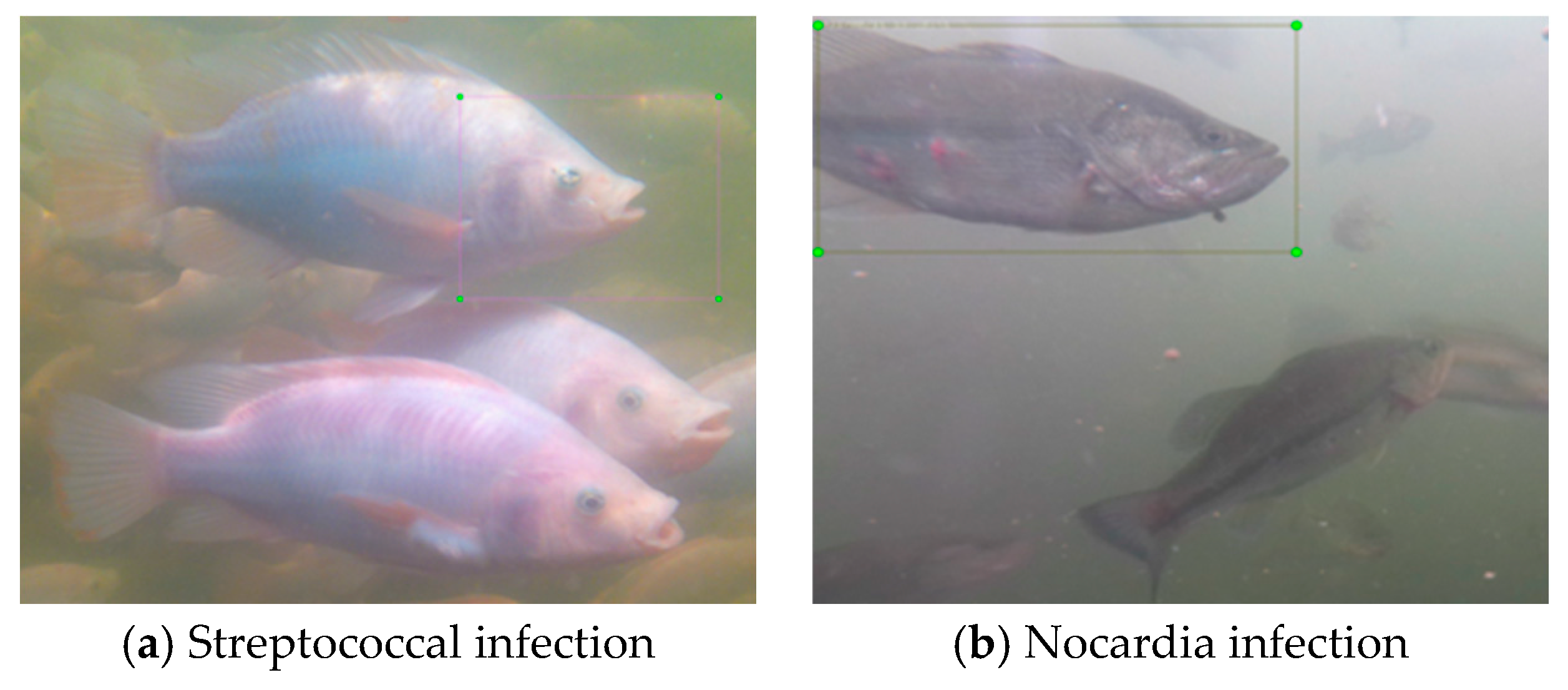
2.1. Dataset
2.2. The Diseased Fish Identification Algorithms
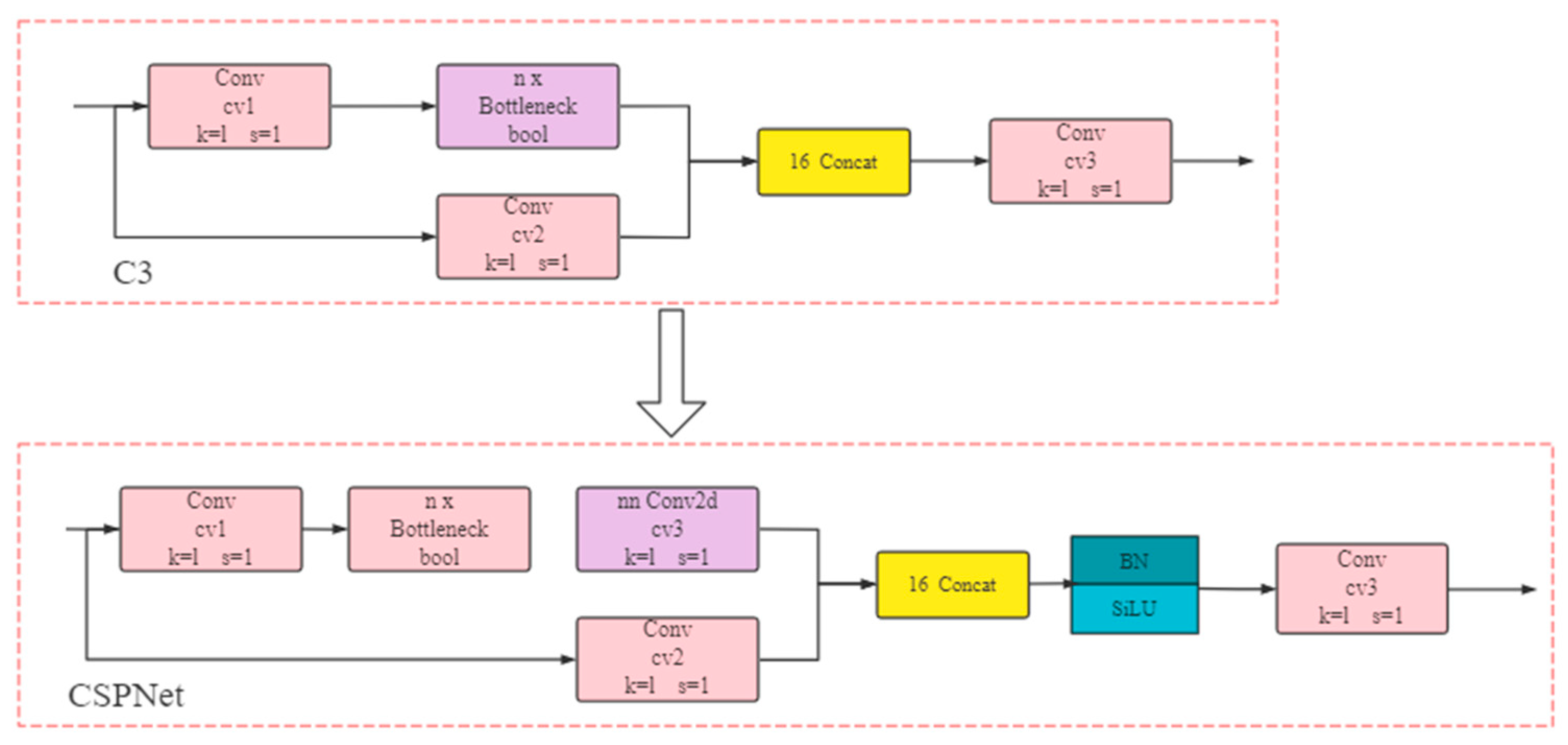
- The implementation of lightweight computing is key to the industrial deployment of algorithms. In order to achieve lightweight computing, the C3 component is used instead of the CSPNet [29] component of the YOLOV5 model in this study. As shown in Figure 5, compared to the CSPNet component, the C3 component removes one convolution operation after the skip connect, and splices directly with another branch of the input image after one convolution operation. The C3 component has a similar effect to the CSPNet component, but with a simpler structure.
- 2.
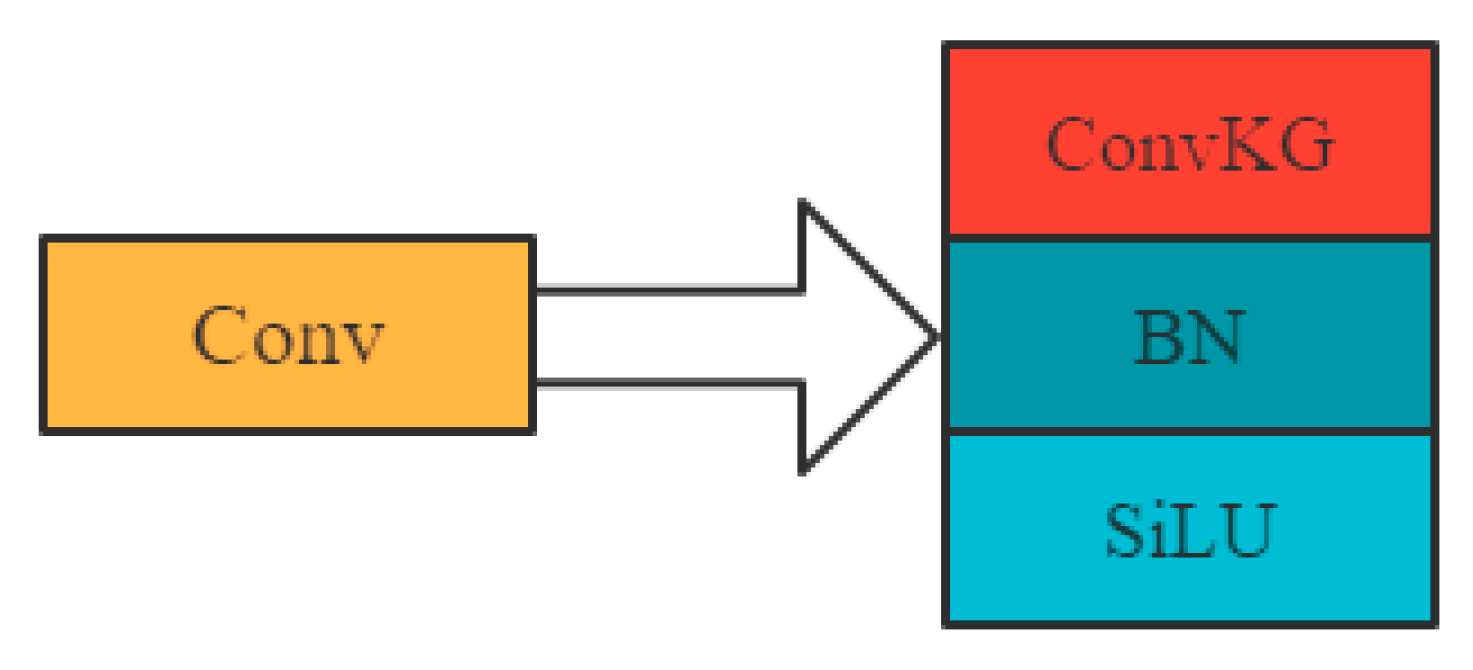
