Physiological and Transcriptomic Response of Asiatic Hard Clam Meretrix meretrix to the Harmful Alga Heterosigma akashiwo
Abstract
1. Introduction
2. Materials and Methods
2.1. Heterosigma Akashiwo Culture
2.2. Meretrix Meretrix Maintenance
2.3. Exposure Experiments and Sample Collection
2.4. Measurements of the Antioxidant Enzyme Activities
2.5. RNA Sequencing, De Novo Transcriptome Assembly, and Functional Annotation
2.6. Transcriptome Assembly and Annotation of Differentially Expressed Genes (DEGs)
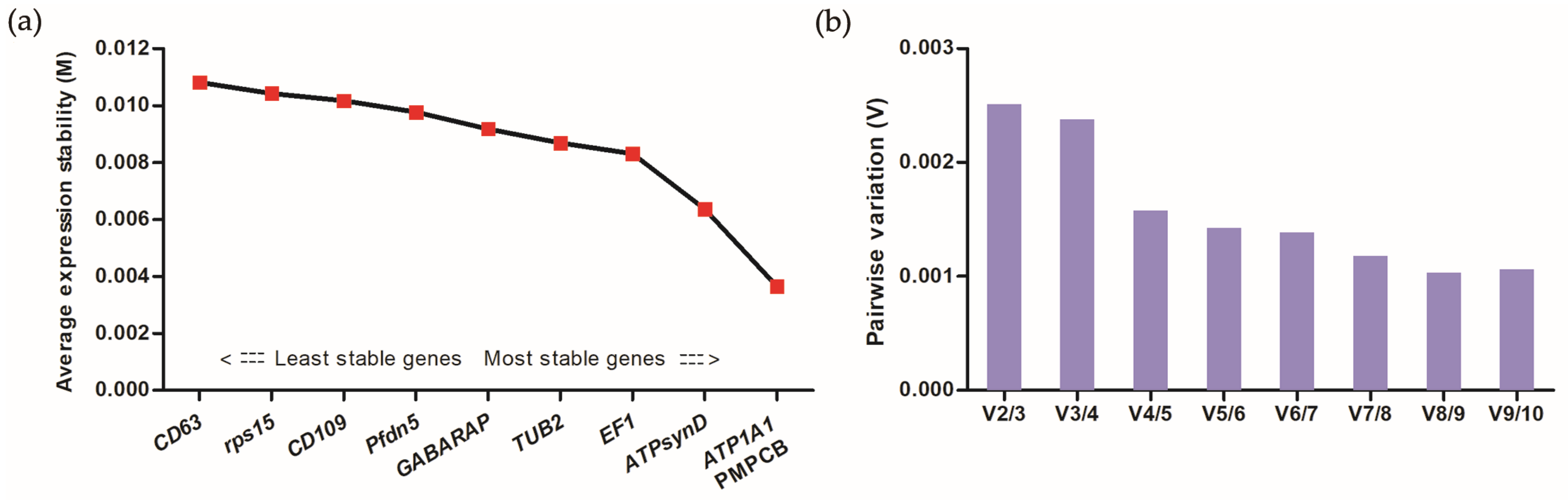
2.7. Identification of Core Genes and Verification of DEGs
3. Results
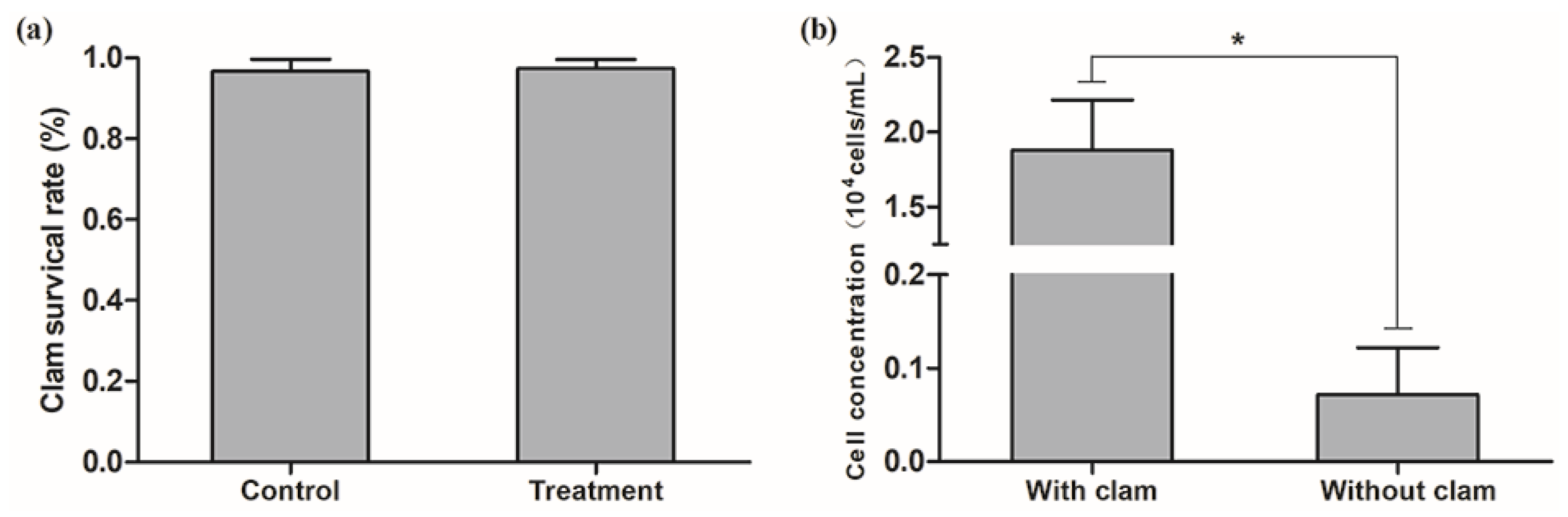
3.1. Exposure of Clams to H. akashiwo
3.2. Effect of H. akashiwo on Clam Antioxidant Functions
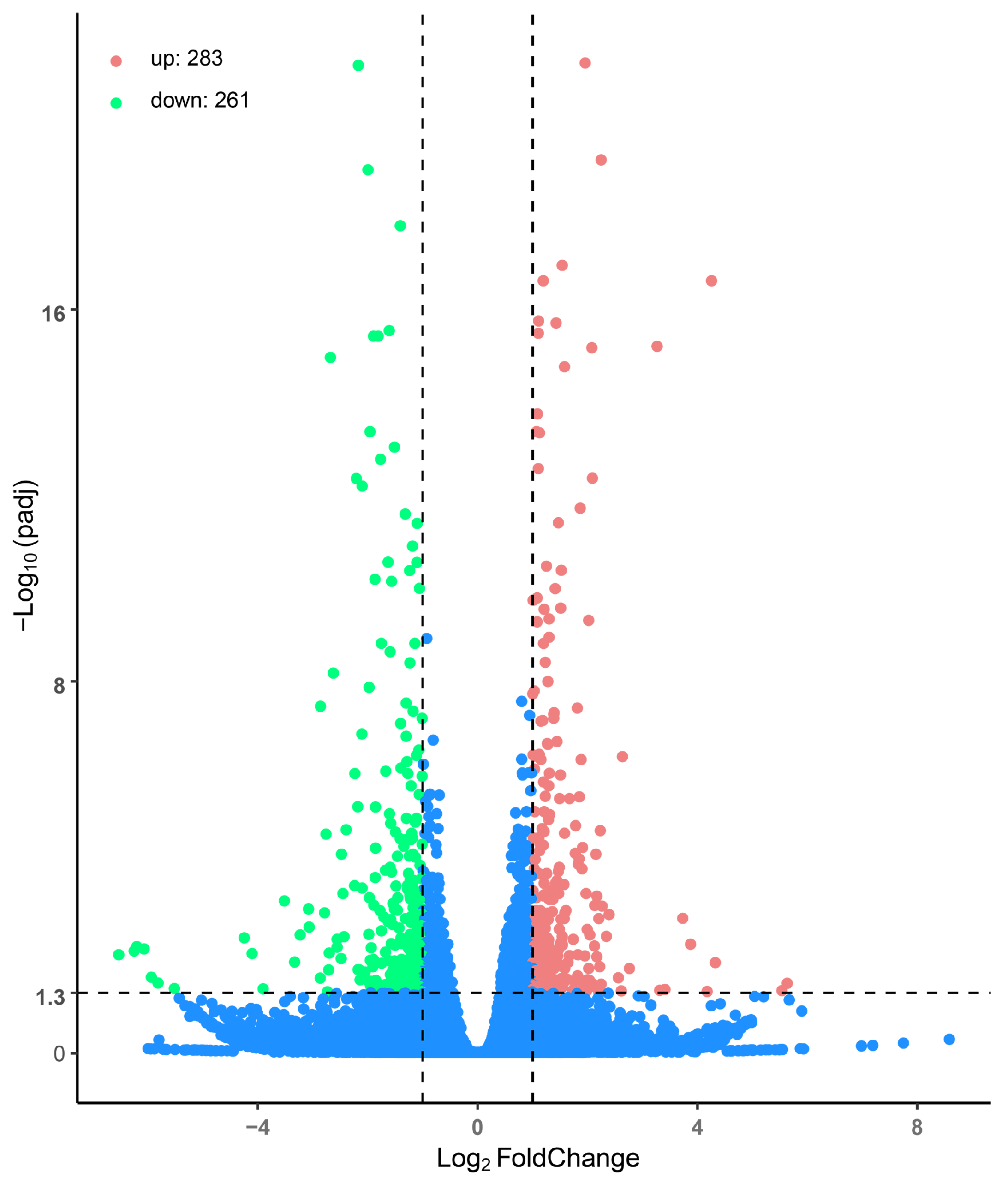
3.3. Transcriptome Profiling and DEG Identification
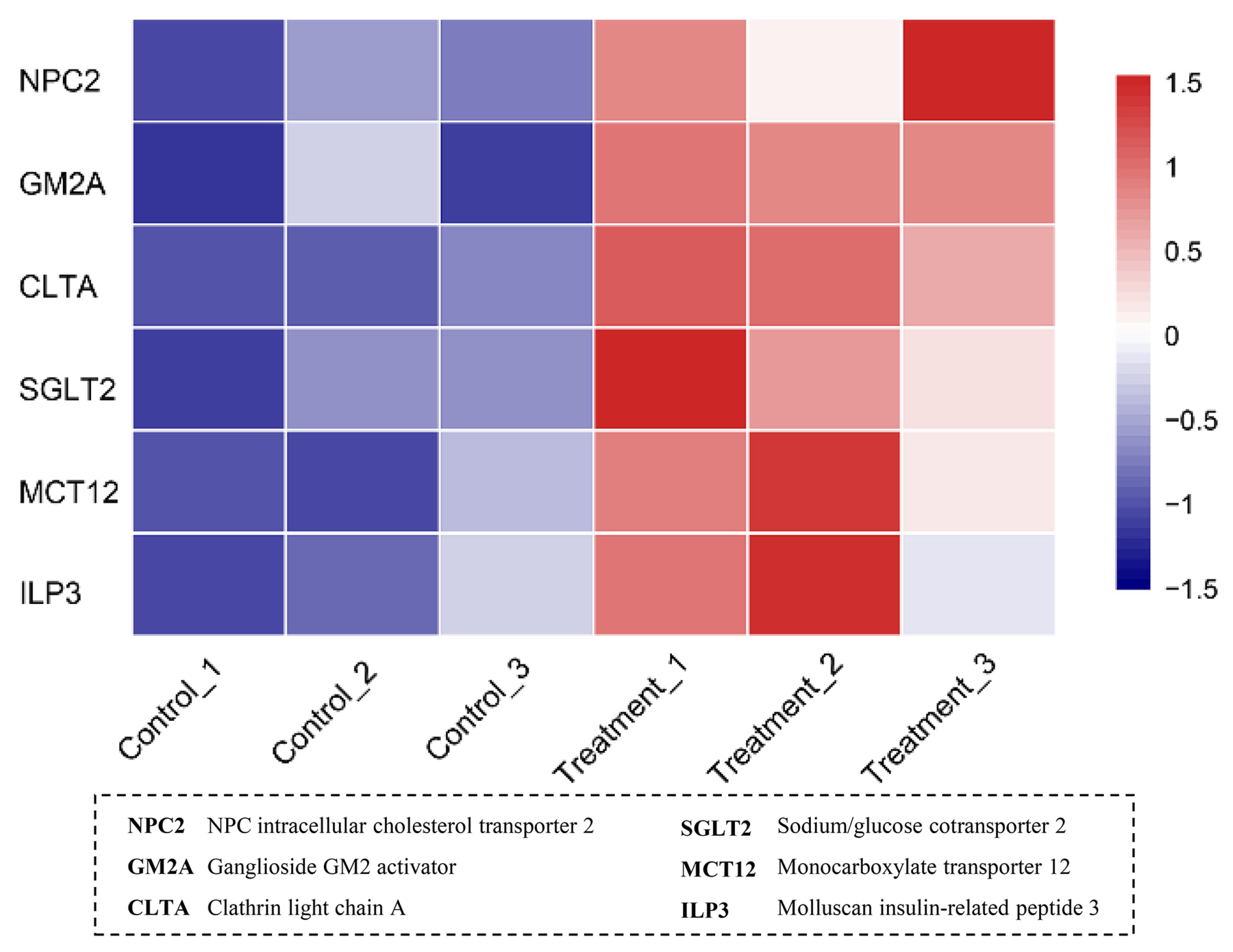
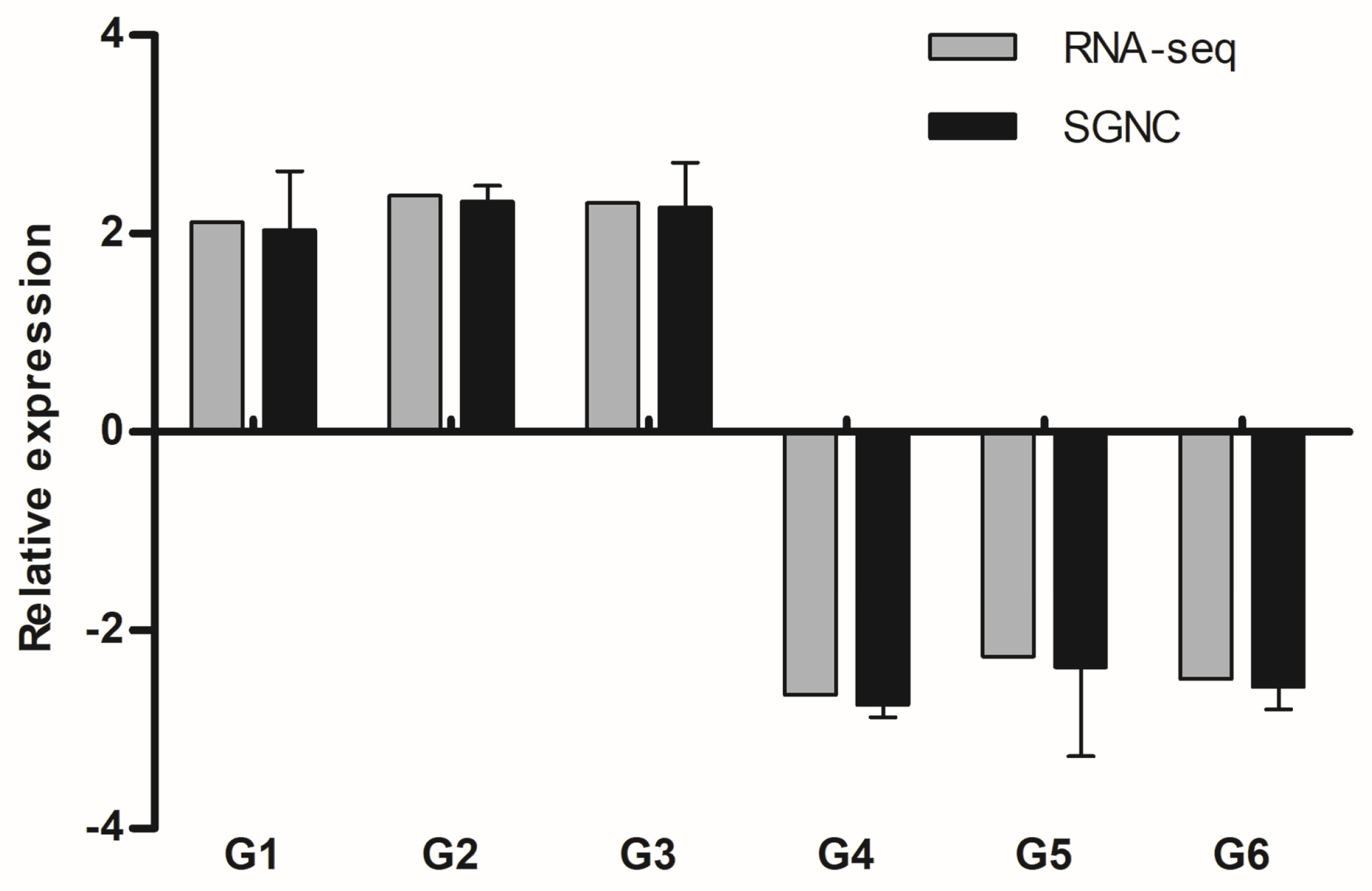
3.4. Verification of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Falkowski, P.G.; Barber, R.T.; Smetacek, V. Biogeochemical controls and feedbacks on ocean primary production. Science 1998, 281, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Cembella, A.D.; Hallegraeff, G.M. Progress in understanding harmful algal blooms: Paradigm shifts and new technologies for research, monitoring, and management. Ann. Rev. Mar. Sci. 2012, 4, 143–176. [Google Scholar] [CrossRef] [PubMed]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.O.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Anderson, D.M. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Li, X.-D.; Tan, Z.-J.; Yu, R.-C.; Zou, J.-Z. Toxic effects, mechanisms, and ecological impacts of harmful algal blooms in China. Harmful Algae 2022, 111, 102148. [Google Scholar] [CrossRef]
