Abstract
The complete mitochondrial genome of Chirolophis wui (Wang and Wang, 1935) was sequenced using the Illumina platform. The genome sequence is 16,522 bp in length with 54% A+T content and contains 13 protein coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and 1 control region (D-loop). The H-strand contains 28 genes (12 PCGs, 14 tRNAs, and 2 rRNAs), whereas the L-strand accommodates 9 genes (ND6 and 8 tRNAs). The nucleotide composition of the mitochondrial genome of C. wui is AT-biased, accounting for 54.0%, with an AT skew value of −0.0556 and a GC skew value of −0.2043. The majority of PCGs utilized the start codon, ATG, while only one gene, COI, utilized the alternative start codon, GTG. Of the 13 PCGs, 6 genes used the termination codon (TAA or TGA), whereas 7 genes used the incomplete termination codon (T or TA). Among the 22 tRNA genes, the tRNA-Leu and tRNA-Ser were found in duplicates. A phylogenetic tree was constructed using 10 complete mitochondrial genome sequences and indicated that C. wui has a very close relationship with C. japonicus and other species in the family Stichaeidae, with a high supporting bootstrap value. This study can provide valuable information for future evolutionary studies on C. wui and Stichaeidae.
Keywords:
Chirolophis wui; Stichaeidae; mitogenome; next-generation sequencing; phylogenetic analysis Key Contribution:
In this study; the whole mitogenome sequence of C. wui was completed. In addition; the phylogenetic relationship within the Stichaeidae family was investigated.
1. Introduction
The mitochondrion is a type of organelle that is capable of directly converting organic materials into energy, which is then used to sustain many biological processes that occur within a cell [1,2,3]. Mitochondria have semiconservatively self-replicating, closed circular, double-stranded mitochondrial DNA [4,5]. The mitochondrial genomes of vertebrates are small, at around 14–26 kb on average, and the order of encoded genes is highly conserved [6,7,8]. The mitochondrial genome of vertebrates typically consists of 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 2 noncoding regions: the control regions and the origin of L-strand replication [9]. The complete mitochondrial genomes have been shown to be useful genetic markers in detecting and differentiating distinct or similar, and even concealed, species within closely related taxa [10,11].
The sequencing of fish mitogenomes has been made possible by recent advancements in molecular biology analytical techniques, resulting in a clear comprehension of the structure of fish mitogenomes (which are 16–18 kb in size) [1,12]. Fish mitogenomes also have highly conserved protein-coding genes, transfer RNAs, ribosomal RNAs, and noncoding regions, although the gene spacing and length vary between species [9,13]. The mitogenome has been a prominent molecular guide in the study of the phylogeny and evolution of fish since the determination of the mtDNA sequences of several fish species [1,14,15].
The family Stichaeidae comprises six different subfamilies: Azygopterinae, Chirolophinae, Lumpeninae, Opisthocentrinae, Stichaeinae, and Xiphisterinae. The family Stichaeidae inhabits the North Pacific, North Atlantic, and Arctic oceans, with the majority of species inhabiting the North Pacific. They are coastal fishes found in the intertidal zone and shallow bays beneath rocks and in algae. They can be found on the outer continental shelf at depths of more than 250 m [16,17,18]. The subfamily Chirolophinae contains four genera: Bryozoichthys, Chirolophis, Gymnoclinus, and Sodatovia [16,19]. The body is moderately elongate and relatively robust. The head is uncovered, and the body is coated with tiny scales. The head, anterior part of the body, and first few dorsal fins have spines with dermal appendages. The anal fin has one weakly developed spine. The pectoral fins are large, and the pelvic fins have one spine and 2–4 soft rays [16,19]. There are eight species in the genus Chirolophis, including C. ascanii, C. decorates, C. japonicus, C. nuagtor, C. saitone, C. snyderi, C. tarsodes, and C. wui (http://www.fishbase.se/search.php, accessed on 10 January 2022) [20]. Chirolophis wui (Wang and Wang, 1935) has only been found in the Republic of Korea, China, and Japan [21,22]. However, molecular studies are limited in comparison to morphological and environmental investigations.
To date, the mitochondrial genome of C. wui has not been studied. As a result, the complete mitochondrial genome of C. wui was sequenced in this study, and their phylogenetic relationship with other Stichaeidae species was investigated. This study can help with future evolutionary studies on C. wui and Stichaeidae.