- As fish activity photos are taken underwater, the scenes in the photos often change with water quality, lighting and fish conditions, resulting in varying degrees of variation and obscuration of fish features, and the original YOLOV5 backbone network is unable to extract clear features. In order to reduce the impact of underwater complexities, this study improves the generalization ability of the network by expanding the training samples and replacing all 3 × 3 convolutional kernels in the backbone network with convolutional kernel group (Conv KG), to enhance the network’s ability to extract features from the photographed fish. Conv KG consists of three parallel 3 × 3, 1 × 3 and 3 × 1 convolutional kernels, which convolve the input image in the same steps to produce feature maps of the same size and number of channels, respectively, and the corresponding feature maps are summed to obtain the output features, as shown in Figure 6. The three parallel convolutional kernels enhance the network’s ability to extract fish surface features.
- 3.
- Due to the variable underwater environment and the large variation in the number of fish in fish photos, which interferes with the original YOLOV5 detection algorithm, this study adds the attention mechanism module, convolutional block attention module (CBAM), to the YOLOV5 network (Figure 7). CBAM is a simple, but effective feed-forward convolutional neural network attention module, which combines the channel attention module (CAM) and spatial attention module (SAM) [30]. Given an intermediate feature map, our module sequentially inferred the attention of an image along two independent dimensions, channel and space, and then multiplied the attention map by the input feature map for adaptive feature refinement. Because CBAM is a lightweight, general-purpose module, it can be seamlessly integrated into any convolutional neural network architecture with negligible overhead, and can be trained end-to-end with the underlying convolutional neural network.
2.3. Experimental Environment
2.4. Performance Evaluation Metrics
3. Results
3.1. Training Result
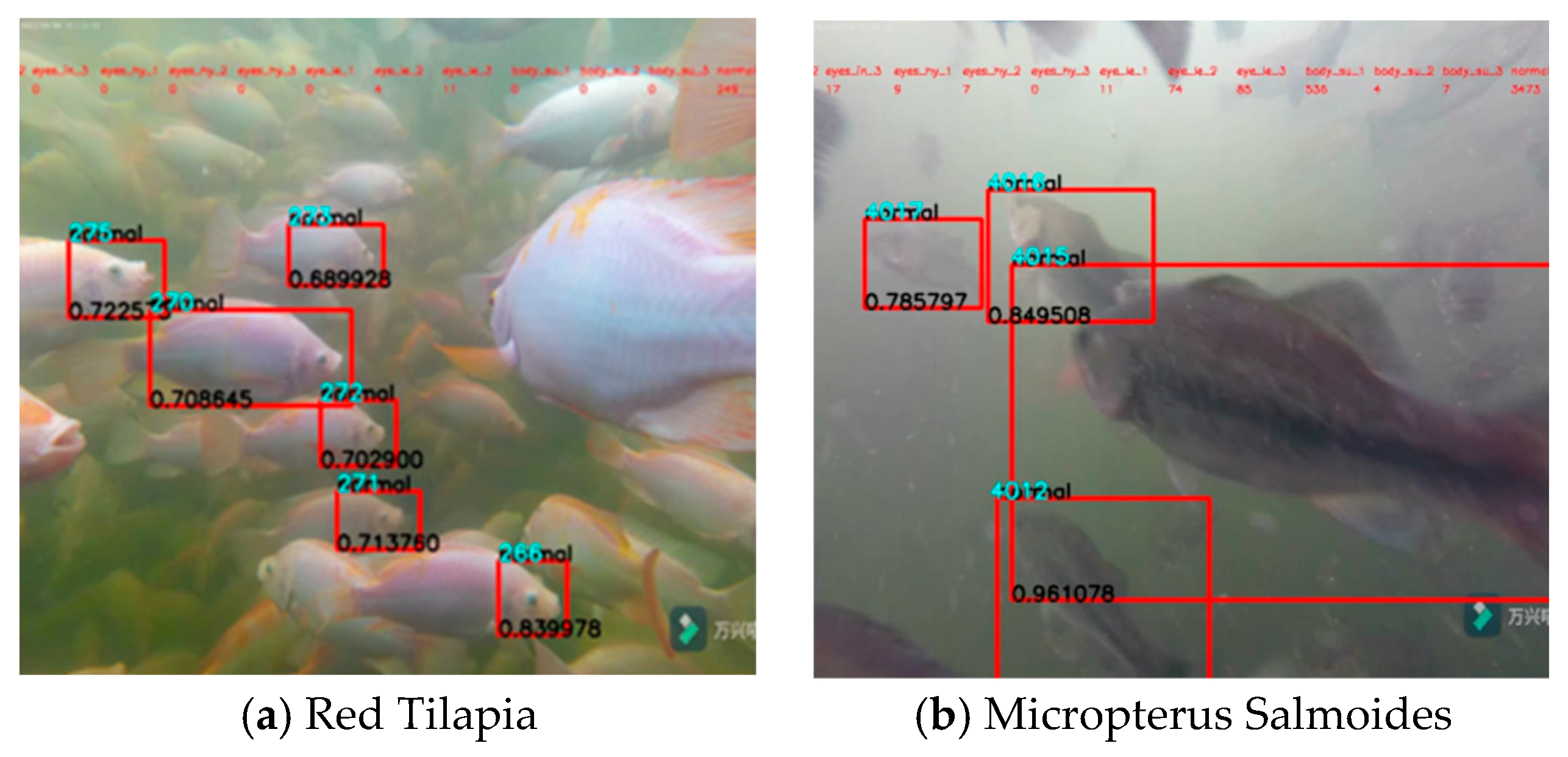