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef]
- Jiang, T.; Tong, M.; Qi, Y. Colors for early warning of harmful algal blooms and hazard classification and grading. Acta Ecol. Sin. 2006, 26, 2035–2040. [Google Scholar]
- Nakamura, A.; Okamoto, T.; Komatsu, N.; Ooka, S.; Oda, T.; Ishimatsu, A.; Muramatsu, T. Fish mucus stimurates the generation of superoxide anion by Chattonella marina and Heterosigma akashiwo. Fisheries. Sci. 1998, 64, 866–869. [Google Scholar] [CrossRef]
- Twiner, M.J.; Dixon, S.J.; Trick, C.G. Toxic effects of Heterosigma akashiwo do not appear to be mediated by hydrogen peroxide. Limnol. Oceanogr. 2001, 46, 1400–1405. [Google Scholar] [CrossRef]
- Ji, N.; Lin, L.; Li, L.; Yu, L.; Zhang, Y.; Luo, H.; Li, M.; Shi, X.; Wang, D.Z.; Lin, S. Metatranscriptome analysis reveals environmental and diel regulation of a Heterosigma akashiwo (Raphidophyceae) bloom. Environ. Microbiol. 2018, 20, 1078–1094. [Google Scholar] [CrossRef]
- Honjo, T. Overview on bloom dynamics and physiological ecology of Heterosigma akashiwo. In Toxic Phytoplankton Blooms in the Sea; Smayda, T., Shimizu, Y., Eds.; Elsevier: Amsterdam, The Netherlands, 1993; pp. 33–41. [Google Scholar]
- Twiner, M.J.; Trick, C.G. Possible physiological mechanisms for production of hydrogen peroxide by the ichthyotoxic flagellate Heterosigma akashiwo. J. Plankton. Res. 2000, 22, 1961–1975. [Google Scholar] [CrossRef]
