Pitx1 Enhancer Variants in Spined and Spine-Reduced Subarctic European Sticklebacks
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Morphology and Computation of Pelvic Scores
2.3. DNA Sequencing and Fragment Analysis
2.3.1. PelA Sequence Analyses
2.3.2. PelA Fragment Analyses
2.3.3. PelB Sequence Analyses
2.3.4. Identification of Single Nucleotide Polymorphisms (SNPs)
3. Results
3.1. Morphology, Pelvic Scores, and Ca2+ Concentration
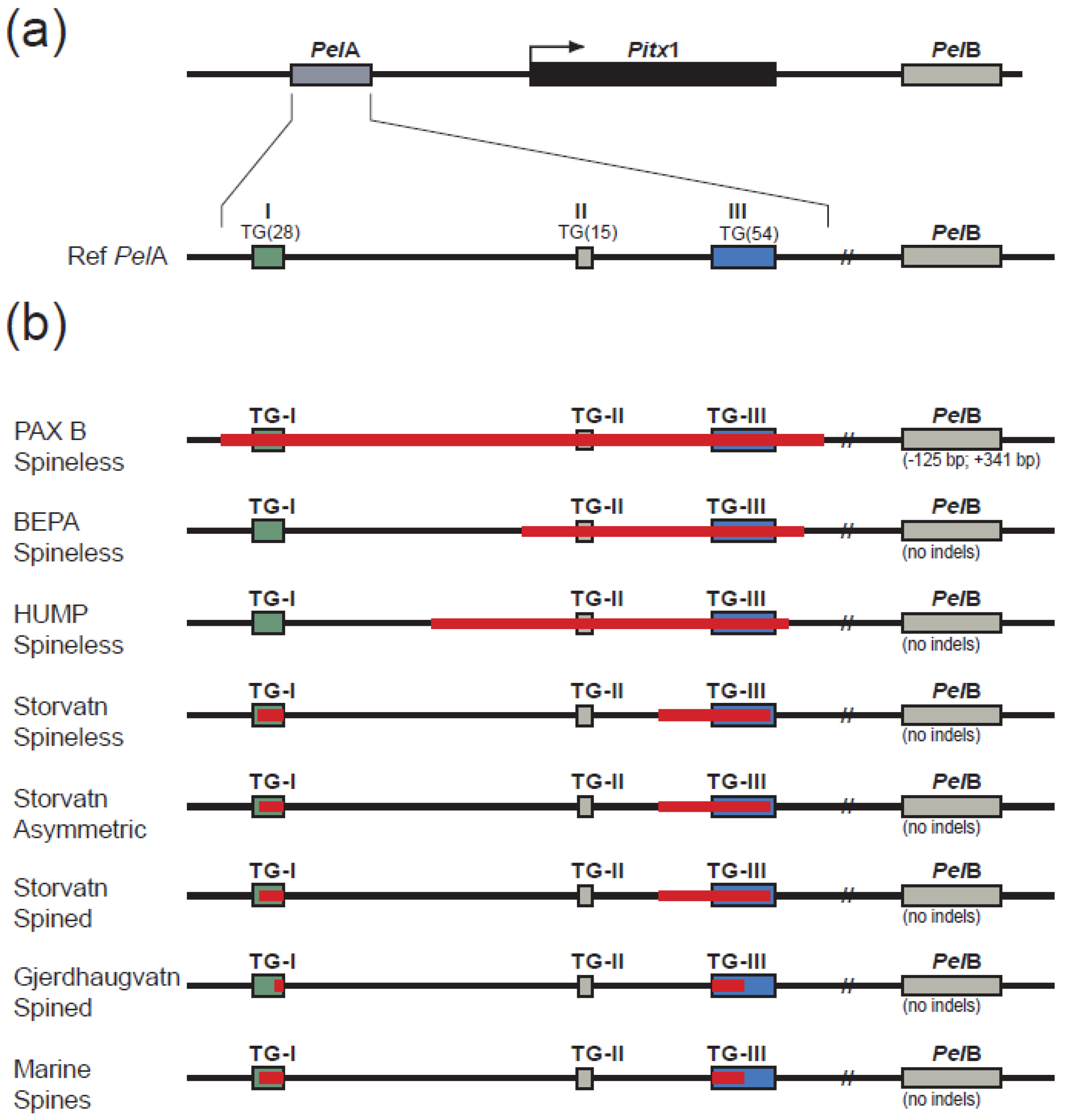
3.2. Allelic Variation of PelA
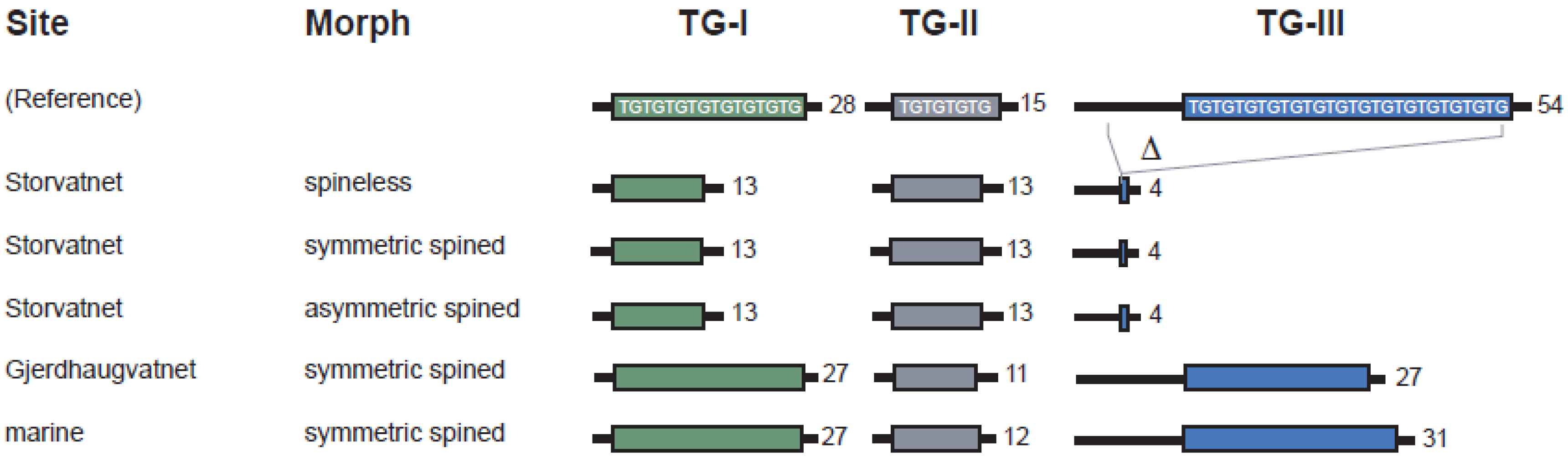
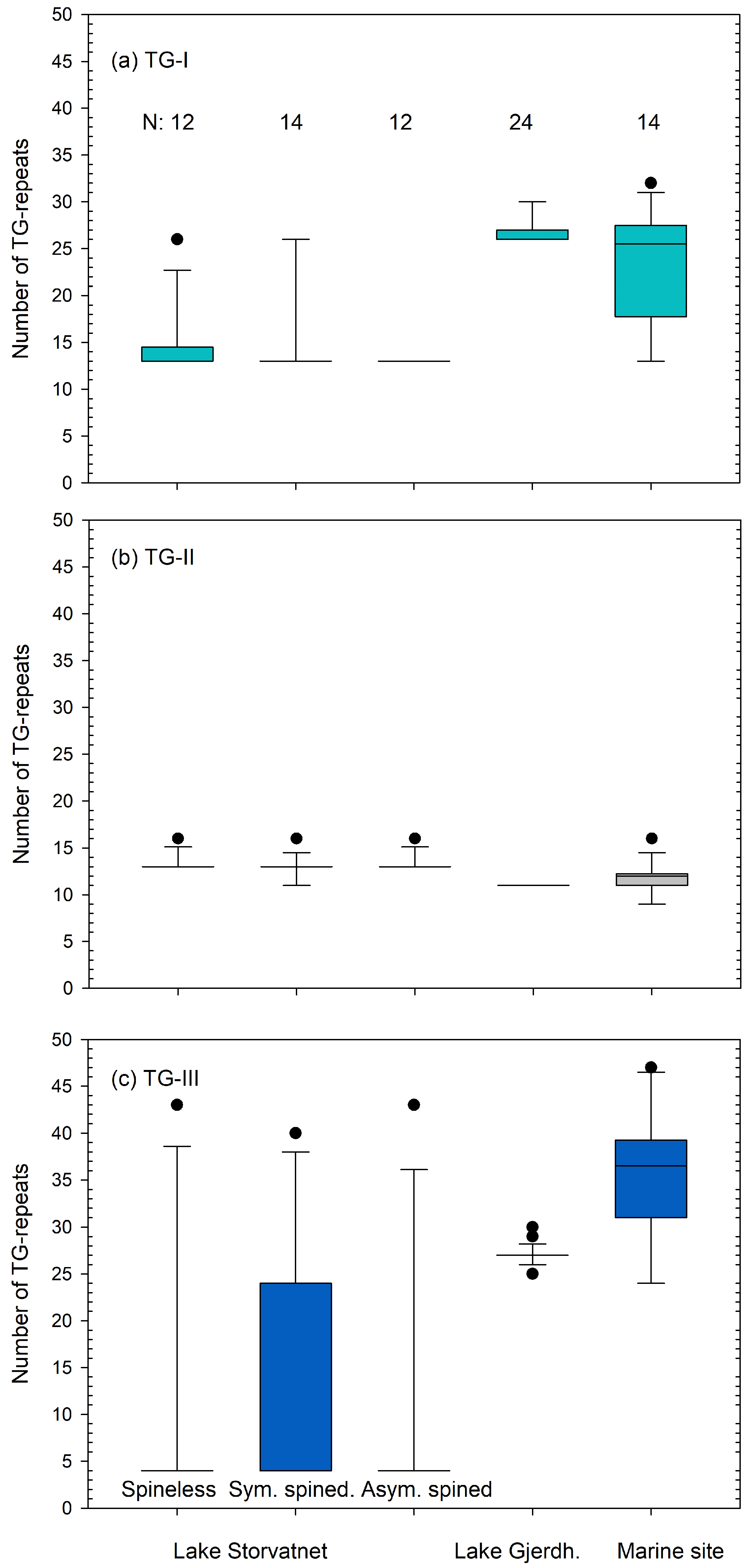
3.2.1. TG-Repeat Array I
3.2.2. TG-Repeat Array II
3.2.3. TG-Repeat Array III
3.2.4. Comparing Haplotypes of Spineless and Spined Sticklebacks from Lake Storvatnet
3.3. Allelic Variation of PelB
3.4. Sequence Alignments of PelA and PelB
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Futuyma, D.J. Evolutionary Biology, 2nd ed.; Sinauer: Sunderland, MA, USA, 1986. [Google Scholar]
- Schluter, D.; Conte, G.L. Genetics and ecological speciation. Proc. Natl. Acad. Sci. USA 2009, 106, 9955–9962. [Google Scholar] [CrossRef]
- Barrett, D.H.; Schluter, D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008, 23, 38–44. [Google Scholar] [CrossRef]
- Innan, H.; Kim, Y. Pattern of polymorphism after strong selection in a domestication event. Proc. Natl. Acad. Sci. USA 2004, 101, 10667–10672. [Google Scholar] [CrossRef]
- Schluter, D.; Clifford, E.A.; Nemethy, M.; McKinnon, J.S. Parallel evolution and inheritance of quantitative traits. Am. Nat. 2004, 163, 809–822. [Google Scholar] [CrossRef]
- Arendt, J.; Reznick, D. Convergence and parallelism reconsidered: What have we learned about genetics of adaptations? Trend Ecol. Evol. 2007, 23, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Little, C. The Terrestrial Invasion: An Ecophysiological Approach to the Origins of Land Animals; Cambridge University Press: Cambridge, UK, 1990. [Google Scholar]