3.2. Identification Results under Various Varieties
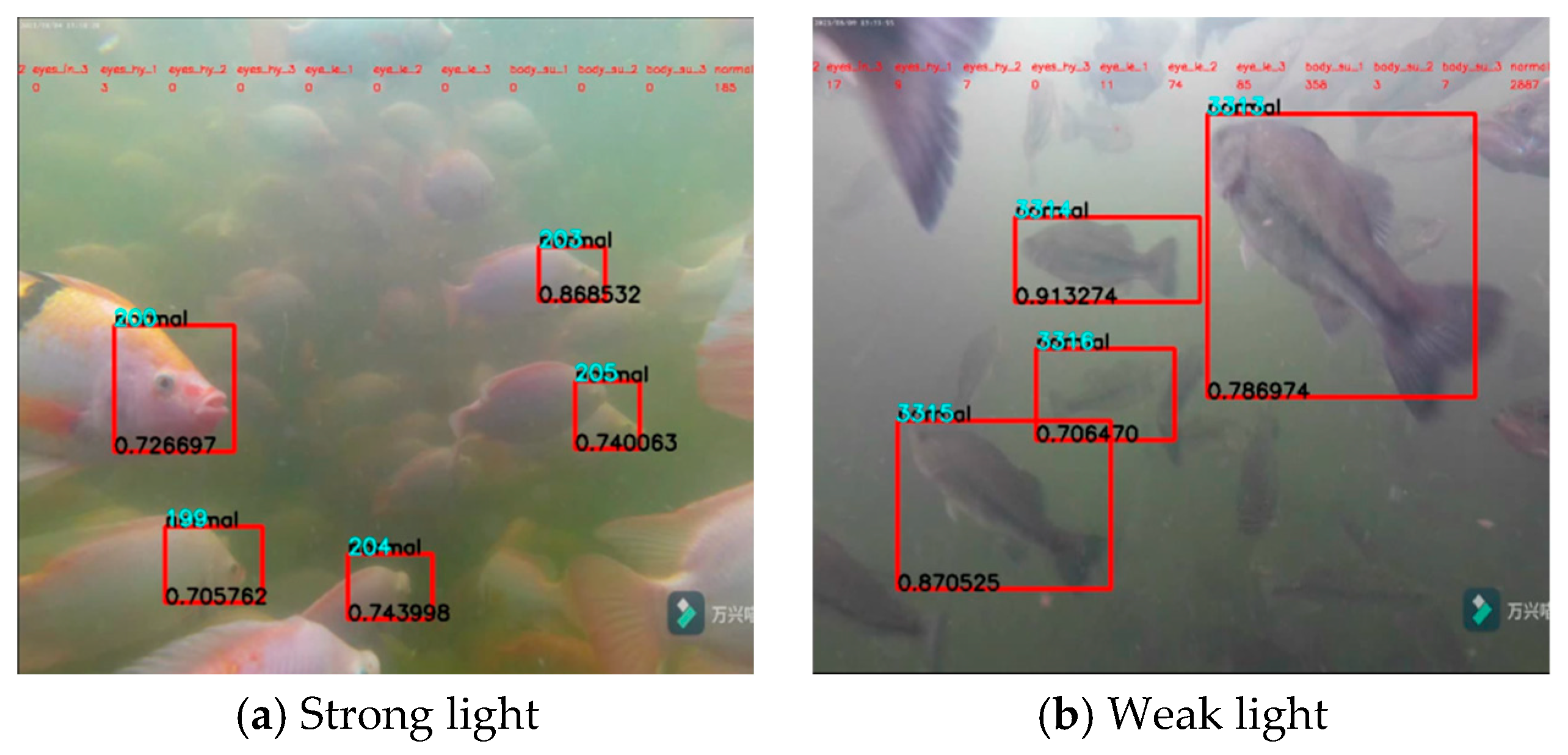
3.3. Identification Results under Various Environments
3.4. Contrast to Mainstream Target Detection Networks
3.5. Ablation Experiment
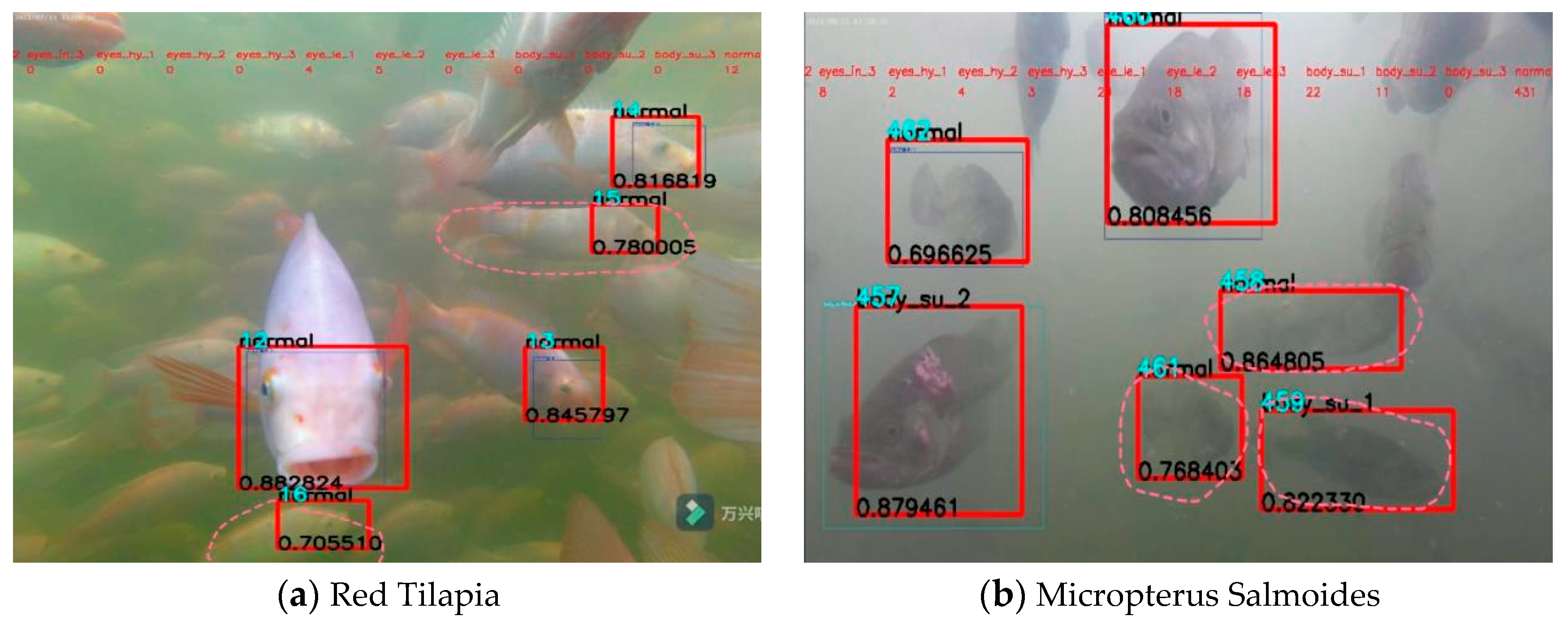
3.6. Comparison of before and after Improvement
4. Discussion
4.1. Robustness of the Method
4.2. Comparison with Other Methods
4.3. Contribution to the Detection of Fish Diseases
5. Conclusions
- The algorithm used in this paper is a target recognition algorithm. It does not actually support multi-target dynamic tracking. As a result, it is unable to track fish movements, and the information collected is too simple to provide a comprehensive picture of their health.