- Twiner, M.J.; Chidiac, P.; Dixon, S.J.; Trick, C.G. Extracellular organic compounds from the ichthyotoxic red tide alga Heterosigma akashiwo elevate cytosolic calcium and induce apoptosis in Sf9 cells. Harmful Algae 2005, 4, 789–800. [Google Scholar] [CrossRef]
- Basti, L.; Hégaret, H.; Shumway, S.E. Harmful algal blooms and shellfish. In Harmful Algal Blooms: A Compendium Desk Reference; Shumway, S.E., Burkholder, J.M., Morton, S.L., Eds.; Wiley Online Library: Chichester, UK, 2018; pp. 135–190. [Google Scholar]
- Keppler, C.J.; Hoguet, J.; Smith, K.; Ringwood, A.H.; Lewitus, A.J. Sublethal effects of the toxic alga Heterosigma akashiwo on the southeastern oyster (Crassostrea virginica). Harmful Algae 2005, 4, 275–285. [Google Scholar] [CrossRef]
- Basti, L.; Nagai, K.; Go, J.; Okano, S.; Oda, T.; Tanaka, Y.; Nagai, S. Lethal effects of ichthyotoxic raphidophytes, Chattonella marina, C. antiqua, and Heterosigma akashiwo, on post-embryonic stages of the Japanese pearl oyster, Pinctada fucata martensii. Harmful Algae 2016, 59, 112–122. [Google Scholar] [PubMed]
- Keppler, C.J.; Lewitus, A.J.; Ringwood, A.H.; Hoguet, J.; Staton, T. Sublethal cellular effects of short-term raphidophyte and brevetoxin exposures on the eastern oyster Crassostrea virginica. Mar. Ecol. Prog. Ser. 2006, 312, 141–147. [Google Scholar] [CrossRef]
- Basti, L.; Go, J.; Okano, S.; Higuchi, K.; Nagai, S.; Nagai, K. Sublethal and antioxidant effects of six ichthyotoxic algae on early-life stages of the Japanese pearl oyster. Harmful Algae 2021, 103, 102013. [Google Scholar] [CrossRef]