2. Materials and Methods
2.1. Sample and DNA Extraction
An individual sample of C. wui was captured from the coast of Taean in the Republic of Korea (36°34′26.27″ N; 126°1′86.3822″ E) (Figure 1), and deposited at the Marine Fish Resources Bank of Korea (MFRBK) in Pukyong National University (PKNU), Busan, Republic of Korea (Dr. Jin-Koo Kim, taengko@pknu.ac.kr) under the voucher number PKU-21087.
Figure 1.
Map showing the sampling location (Red spot). Map was downloaded from d-map (https://d-maps.com/carte.php?num_car=6021&lang=en, accessed on 10 January 2022).
Using the DNeasy Blood and Tissue Kit (Qiagen, Germany), total genomic DNA was isolated from the muscle tissue according to the manufacturer’s recommendations. The quality and purity of gDNA were evaluated using a NanoDrop 3300 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The gDNA was stored at −20 ℃ for further analysis.
2.2. Next-Generation Sequencing and Mitogenome Assembly
The TrueSeq Nano DNA Kit was used to create the DNA library, which was then sequenced on the Illumina platform using 150 bp paired-end reads (Illumina, HiSeq 2500, San Diego, CA, USA) at Macrogen (Daejeon, Republic of Korea). For the library, a total of 67,988,441 clean reads of each direction were produced. Using Trimmomatic [23], adapter sequences and low-quality reads were deleted to reduce analytical bias. The overall quality of the produced sequencing reads was verified with FastQC v0.11.5 (Babraham Institute, Bioinformatics) [24]. Mitogenome assembly was accomplished de novo using various κ-mers and the SPAdes v3.13.0 tool [25]. The filtered Illumina reads and reconstructed mitogenome were submitted to BioProject, the Sequence Read Archive (SRA), and GenBank under the corresponding accession numbers PRJNA855310, SRR19965989, and OP388414, respectively.
2.3. Mitogenome Annotation, Sequence Analysis, and Phylogenetic Analysis
The mitogenome of C. wui was analyzed for annotation using the MITOS server (http://mitos.bioinf.uni-leipzig.de/index.py, accessed on 10 January 2022) [26]. The PCGs were validated using the ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 10 January 2022) following translation into the predicted amino acids according to the vertebrate mitochondrial genetic code. Based on the mitochondrial code of vertebrates, the codon usage in PCGs was predicted via the Codon Usage web server [27]. The Tandem Repeat Finder v4.09 web server was used to look into the number of repetitions in the region [28]. MEGA-X v10.2.4 was used to determine the nucleotide composition of the D-loop, rRNAs, tRNAs, PCGs, and mitogenome of C. wui [29]. The following formulae were used to determine the asymmetry in the mitogenome base composition. In terms of the four nucleotides (A, T, G, and C), the skews were calculated as follows: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C) [30]. The MITOS server [26] and ARWEN server [31] were used to estimate the secondary structures of tRNA and rRNA genes.
To determine the phylogenetic position of C. wui within Stichaeidae, 9 mitogenomes of Stichaeidae species were downloaded from GenBank, and complete sequences of Cottus szanaga species were used as an outgroup (Table 1). The complete mitochondrial genomes of these species were aligned using ClustalW [32], maximum-likelihood (Tamaru-Nei model) [33] analysis was conducted using MEGA XI v11.0.8 [34], and the tree topology was evaluated with 1000 bootstrap replicates.

Table 1.
The list of mitogenomes used in this study.
3. Results and Discussion
3.1. Mitochondrial Genome Assembly, Annotation, and Sequence Analysis
A total of 135.9 million reads (Table S1) were produced with the next-generation sequencing of C. wui, and after eliminating the adapter sequences and low-quality reads, the remaining 113.1 million reads (Table S2) were appropriate for the genomic assembly methods.
The size of the complete mitochondrial genome of C. wui was 16,522 bp, and the data were deposited in NCBI GenBank (OP388414) (Figure 2). The mitogenome of C. wui is longer than those of C. japonicus (16,521 bp) and C. ascanii (16,520 bp), as well as Opisthocentrus zonope (16,518 bp), O. ocellatus (16,517 bp), and O. tenuis (16,515 bp) of the Opisthocentrinae subfamily. However, it is shorter than the Stichaeinae subfamily members Stichaeus fuscus (16,529 bp), S. grigorjewi (16,532 bp), and S. nozawae (16,530 bp).

Figure 2.