- Lee, C.E.; Bell, M.A. Causes and consequences of recent freshwater invasions by saltwater animals. Trends Ecol. Evol 1999, 14, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Reimchen, T.E. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus: An adaptation to predators? Can. J. Zool. 1980, 58, 1232–1244. [Google Scholar] [CrossRef]
- Giles, N. The possible role of environmental calcum levels during the evolution of phenotypic diversity in Outer Hebridean populations of the three-spined stickleback, Gasterosteus aculeatus. J. Zool. 1983, 199, 535–544. [Google Scholar] [CrossRef]
- Baumgartner, J.V.; Bell, M.A. Lateral plate morph variation in California populations of the threespine stickleback, Gasterosteus aculeatus. Evolution 1984, 38, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.A.; Orti, G.; Walker, J.A.; Koenings, J.P. Evolution of pelvic reduction in threespine stickleback fish: A test of competing hypotheses. Evolution 1993, 47, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Barrett, R.D.H.; Paccard, A.; Healy, T.M.; Bergek, S.; Schulte, P.M.; Schluter, D.; Rogers, S.M. Rapid evolution of cold tolerance in stickleback. Proc. R. Soc. Lond. B Biol. Sci. 2011, 278, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, W.E.; Bell, M.A. Twenty years of body shape evolution in a threespine stickleback population adapting to a lake environment. Biol. J. Linn. Soc. Lond. 2012, 105, 817–831. [Google Scholar] [CrossRef]
- Shapiro, M.D.; Marks, M.E.; Peichel, C.L.; Blackman, B.K.; Nereng, K.S.; Jonsson, B.; Schluter, D.; Kingsley, D.M. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 2004, 428, 717–723. [Google Scholar] [CrossRef]
- Klepaker, T.O.; Østbye, K. Pelvic anti-predator armour reduction in Norwegian populations of the threespine stickleback: A rare phenomenon with adaptive implications? J. Zool. 2008, 26, 81–88. [Google Scholar] [CrossRef]