- Mat, A.M.; Klopp, C.; Payton, L.; Jeziorski, C.; Chalopin, M.; Amzil, Z.; Tran, D.; Wikfors, G.H.; Hégaret, H.; Soudant, P. Oyster transcriptome response to Alexandrium exposure is related to saxitoxin load and characterized by disrupted digestion, energy balance, and calcium and sodium signaling. Aquat. Toxicol. 2018, 199, 127–137. [Google Scholar] [CrossRef]
- Liu, B.; Dong, B.; Tang, B.; Zhang, T.; Xiang, J. Effect of stocking density on growth, settlement and survival of clam larvae, Meretrix meretrix. Aquaculture 2006, 258, 344–349. [Google Scholar] [CrossRef]
- Xu, N.; Wang, M.; Tang, Y.; Zhang, Q.; Duan, S.; Gobler, C. Acute toxicity of the cosmopolitan bloom-forming dinoflagellate Akashiwo sanguinea to finfish, shellfish, and zooplankton. Aquat. Microb. Ecol. 2017, 80, 209–222. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Ryther, J.H. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic. Acids. Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Bio. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ji, N.; Wang, J.; Zhang, Z.; Chen, L.; Xu, M.; Yin, X.; Shen, X. Transcriptomic response of the harmful algae Heterosigma akashiwo to polyphosphate utilization and phosphate stress. Harmful Algae 2022, 117, 102267. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef]
- Alexander, H.; Jenkins, B.D.; Rynearson, T.A.; Dyhrman, S.T. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. USA 2015, 112, E2182–E2190. [Google Scholar] [CrossRef] [PubMed]