Sample image and the mitochondrial genome map of Chirolophis wui. (a) Specimen reference image; (b) the mitochondrial genome of C. wui. Genes outside the circle are transcribed in a clockwise direction and those inside in a counterclockwise direction.
The mitogenome of C. wui consisted of 13 PCGs, 2 rRNA genes, 22 tRNA genes, and 1 control region (D-loop) (Figure 3, Table 2). Only ND6 and 8 tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) were encoded on the L-strand of the 37 mitochondrial genes; all of the other genes were encoded on the H-strand. As in the typical vertebrate mitochondrial genome, the 12S and 16S rRNA genes of C. wui are located between the tRNA-Phe and tRNA-Leu genes, with the tRNA-Val gene in between. Regarding the results of comparative mitochondrial genome analysis with C. japonicus and C. ascanii, they were of the same composition and order as C. wui.

Figure 3.
Graphical representation of nucleotide composition contents and AT and GC skew. They should be listed as: (a) nucleotide composition AT% and GC%; (b) AT and GC skew.

Table 2.
Gene annotations of the complete mitochondrial genome of C. wui.
The mitogenome of C. wui is 54.0% AT-biased, with 25.5% A, 28.5% T, 18.3% G, and 27.7% C, which is also similar to those of Stichaeidae species (C. japonicus, 54.1%; C. ascanii, 53.8%; O. zonope, 55.4%; S. fuscus, 53.8%). As measured by the GC skew value of −0.2043, the nucleotide composition is heavily biased toward C and somewhat biased toward A and T (with the AT skew value of −0.0556) (Table 2 and Table 3).

Table 3.
General metrics of nucleotide composition of C. wui.
Six gene junctions have overlaps of a combined 25 bp: tRNA-Ile-tRNA-Met (overlap = 1 bp), tRNA-Met-ND2 (overlap = 1 bp), ATPase 8-ATPase 6 (overlap = 10 bp), ND4L-ND4 (overlap = 8 bp), ND5-ND6 (overlap = 4 bp), and tRNA-Thr-tRNA-Pro (overlap = 1). Furthermore, in a total of 10 gene junctions, short intergenic gaps between 1 and 38 bp in length were detected (Table 2).
3.2. Protein Coding Genes
A total of 13 PCGs were annotated in the mitogenome of C. wui (Figure 3, Table 4). Of 13 PCGs, 12 genes start with the conventional initiation codons ATG (ND1, ND2, COII, ATPase 8, ATPase 6, COIII, ND3, ND4L, ND4, ND5, ND6, and CytB). As previously reported [35], COI possessed an alternate putative start codon (GTG). Six PCGs had a complete and typical stop codon at the end. Five genes (COI, ATPase 8, ND4L, ND5, and ND6) ended with TAA, and one gene ended with TAG (ND1). Seven PCGs ended with an incomplete stop codon. Four genes (COII, ND3, ND4, and CytB) ended with T, and three genes (ND2, ATPase 6, and COIII) ended with TA. It is commonly known that the mitochondrial genome contains incomplete stop codons [36,37]. Due to the inclusion of A throughout the RNA processing process, it is assumed that the incomplete termination codon will become the complete stop codon [38].

Table 4.
Codon usage analysis of PCGs in the mitochondrial genome of C. wui.
PCGs range in size from 168 bp for ATPase 8 to 1839 bp for the longest PCG, ND5. As a short gene that is under little selective pressure and shows a great deal of variability at the amino acid and nucleotide levels, ATPase 8 is notoriously difficult to discover [39].
CTT (Leu, N = 227 times used, 5.97%), ATT (Ile, N = 169 times used, 4.45%), GCC (Ala, N = 145 times used, 3.82%), TTT (Phe, N = 139 times used, 3.66%), and CTC (Ala, N = 131 times used, 3.45%) were the most commonly utilized codons in the PCGs of C. wui mitochondrial genome. Codons such as CGG (Arg, N = 11 times used, 0.29% of the total) and TGT (Cys, N = 8 times used, 0.21% of the total) are used less frequently but nonetheless occur. In addition, serine-encoding codons such as AGG and AGA have never been implemented (Table 4).