- Fang, B.; Kemppainen, P.; Momigliano, P.; Feng, X.; Merilä, J. On the causes of geographically heterogeneous parallel evolution in sticklebacks. Nat. Ecol. Evol. 2020, 4, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Bell, M.A.; Orti, G. Pelvic reduction in threespine stickleback from Cook Inlet Lakes: Geographical distribution and intrapopulation variation. Copeia 1994, 2, 314–325. [Google Scholar] [CrossRef]
- Wootton, R.J. The Biology of the Sticklebacks; Academic Press: London, UK, 1976. [Google Scholar]
- Klepaker, T. Morphological changes in a marine population of threespine stickleback, Gasterosteus aculeatus, recently isolated in fresh water. Can. J. Zool. 1993, 71, 1251–1258. [Google Scholar] [CrossRef]
- Roberts Kingman, G.A.; Vyas, D.N.; Jones, F.C.; Brady, S.D.; Chen, H.I.; Reid, K.; Milhaven, M.; Bertino, T.S.; Aguirre, W.E.; Heins, D.C.; et al. Predicting future from past: The genomic basis of recurrent and rapid stickleback evolution. Sci. Adv. 2021, 7, eabg5285. [Google Scholar] [CrossRef]
- Reimchen, T.E. Predators and morphological evolution in threespine stickleback. In The Evolutionary Biology of the Threespine Stickleback; Bell, M.A., Foster, S.A., Eds.; Oxford University Press: Oxford, UK, 1994; pp. 240–276. [Google Scholar]
- Hagen, D.W.; Gilbertson, L.G. Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America. Evolution 1972, 26, 32–51. [Google Scholar] [CrossRef]
- Moodie, G.E.E. Predation, natural selection and adaptation in an unusual threespine stickleback. Heredity 1972, 28, 115–167. [Google Scholar] [CrossRef]
- Bell, M.A.; Francis, G.W.; Havens, A.C. Pelvic reduction and its directional asymmetry in threespine sticklebacks from the Cook Inlet Region, Alaska. Copeia 1985, 2, 437–444. [Google Scholar] [CrossRef]