- Mardones, J.I.; Paredes-Mella, J.; Flores-Leñero, A.; Yarimizu, K.; Godoy, M.; Artal, O.; Corredor-Acosta, A.; Marcus, L.; Cascales, E.; Espinoza, J.P. Extreme harmful algal blooms, climate change, and potential risk of eutrophication in Patagonian fjords: Insights from an exceptional Heterosigma akashiwo fish-killing event. Prog. Oceanogr. 2023, 210, 102921. [Google Scholar] [CrossRef]
- O’Halloran, C.; Silver, M.W.; Holman, T.R.; Scholin, C.A. Heterosigma akashiwo in central California waters. Harmful Algae 2006, 5, 124–132. [Google Scholar] [CrossRef]
- Chang, F.H.; Anderson, C.; Boustead, N.C. First record of a Heterosigma (Raphidophyceae) bloom with associated mortality of cage-reared salmon in Big Glory Bay, New Zealand. New Zeal. J. Mar. Freshwater Res. 1990, 24, 461–469. [Google Scholar] [CrossRef]
- Astuya, A.; Ramírez, A.E.; Aballay, A.; Araya, J.; Silva, J.; Ulloa, V.; Fuentealba, J. Neurotoxin-like compounds from the ichthyotoxic red tide alga Heterosigma akashiwo induce a TTX-like synaptic silencing in mammalian neurons. Harmful Algae 2015, 47, 1–8. [Google Scholar] [CrossRef]
- Taga, S.; Yamasaki, Y.; Kishioka, M. Dietary effects of the red-tide raphidophyte Heterosigma akashiwo on growth of juvenile Manila clams, Ruditapes philippinarum. Plankton Benthos Res. 2013, 8, 102–105. [Google Scholar] [CrossRef]
- Galimany, E.; Rose, J.M.; Alix, J.; Dixon, M.S.; Wikfors, G.H. Responses of the ribbed mussel, Geukensia demissa, to the harmful algae Aureococcus anophagefferens and Heterosigma akashiwo. J. Mollus. Stud. 2014, 80, 123–130. [Google Scholar] [CrossRef]