3.3. Ribosomal RNA, Transfer RNA Genes, and Control Region
In C. wui, the length of the ribosomal RNA genes was 2641 bp (15.98% of the total mitogenome). The sizes of the 12S rRNA and 16s rRNA genes were 948 bp and 1693 bp, respectively. The overall base composition of the 12S rRNA was T = 21.7%, C = 25.5%, A = 30.1%, G = 2.7%, and AT = 51.8%. The 12S rRNA gene showed a weakly positive AT skew (0.1622) and GC skew (−0.0581) compared with the AT skew (0.1554) and GC skew (−0.0419) of the 16s RNA gene.
The mitochondrial genome of C. wui was analyzed using the MITOS website, and 22 tRNA-encoding genes were found (Figure 4). The length of 22 tRNAs ranged from 66 bp (tRNA-Cys) to 74 bp (tRNA-Leu-1 and tRNA-Lys). The tRNAs of leucine (Leu) and serine (Ser) existed in two copies with different anticodons. The existence of these tRNAs in the mitochondrial genome as numerous copies has been proven in various vertebrates [11,35,40,41]. The sequences of all tRNA genes of C. wui were folded into a canonical cloverleaf secondary structure consisting of an amino acid accepter (AA) arm, a dihydrouridine (DHU) arm, an anticodon (AC) arm, a TΨC arm, and variable (V) arms [10,11,42]. The tRNA-Ser-1 (AGT) and tRNA-Cys (GCA) had an incorrect DHU arm, which have been reported in several fish [11,43]. All tRNAs of C. wui had AT skew values of 0.1073, and GC skew values of −0.0746.

Figure 4.
Inferred secondary structures of 23 tRNAs from C. wui.
Located between the tRNA-Pro and tRNA-Phe, the C. wui CR (D-loop) was 851 bp in length (Table 2). The D-loop had a total of T = 28.6%, C = 21.7%, A = 30.6%, and G = 19.2% by base composition. In addition, the AT skew value was 0.0338, whereas the GC skew value was −0.0611 (Table 3, Figure 3). As a result of frequent insertions/deletions and substitutions of nucleotides, it is known that this region exhibits substantial length variation [44]. The tandem repeat sequences were not detected in the D-loop region in C. wui using the Tandem Repeats Finder web server [28]. Although the exact purpose of the control region (D-loop) is unknown, it is anticipated that it will be crucial to the replication and transcription of the genome [11].
3.4. Phylogenetic Analysis
The phylogenetic tree was constructed using the mitogenome sequences from ten species, including C. wui and eight additional Stichaeidae species, and one Cottage species, Cottus szanaga (KX762050) [45], as an outgroup (Figure 5). Table 1 lists the mitogenomes that were analyzed in this study and their corresponding accession numbers.
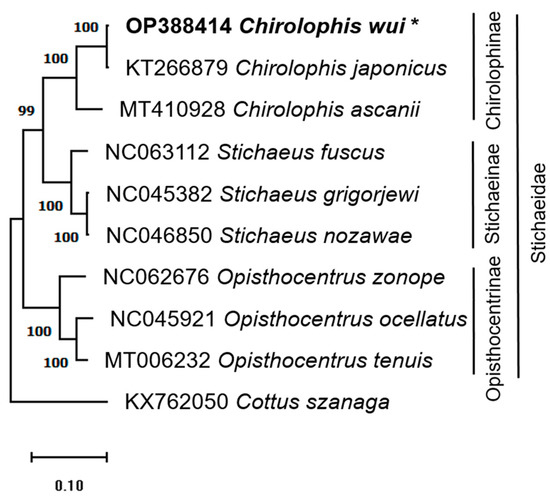
Figure 5.
The phylogenetic tree was obtained using the complete mitochondrial genome sequences from ten species (nine from the Stichaeidae family and Cottus szanaga from the Cottidae family as an outgroup) and 1000 bootstrap repetitions using the maximum-likelihood approach. The numbers on the branches represent the bootstrap values, and the star next to Chirolophis wui denotes the species used in this research.
The taxa of the nine Stichaeidae species were well clustered, and C. szanaga was distinct from the Stichaeidae. Within Stichaeidae, the phylogeny was shown by the Chirolophinae, Stichaeinae, and Opisthocentrinae. Within each of the three subfamilies, there was only one genus (Chirolophis, Stichaeus, and Opisthocentrus). The phylogenetic analysis showed that C. wui is most closely related to C. japonicus. C. wui and C. japonicus are sisters of C. ascanii.
The results presented in this study would play an important role in the investigation of the phylogenetic relationships and taxonomy of the family Stichaeidae.