- Klepaker, T.; Østbye, K.; Bell, M.A. Regressive evolution of the pelvic complex in stickleback fishes: A study of convergent evolution. Evol. Ecol. Res. 2013, 15, 413–435. [Google Scholar]
- McPhail, J.D. Ecology and evolution of sympatric sticklebacks (Gasterosteus): Evidence for a species-pair in Paxton Lake, Texada Island, British Columbia. Can. J. Zool. 1992, 70, 361–369. [Google Scholar] [CrossRef]
- Chan, Y.F.; Marks, M.E.; Jones, F.C.; Villarreal, G., Jr.; Shapiro, M.D.; Brady, S.D.; Southwick, A.M.; Absher, D.M.; Grimwood, J.; Schmutz, J.; et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 2010, 327, 302–305. [Google Scholar] [CrossRef]
- Peichel, C.L.; Nereng, K.S.; Ohgi, K.A.; Cole, B.L.E.; Colosimo, P.F.; Buerkle, C.A.; Schluter, D.; Kingsley, D.M. The genetic architecture of divergence between threespine stickleback species. Nature 2001, 414, 901–905. [Google Scholar] [CrossRef]
- Reimchen, T.E.; Bergstrom, C.; Nosil, P. Natural selection and the adaptive radiation of Haida Gwaii stickleback. Evol. Ecol. Res. 2013, 15, 241–269. [Google Scholar]
- Coyle, S.M.; Huntingford, F.A.; Peichel, C.L. Parallel evolution of Pitx1 underlies pelvic reduction in Scottish threespine stickleback (Gasterosteus aculeatus). J. Hered. 2007, 98, 581–586. [Google Scholar] [CrossRef]
- Marchinko, K.B. Predation’s role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution 2009, 63, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.; Lucek, K.; Haesler, M.; Seehausen, O.; Sivasundar, A. Little evidence for a selective advantage of armour-reduced threespined stickleback individuals in an invertebrate predation experiment. Evol. Ecol. 2012, 26, 1293–1309. [Google Scholar] [CrossRef]
- Cresko, W.A.; Amores, A.; Wilson, C.; Murphy, J.; Currey, M.; Phillips, P.; Bell, M.A.; Kimmel, C.B.; Postlethwait, J.H. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA 2004, 101, 6050–6055. [Google Scholar] [CrossRef]