- Hegaret, H.; Wikfors, G.H.; Shumway, S.E. Diverse feeding responses of five species of bivalve mollusc when exposed to three species of harmful algae. J. Shellfish. Res. 2007, 26, 549–559. [Google Scholar] [CrossRef]
- Haque, S.M.; Onoue, Y. Effects of salinity on growth and toxin production of a noxious phytoflagellate, Heterosigma akashiwo (Raphidophyceae). Bot. Mar. 2002, 45, 356–365. [Google Scholar] [CrossRef]
- Fredrickson, K.A.; Strom, S.L.; Crim, R.; Coyne, K.J. Interstrain variability in physiology and genetics of Heterosigma akashiwo (Raphidophyceae) from the West coast of North America. J. Phycol. 2011, 47, 25–35. [Google Scholar] [CrossRef]
- Braga, A.C.; Pereira, V.; Marçal, R.; Marques, A.; Guilherme, S.; Costa, P.R.; Pacheco, M. DNA damage and oxidative stress responses of mussels Mytilus galloprovincialis to paralytic shellfish toxins under warming and acidification conditions–Elucidation on the organ-specificity. Aquat. Toxicol. 2020, 228, 105619. [Google Scholar] [CrossRef]
- Hanquet, A.-C.; Jouaux, A.; Heude, C.; Mathieu, M.; Kellner, K. A sodium glucose co-transporter (SGLT) for glucose transport into Crassostrea gigas vesicular cells: Impact of alimentation on its expression. Aquaculture 2011, 313, 123–128. [Google Scholar] [CrossRef]
- Li, Y.; Fu, H.; Zhang, F.; Ren, L.; Tian, J.; Li, Q.; Liu, S. Identification, characterization, and expression profiles of insulin-like peptides suggest their critical roles in growth regulation of the Pacific oyster, Crassostrea gigas. Gene 2021, 769, 145244. [Google Scholar] [CrossRef]