4. Conclusions
In conclusion, the current study presented the first complement mitogenome assembly and annotation of C. wui. We described the characterization of the mitochondrial genome of C. wui using various genetic and phylogenetic research approaches. These results can help to advance understanding and collect fundamental genetic data for the Stichaeidae family.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/fishes8030165/s1, Table S1: Summary of Chirolophis wui mitogenome data produced/ stats during de novo assembly analysis in Illumina platform using SPAdes 3.13.0 assembly method, Table S2: Chirolophis wui mitogenome overall self-mapping stats.
Author Contributions
Y.-S.L., M.P.P., K.R.M. and J.-O.K. performed the experiments, analyzed the data, were involved in certain tools for analysis and drafting of the paper, and approved the final draft. Y.B.S., Y.-J.L., K.R.M. and J.-K.K. were involved in certain tools for analysis, organizing the results, and preparing figures. G.-D.K. was involved in the conception and design of the work, funding acquisition, revising it critically for intellectual content, and the final approval of the version to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This research was part of the project titled ‘Development of Discrimination Method and On-Site Kit for Geographical Origin of Fishery Product (Project No. 20200425)’, funded by the Ministry of Ocean and Fisheries, Republic of Korea.
Institutional Review Board Statement
The sample used for this study was the dead body of a fish and, as per the animal experimental ethics in the Republic of Korea (Standard operating guideline; IACUC—Institutional Animal Care and Use Committee, Book no. 11-1543061-000457-01, effective from December 2020), we did not require approval from the Ethics Committee.
Informed Consent Statement
Not applicable.
Data Availability Statement
The mitogenome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under accession number OP388414. The associated BioProject, SRA, and the complement mitogenome numbers are PRJNA855310, SRR19965989, and OP388414, respectively.
Acknowledgments
This research was conducted using a fish specimen provided by the Marine Fish Resources Bank of Korea (MFRBK) and Pukyong National University (PKNU).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sun, C.-H.; Liu, H.-Y.; Xu, N.; Zhang, X.-L.; Zhang, Q.; Han, B.-P. Mitochondrial genome structures and phylogenetic analyses of two tropical characidae fishes. Front. Genet. 2021, 12, 627402. [Google Scholar] [CrossRef]
- Parhi, J.; Tripathy, P.S.; Priyadarshi, H.; Mandal, S.C.; Pandey, P.K. Diagnosis of mitogenome for robust phylogeny: A case of Cypriniformes fish group. Gene 2019, 713, 143967. [Google Scholar] [CrossRef]
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, C.A.; Saunders, N.C. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Ann. Rev. Ecol. System. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Prosdocimi, F.; de Carvalho, D.C.; de Almeida, R.N.; Beheregaray, L.B. The complete mitochondrial genome of two recently derived species of the fish genus Nannoperca (Perciformes, Percichthyidae). Mol. Biol. Rep. 2012, 39, 2767–2772. [Google Scholar] [CrossRef]
- Castro Paz, F.P.; Batista, J.d.S.; Porto, J.I.R. DNA barcodes of rosy tetras and allied species (Characiformes: Characidae: Hyphessobrycon) from the Brazilian Amazon basin. PLoS ONE 2014, 9, e98603. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Tzeng, C.-S.; Hui, C.-F.; Shen, S.-C.; Huang, P. The complete nucleotide sequence of the Crossostoma lacustre mitochondrial genome: Conservation and variations among vertebrates. Nucleic Acids Res. 1992, 20, 4853–4858. [Google Scholar] [CrossRef]
- Billington, N.; Hebert, P.D. Mitochondrial DNA diversity in fishes and its implications for introductions. Can. J. Fish. Aquat. Sci. 1991, 48, 80–94. [Google Scholar] [CrossRef]
- Alam, M.T.; Petit, R.A., III; Read, T.D.; Dove, A.D. The complete mitochondrial genome sequence of the world’s largest fish, the whale shark (Rhincodon typus), and its comparison with those of related shark species. Gene 2014, 539, 44–49. [Google Scholar] [CrossRef]