- Kingman, G.A.R.; Lee, D.; Jones, F.C.; Desmet, D.; Bell, M.A.; Kingsley, D.M. Longer or shorter spines: Reciprocal trait evolution in stickleback via triallelic regulatory changes in Stanniocalcin2a. Proc. Natl. Acad. Sci. USA 2021, 118, e2100694118. [Google Scholar] [CrossRef] [PubMed]
- Petit, F.; Sears, K.E.; Ahituv, N. Limb development: A paradigm of gene regulation. Nat. Rev. Genet. 2017, 18, 245–258. [Google Scholar] [CrossRef]
- Thompson, A.C.; Capellini, T.D.; Guenther, C.A.; Chan, Y.F.; Infante, C.R.; Menke, D.B.; Kingsley, D.M. A novel enhancer near the Pitx1 gene influences development and evolution of pelvic appendages in vertebrates. eLife 2018, 7, e38555. [Google Scholar] [CrossRef]
- Liu, R.; Liu, H.; Chen, X.; Kirby, M.; Brown, P.O.; Zhao, K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 2001, 106, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.T.; Wang, G.; Thompson, A.C.; Wucherpfennig, J.I.; Reimchen, T.E.; MacColl, A.D.C.; Schluter, D.; Bell, M.A.; Vasquez, K.M.; Kingsley, D.M. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 2019, 363, 81–84. [Google Scholar] [CrossRef]
- Kratochwil, C.F.; Meyer, A. Fragile DNA contributes to repeated evolution. Genome Biol. 2019, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Cole, N.J.; Tanaka, M.; Prescott, A.; Tickle, C. Expression of limb initiation genes and clues to the morphological diversification of threespine stickleback. Curr. Biol. 2003, 13, R951–R952. [Google Scholar] [CrossRef] [PubMed]
- Tickle, C.; Cole, N.J. Morphological diversity: Taking the spine out of three-spine stickleback. Curr. Biol. 2004, 14, R422–R424. [Google Scholar] [CrossRef]
- Klepaker, T.; Ostbye, K.; Bernatchez, L.; Vollestad, L.A. Spatio-temporal patterns in pelvic reduction in threespine stickleback (Gasterosteus aculeatus L.) in Lake Storvatnet. Evol. Ecol. Res. 2012, 14, 169–191. [Google Scholar]