- The dataset used in this study for two common fish diseases is not sufficient for practical application. More species of fish disease behavior should be collected to supplement the dataset.
- In future work, we will use stereo and multispectral cameras to capture images, to reduce the impact of illumination, water quality and refraction on image quality. In addition, we have found a number of methods regarding geometric features that can be used to describe the size, weight and swimming speed of the fish, to construct a more accurate estimation model of the health of the fish.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ai, L.; Ma, B.; Shao, S.; Zhang, L.; Zhang, L. Heavy metals in Chinese freshwater fish: Levels, regional distribution, sources and health risk assessment. Sci. Total Environ. 2022, 853, 158455. [Google Scholar] [CrossRef]
- Fang, J.; Fabinyi, M. Characteristics and Dynamics of the Freshwater Fish Market in Chengdu, China. Front. Sustain. Food Syst. 2021, 5, 638997. [Google Scholar] [CrossRef]
- Fluet-Chouinard, E.; Funge-Smith, S.; McIntyre, P.B. Global hidden harvest of freshwater fish revealed by household surveys. Proc. Natl. Acad. Sci. USA 2018, 115, 7623–7628. [Google Scholar] [CrossRef]
- Zhang, Y. Freshwater biodiversity conservation in China: Progress in the Yangtze River basin. Aquat. Conserv.-Mar. Freshw. Ecosyst. 2022, 32, 1565–1570. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Antimicrobial resistance in aquaculture: Current knowledge and alternatives to tackle the problem. Int. J. Antimicrob. Agents 2018, 52, 135–143. [Google Scholar] [CrossRef]
- Bulfon, C.; Volpatti, D.; Galeotti, M. Current research on the use of plant-derived products in farmed fish. Aquac. Res. 2015, 46, 513–551. [Google Scholar] [CrossRef]
- Boyd, C.E.; D’Abramo, L.R.; Glencross, B.D.; Huyben, D.C.; Juarez, L.M.; Lockwood, G.S.; McNevin, A.A.; Tacon, A.G.J.; Teletchea, F.; Tomasso, J.R.; et al. Achieving sustainable aquaculture: Historical and current perspectives and future needs and challenges. J. World Aquac. Soc. 2020, 51, 578–633. [Google Scholar] [CrossRef]
- Assefa, A.; Abunna, F. Maintenance of Fish Health in Aquaculture: Review of Epidemiological Approaches for Prevention and Control of Infectious Disease of Fish. Vet. Med. Int. 2018, 2018, 5432497. [Google Scholar] [CrossRef]
- Chen, C.-H.; Chen, L.-H.; Chen, C.-Y. Automatic Fish Segmentation and Recognition in Taiwan Fish Market using Deep Learning Techniques. J. Imaging Sci. Technol. 2021, 65, 40403-1–40403-10. [Google Scholar] [CrossRef]
- Rauf, H.T.; Lali, M.I.U.; Zahoor, S.; Shah, S.Z.H.; Rehman, A.U.; Bukhari, S.A.C. Visual features based automated identification of fish species using deep convolutional neural networks. Comput. Electron. Agric. 2019, 167, 105075. [Google Scholar] [CrossRef]
- Qi, C.; Chang, J.; Zhang, J.; Zuo, Y.; Ben, Z.; Chen, K. Medicinal Chrysanthemum Detection under Complex Environments Using the MC-LCNN Model. Plants 2022, 11, 838. [Google Scholar] [CrossRef]
- Maloy, H.; Aamodt, A.; Misimi, E. A spatio-temporal recurrent network for salmon feeding action recognition from underwater videos in aquaculture. Comput. Electron. Agric. 2019, 167, 105087. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Wang, Y.; Zhao, Z.; Liu, J.; Liu, Y.; Sun, C.; Zhou, C. Automatic Fish Population Counting by Machine Vision and a Hybrid Deep Neural Network Model. Animals 2020, 10, 364. [Google Scholar] [CrossRef]