- Deng, Y.-P.; Li, R.; Wang, H.-M.; Liu, G.-H.; Tu, Y. Complete Mitochondrial Genome of Contracaecum Sp. (Nematoda: Ascarididae) from Night Herons in China. J. Nematol. 2022, 54, 20220048. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.I.; Baek, J.Y.; Kim, M.J.; Jeong, H.C.; Kim, K.-G.; Bae, C.H.; Han, Y.S.; Jin, B.R.; Kim, I. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol. Cells 2009, 28, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. Origin and evolution of mitochondrial DNA. Annu. Rev. Cell Biol. 1989, 5, 25–50. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Z.; Gao, J.; Zhao, Y.; Sun, P.; Lu, K. The complete mitochondrial genome of Indonesian tiger fish Datnioides microlepis (Bleeker 1854). Mitochondrial DNA B Resour. 2016, 1, 328–329. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Liu, S.; Luo, H.; Lin, Q. Population genetic structure and phylogenetic analysis of gray’s pipefish, Halicampus grayi in the South China Sea. Genes Genom. 2020, 42, 155–164. [Google Scholar] [CrossRef]
- Mecklenburg, C. Family Stichaeidae Gill 1864-pricklebacks. California Academy of Sciences. Annot. Check List. Fishes 2004, 35, 1–36. [Google Scholar]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Gill, T. Note on the family of stichaeoids. Proc. Acad. Nat. Sci. Phila. 1864, 16, 208–211. [Google Scholar]
- Jordan, D.S.; Evermann, B.W. The Fishes of North and Middle America: A Descriptive Catalogue of the Species of Fish-Like Vertebrates Found in the Waters of North America, North of the Isthmus of Panama; US Government Printing Office: Washington, DC, USA, 1896.
- Liu, L.; Liu, Q.; Gao, T. Genome-wide survey reveals the phylogenomic relationships of Chirolophis japonicus Herzenstein, 1890 (Stichaeidae, Perciformes). ZooKeys 2022, 1129, 52–72. [Google Scholar] [CrossRef]
- Nakabo, T. Fishes of Japan with Pictorial Keys to the Species, English Edition; Tokai University Press: Tokyo, Japan, 2002. [Google Scholar]
- Kim, I.-S.; Choi, S.-H. Fishes of the Southwestern Coast of Korea. Anim. Syst. Evol. Divers. 1998, 14, 135–157. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef]
- Laslett, D.; Canbäck, B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008, 24, 172–175. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Molecular Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, M.; Wang, S.; Gu, Z.; Wang, A.; Liu, C.; Yang, Y.; Liu, S. The Complete Mitochondrial Genome of Hyotissa hyotis (Bivalvia: Gryphaeidae) Reveals a Unique Gene Order within Ostreoidea. Fishes 2022, 7, 317. [Google Scholar] [CrossRef]
- Yu, H.; Li, Q. Complete mitochondrial DNA sequence of Crassostrea nippona: Comparative and phylogenomic studies on seven commercial Crassostrea species. Mol. Biol. Rep. 2012, 39, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xu, X.; Yu, Z.; Wei, Z.; Xia, J. Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol. Phylogenet. Evol. 2010, 57, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, X.; Li, L.; Xu, X.; Xia, J.; Yu, Z. New features of Asian Crassostrea oyster mitochondrial genomes: A novel alloacceptor tRNA gene recruitment and two novel ORFs. Gene 2012, 507, 112–118. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-H.; Sun, P.-Y.; Lao, Y.-L.; Wu, T.; Zhang, Y.-N.; Huang, Q.; Zhang, Q. Mitogenome of a monotypic genus, Oliotius Kottelat, 2013 (Cypriniformes: Cyprinidae): Genomic characterization and phylogenetic position. Gene 2023, 851, 147035. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wilson, B.; Kumar, P.; Dutta, A. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu. Rev. Genet. 2020, 54, 47–69. [Google Scholar] [CrossRef]
- Kundu, S.; Binarao, J.D.; De Alwis, P.S.; Kim, A.R.; Lee, S.-R.; Andriyono, S.; Gietbong, F.Z.; Kim, H.-W. First Mitogenome of Endangered Enteromius thysi (Actinopterygii: Cypriniformes: Cyprinidae) from Africa: Characterization and Phylogeny. Fishes 2022, 8, 25. [Google Scholar] [CrossRef]
- Zhang, D.-X.; Hewitt, G.M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Balakirev, E.S.; Saveliev, P.A.; Ayala, F.J. Complete mitochondrial genomes of the Cherskii’s sculpin Cottus czerskii and Siberian taimen Hucho taimen reveal GenBank entry errors: Incorrect species identification and recombinant mitochondrial genome. Evol. Bioinform. 2017, 13, 1176934317726783. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).