- Bell, M.A.; Khalef, V.; Travis, M.P. Directional asymmetry of pelvic vestiges in threespine stickleback. J. Exp. Zool. B Mol. Dev. Evol. 2007, 308, 189–199. [Google Scholar] [CrossRef]
- Hindar, A.; Garmo, Ø.; Austnes, K.; Sample, J.E. Nasjonal innsjøundersøkelse 2019; Rapport L.Nr. 7530-2020; Norwegian Institute for Water Research: Oslo, Norway, 2020. (In Norwegian) [Google Scholar]
- Fang, B.; Merilä, J.; Ribeiro, F.; Alexandre, C.M.; Momigliano, P. Worldwide phylogeny of three-spined sticklebacks. Mol. Phylogenet. Evol. 2018, 127, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Fang, B.; Kemppainen, P.; Momigliano, P.; Merilä, J. Population Structure Limits Parallel Evolution in Sticklebacks. Mol. Biol. Evol. 2021, 38, 4205–4221. [Google Scholar] [CrossRef] [PubMed]
- Kemppainen, P.; Li, Z.; Rastas, P.; Löytynoja, A.; Fang, B.; Yang, J.; Guo, B.; Shikano, T.; Merilä, J. Genetic population structure constrains local adaptation in sticklebacks. Mol. Ecol. 2021, 30, 1946–1961. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Bassham, S.; Etter, P.D.; Stiffler, N.; Johnson, E.A.; Cresko, W.A. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010, 6, e1000862. [Google Scholar] [CrossRef]
- Karlsen, B.O.; Klingan, K.; Emblem, Å.; Jørgensen, T.E.; Jueterbock, A.; Furmanek, T.; Hoarau, G.; Johansen, S.D.; Nordeide, J.T.; Moum, T. Genomic divergence between the migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 2013, 22, 5098–5111. [Google Scholar] [CrossRef] [PubMed]

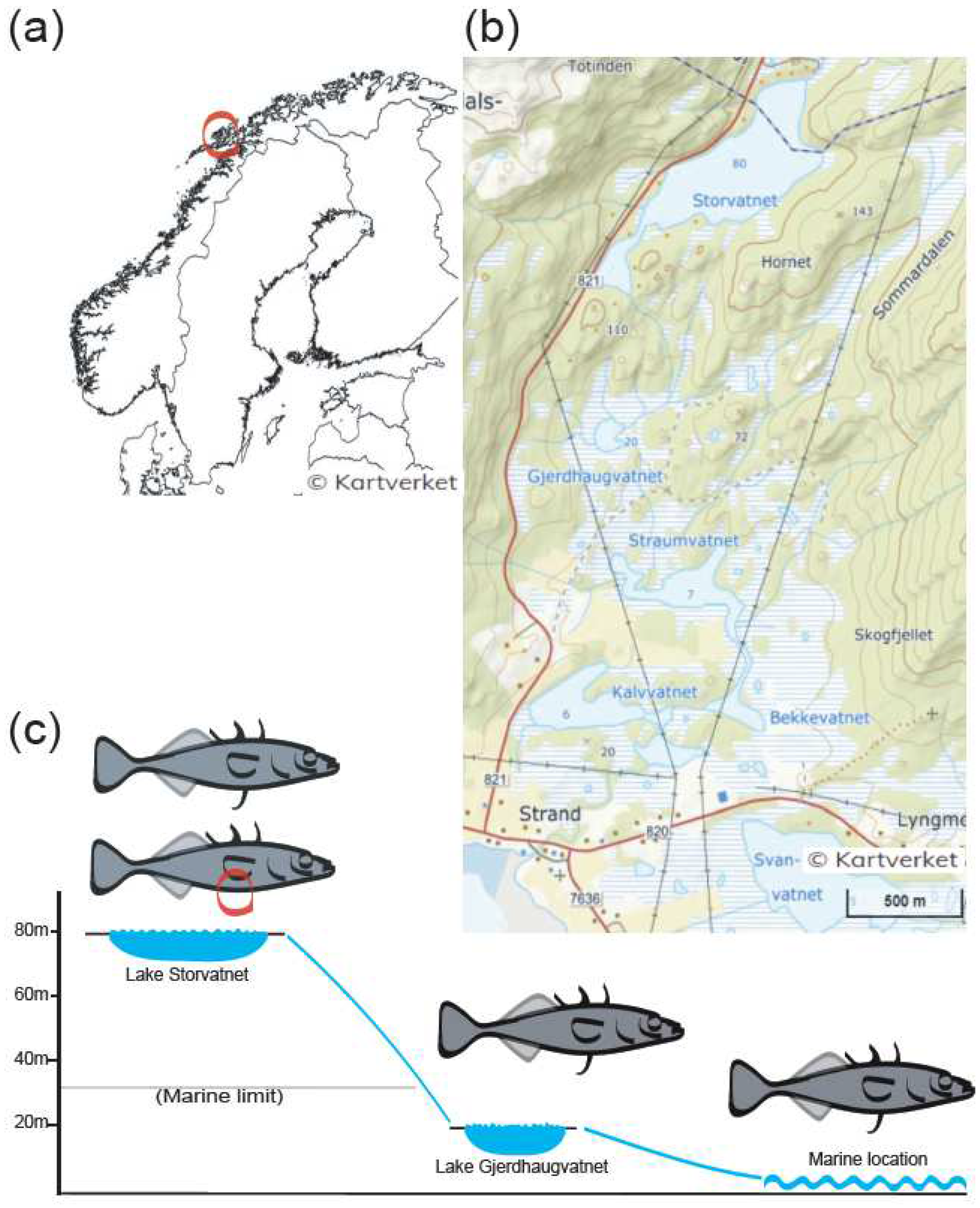
| Site | N | Spineless | Symmetric Spined | Asymmetric | Spine Length (cm) (Mean ± Sd) | Body Length (cm) (Mean ± Sd) | |
|---|---|---|---|---|---|---|---|
| Right-Biased | Left-Biased | ||||||
| Storvatnet | 304 | 92 (30%) | 113 (37%) | 29 (10%) | 70 (23%) | 0.26 ± 0.100 | 4.7 ± 0.60 |