- Labao, A.B.; Naval, P.C., Jr. Cascaded deep network systems with linked ensemble components for underwater fish detection in the wild. Ecol. Inform. 2019, 52, 103–121. [Google Scholar] [CrossRef]
- Roy, A.M.; Bhaduri, J. Real-time growth stage detection model for high degree of occultation using DenseNet-fused YOLOv4. Comput. Electron. Agric. 2022, 193, 106694. [Google Scholar] [CrossRef]
- Fan, S.; Liang, X.; Huang, W.; Zhang, V.J.; Pang, Q.; He, X.; Li, L.; Zhang, C. Real-time defects detection for apple sorting using NIR cameras with pruning-based YOLOV4 network. Comput. Electron. Agric. 2022, 193, 106715. [Google Scholar] [CrossRef]
- Qi, C.; Gao, J.; Pearson, S.; Harman, H.; Chen, K.; Shu, L. Tea chrysanthemum detection under unstructured environments using the TC-YOLO model. Expert Syst. Appl. 2022, 193, 116473. [Google Scholar] [CrossRef]
- Ge, H.; Dai, Y.; Zhu, Z.; Liu, R. A Deep Learning Model Applied to Optical Image Target Detection and Recognition for the Identification of Underwater Biostructures. Machines 2022, 10, 809. [Google Scholar] [CrossRef]
- Cai, K.W.; Miao, X.Y.; Wang, W.; Pang, H.S.; Liu, Y.; Song, J.Y. A modified YOLOv3 model for fish detection based on MobileNetv1 as backbone. Aquac. Eng. 2020, 91, 102117. [Google Scholar] [CrossRef]
- Li, J.; Zhu, K.; Wang, F.; Jiang, F. Deep neural network-based real time fish detection method in the scene of marine fishing supervision. J. Intell. Fuzzy Syst. 2021, 41, 4527–4532. [Google Scholar] [CrossRef]
- Hu, X.; Liu, Y.; Zhao, Z.; Liu, J.; Yang, X.; Sun, C.; Chen, S.; Li, B.; Zhou, C. Real-time detection of uneaten feed pellets in underwater images for aquaculture using an improved YOLO-V4 network. Comput. Electron. Agric. 2021, 185, 106135. [Google Scholar] [CrossRef]
- Jalal, A.; Salman, A.; Mian, A.; Shortis, M.; Shafait, F. Fish detection and species classification in underwater environments using deep learning with temporal information. Ecol. Inform. 2020, 57, 101088. [Google Scholar] [CrossRef]
- Wen, C.; Wen, J.; Li, J.; Luo, Y.; Chen, M.; Xiao, Z.; Xu, Q.; Liang, X.; An, H. Lightweight silkworm recognition based on Multi-scale feature fusion. Comput. Electron. Agric. 2022, 200, 107234. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, Q.; Meng, H.; Gao, D. An underwater target recognition method based on improved YOLOv4 in complex marine environment. Syst. Sci. Control. Eng. 2022, 10, 590–602. [Google Scholar] [CrossRef]
- Prasetyo, E.; Suciati, N.; Fatichah, C. Yolov4-tiny with wing convolution layer for detecting fish body part. Comput. Electron. Agric. 2022, 198, 107023. [Google Scholar] [CrossRef]
- Abinaya, N.S.; Susan, D.; Sidharthan, R.K. Deep learning-based segmental analysis of fish for biomass estimation in an occulted environment. Comput. Electron. Agric. 2022, 197, 106985. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, S.; Zhao, S.; Wang, Q.; Li, D.; Zhao, R. Real-time detection and tracking of fish abnormal behavior based on improved YOLOV5 and SiamRPN plus. Comput. Electron. Agric. 2022, 192, 106512. [Google Scholar] [CrossRef]
- He, K.; Sun, J.; Tang, X. Single Image Haze Removal Using Dark Channel Prior. IEEE Trans. Pattern Anal. Mach. Intell. 2011, 33, 2341–2353. [Google Scholar] [CrossRef]
