Protein Arginine Methyltransferase 5 Is Necessary for Embryonic Development in Medaka Oryzias latipes
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Experimental Animal
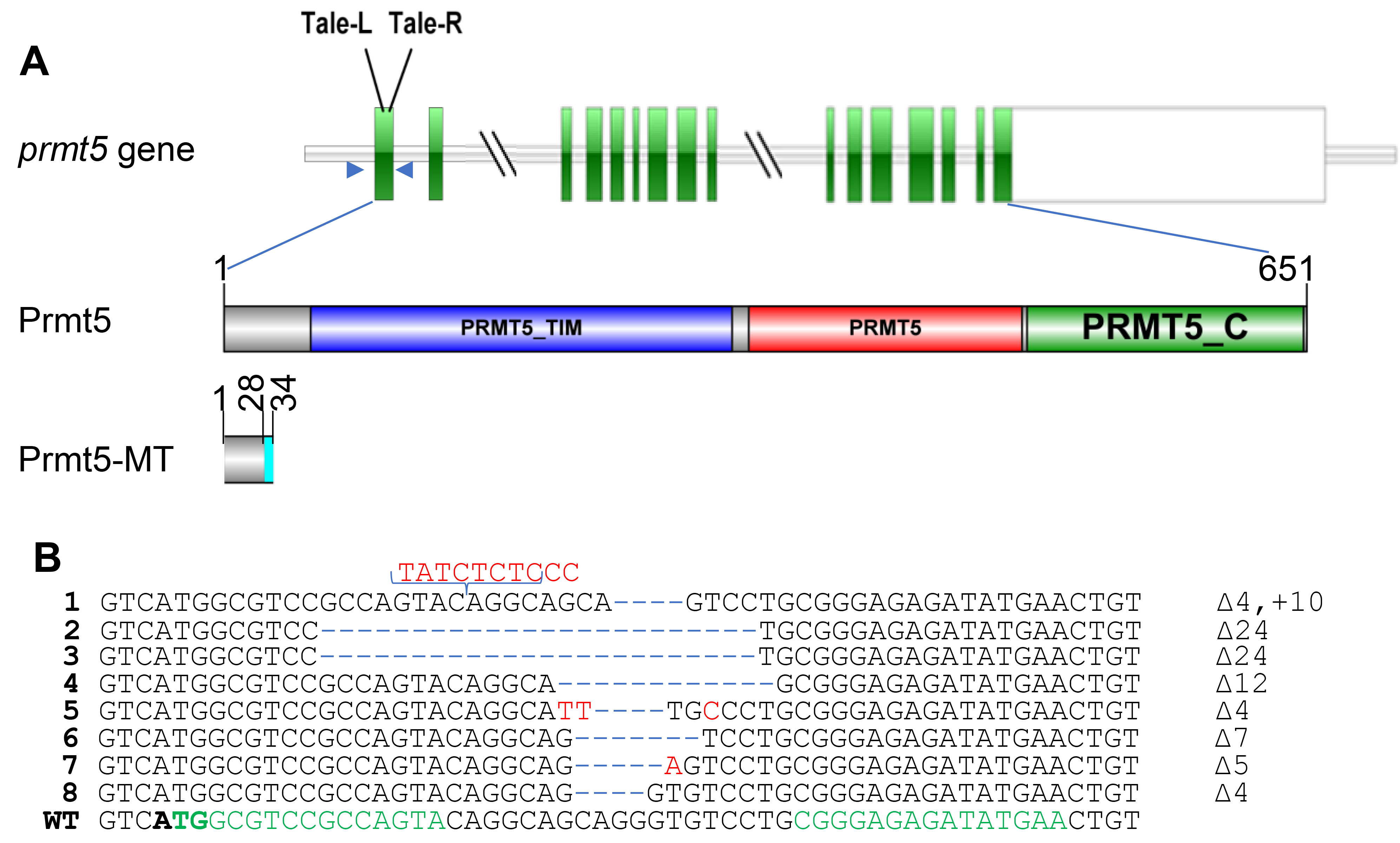
2.3. Gene Editing by TALENs