| Gjerdhaugvatnet | 73 | 0 | 73 | 0 | 0 | 0.37 ± 0.070 | 4.1 ± 0.60 |
| Marine | 50 | 0 | 50 | 0 | 0 | 0.55 ± 0.100 | 4.8 ± 0.70 |
| P.v. 1 | Pelvic Scores |
Combined Pelvic Scores (CPS) | Locations (n) | Remarks | |||
|---|---|---|---|---|---|---|---|
| Left PS | Right PS | Storvatn | Gjerdhaugvatn | Marine | |||
| b | 4 | 4 | 8 | 113 | 73 | 50 | Symmetric spined |
| c | 4 | 4 short | 8 | 35 | 0 | 0 | Left-biased asymmetry |
| d | 4 short | 4 | 8 | 22 | 0 | 0 | Right-biased asymmetry |
| f | 3 | 4 | 7 | 7 | 0 | 0 | Right-biased asymmetry |
| e | 4 | 3 | 7 | 29 | 0 | 0 | Left-biased asymmetry |
| h | 4 | 1 | 5 | 5 | 0 | 0 | Left-biased asymmetry |
| g | 4 | 2 | 6 | 1 | 0 | 0 | Left-biased asymmetry |
| i | 3 | 3 | 6 | 32 | 0 | 0 | Spineless |
| j | 1 | 3 | 4 | 1 | 0 | 0 | Spineless |
| l | 1 | 1 | 2 | 51 | 0 | 0 | Spineless |
| k | 2 | 2 | 4 | 8 | 0 | 0 | Spineless |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adhikari, D.; Hanssen, I.K.; Johansen, S.D.; Moum, T.B.; Nordeide, J.T. Pitx1 Enhancer Variants in Spined and Spine-Reduced Subarctic European Sticklebacks. Fishes 2023, 8, 164. https://doi.org/10.3390/fishes8030164
Adhikari D, Hanssen IK, Johansen SD, Moum TB, Nordeide JT. Pitx1 Enhancer Variants in Spined and Spine-Reduced Subarctic European Sticklebacks. Fishes. 2023; 8(3):164. https://doi.org/10.3390/fishes8030164
Chicago/Turabian StyleAdhikari, Dhurba, Ida K. Hanssen, Steinar D. Johansen, Truls B. Moum, and Jarle T. Nordeide. 2023. "Pitx1 Enhancer Variants in Spined and Spine-Reduced Subarctic European Sticklebacks" Fishes 8, no. 3: 164. https://doi.org/10.3390/fishes8030164
APA StyleAdhikari, D., Hanssen, I. K., Johansen, S. D., Moum, T. B., & Nordeide, J. T. (2023). Pitx1 Enhancer Variants in Spined and Spine-Reduced Subarctic European Sticklebacks. Fishes, 8(3), 164. https://doi.org/10.3390/fishes8030164