- Wang, C.Y.; Liao, H.Y.M.; Wu, Y.H.; Chen, P.Y.; Hsieh, J.W.; Yeh, I.H.; IEEE Comp, S.O.C. CSPNet: A New Backbone that can Enhance Learning Capability of CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 14–19 June 2020; pp. 1571–1580. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the 15th European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Li, D.; Su, H.; Jiang, K.; Liu, D.; Duan, X. Fish Face Identification Based on Rotated Object Detection: Dataset and Exploration. Fishes 2022, 7, 219. [Google Scholar] [CrossRef]
- Deng, Y.; Tan, H.; Tong, M.; Zhou, D.; Li, Y.; Zhu, M. An Automatic Recognition Method for Fish Species and Length Using an Underwater Stereo Vision System. Fishes 2022, 7, 326. [Google Scholar] [CrossRef]




| Collection Name | Red Tilapia | Micropterus Salmoides |
|---|---|---|
| Eye infections | 785 | 0 |
| Surface decay | 0 | 2080 |
| Died | 213 | 0 |
| Normal | 2002 | 920 |
| Summation | 3000 | 3000 |
| Configuration | Parameter |
|---|---|
| CPU | Intel Core i7-9700 K |
| GPU | Nvidia GeForce RTX 3080 Ti × 2 |
| Operating system | Windows 10 |
| Accelerated environment | CUDA10.2 CUDNN7.6.5 |
| Development environment | Visual Studio 2020 |
| Library | Opencv3.4.0 |
| Method | Batch Size | Learning Rate | Epoch | Momentum | Weight Decay |
|---|---|---|---|---|---|
| SSD | 16 | 0.001 | 475 | 0.9 | 0.0005 |
| Faster-RCNN | 16 | 0.001 | 475 | 0.9 | 0.0005 |
| YOLOv3 | 16 | 0.001 | 475 | 0.9 | 0.0005 |
| YOLOv4 | 16 | 0.001 | 475 | 0.9 | 0.0005 |
| YOLOV5 | 16 | 0.01 | 475 | 0.9 | 0.0005 |
| DFYOLO | 16 | 0.01 | 475 | 0.9 | 0.0005 |
| Method | Occupy Memory/MB | FPS | Recall/% | Precision/% | mAP50/% | mAP50:95/% |
|---|---|---|---|---|---|---|
| SSD | 63.2 | 69.73 | 73.01 | 76.81 | 68.24 | 42.03 |
| Faster-RCNN | 77.8 | 57.97 | 83.28 | 86.18 | 79.87 | 51.14 |
| YOLOv3 | 62.1 | 69.44 | 86.12 | 82.38 | 90.99 | 41.10 |
| YOLOv4 | 226 | 78.24 | 93.84 | 93.58 | 93.39 | 59.53 |
| YOLOV5 | 14.5 | 96.43 | 93.76 | 94.36 | 94.52 | 78.53 |
| DFYOLO | 13.6 | 93.21 | 99.31 | 99.75 | 99.38 | 88.09 |
| Baseline Network | Bilateral Filtering | Haze Removal | Image Transformation | Conv KG | CBAM | Recall/% | Precision/% | mAP50/% | mAP50:95/% |
|---|---|---|---|---|---|---|---|---|---|
| YOLOV5 | 93.76 | 94.36 | 94.52 | 78.53 | |||||
| YOLOV5 | √ | 95.81 | 94.40 | 94.58 | 79.28 | ||||
| YOLOV5 | √ | √ | 95.85 | 94.86 | 96.02 | 82.12 | |||
| YOLOV5 | √ | √ | √ | 95.75 | 93.42 | 95.41 | 82.07 | ||
| YOLOV5 | √ | √ | √ | 97.21 | 98.24 | 98.35 | 87.98 | ||
| YOLOV5 | √ | √ | √ | √ | 97.18 | 98.85 | 98.16 | 86.06 | |
| YOLOV5 | √ | √ | √ | √ | √ | 99.31 | 99.75 | 99.38 | 88.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, H.; Zhang, G.; Yang, X.; Wen, L.; Zhao, W. Diseased Fish Detection in the Underwater Environment Using an Improved YOLOV5 Network for Intensive Aquaculture. Fishes 2023, 8, 169. https://doi.org/10.3390/fishes8030169
Wang Z, Liu H, Zhang G, Yang X, Wen L, Zhao W. Diseased Fish Detection in the Underwater Environment Using an Improved YOLOV5 Network for Intensive Aquaculture. Fishes. 2023; 8(3):169. https://doi.org/10.3390/fishes8030169
Chicago/Turabian StyleWang, Zhen, Haolu Liu, Guangyue Zhang, Xiao Yang, Lingmei Wen, and Wei Zhao. 2023. "Diseased Fish Detection in the Underwater Environment Using an Improved YOLOV5 Network for Intensive Aquaculture" Fishes 8, no. 3: 169. https://doi.org/10.3390/fishes8030169
APA StyleWang, Z., Liu, H., Zhang, G., Yang, X., Wen, L., & Zhao, W. (2023). Diseased Fish Detection in the Underwater Environment Using an Improved YOLOV5 Network for Intensive Aquaculture. Fishes, 8(3), 169. https://doi.org/10.3390/fishes8030169