- Kim, H.G. Mitigation and controls of HABs. In Ecology of Harmful Algae; Granéli, E., Turner, J.T., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 327–338. [Google Scholar]
- Galimany, E.; Lunt, J.; Freeman, C.J.; Houk, J.; Sauvage, T.; Santos, L.; Lunt, J.; Kolmakova, M.; Mossop, M.; Domingos, A. Bivalve feeding responses to microalgal bloom species in the Indian River Lagoon: The potential for top-down control. Estuar. Coast. 2020, 43, 1519–1532. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, N.; Xu, M.; Wang, J.; Li, J.; Liu, S.; Yin, X.; Shen, X.; Cai, Y. Physiological and Transcriptomic Response of Asiatic Hard Clam Meretrix meretrix to the Harmful Alga Heterosigma akashiwo. Fishes 2023, 8, 67. https://doi.org/10.3390/fishes8020067
Ji N, Xu M, Wang J, Li J, Liu S, Yin X, Shen X, Cai Y. Physiological and Transcriptomic Response of Asiatic Hard Clam Meretrix meretrix to the Harmful Alga Heterosigma akashiwo. Fishes. 2023; 8(2):67. https://doi.org/10.3390/fishes8020067
Chicago/Turabian StyleJi, Nanjing, Mingyang Xu, Junyue Wang, Junjia Li, Shishi Liu, Xueyao Yin, Xin Shen, and Yuefeng Cai. 2023. "Physiological and Transcriptomic Response of Asiatic Hard Clam Meretrix meretrix to the Harmful Alga Heterosigma akashiwo" Fishes 8, no. 2: 67. https://doi.org/10.3390/fishes8020067
APA StyleJi, N., Xu, M., Wang, J., Li, J., Liu, S., Yin, X., Shen, X., & Cai, Y. (2023). Physiological and Transcriptomic Response of Asiatic Hard Clam Meretrix meretrix to the Harmful Alga Heterosigma akashiwo. Fishes, 8(2), 67. https://doi.org/10.3390/fishes8020067