2.4. Genomic DNA Extraction and Polymerase Chain Reaction (PCR)
2.5. Detection of Mutation by T7E1 (T7 Endonuclease I) or Sequencing
2.6. Detection of the Gene Expression by Quantitative RT-PCR
2.7. Statistical Analysis
3. Results
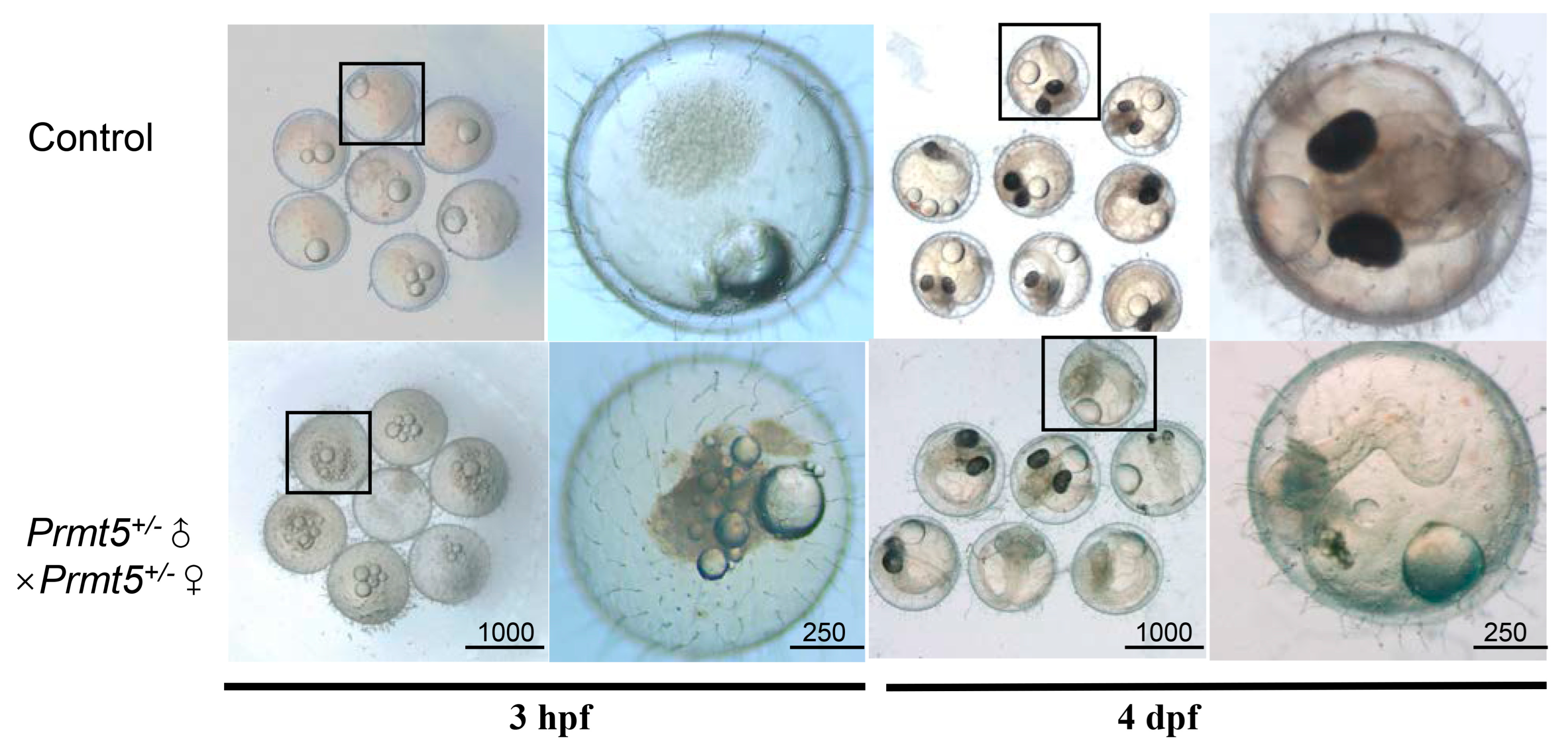
3.1. Mutation of Medaka Prmt5
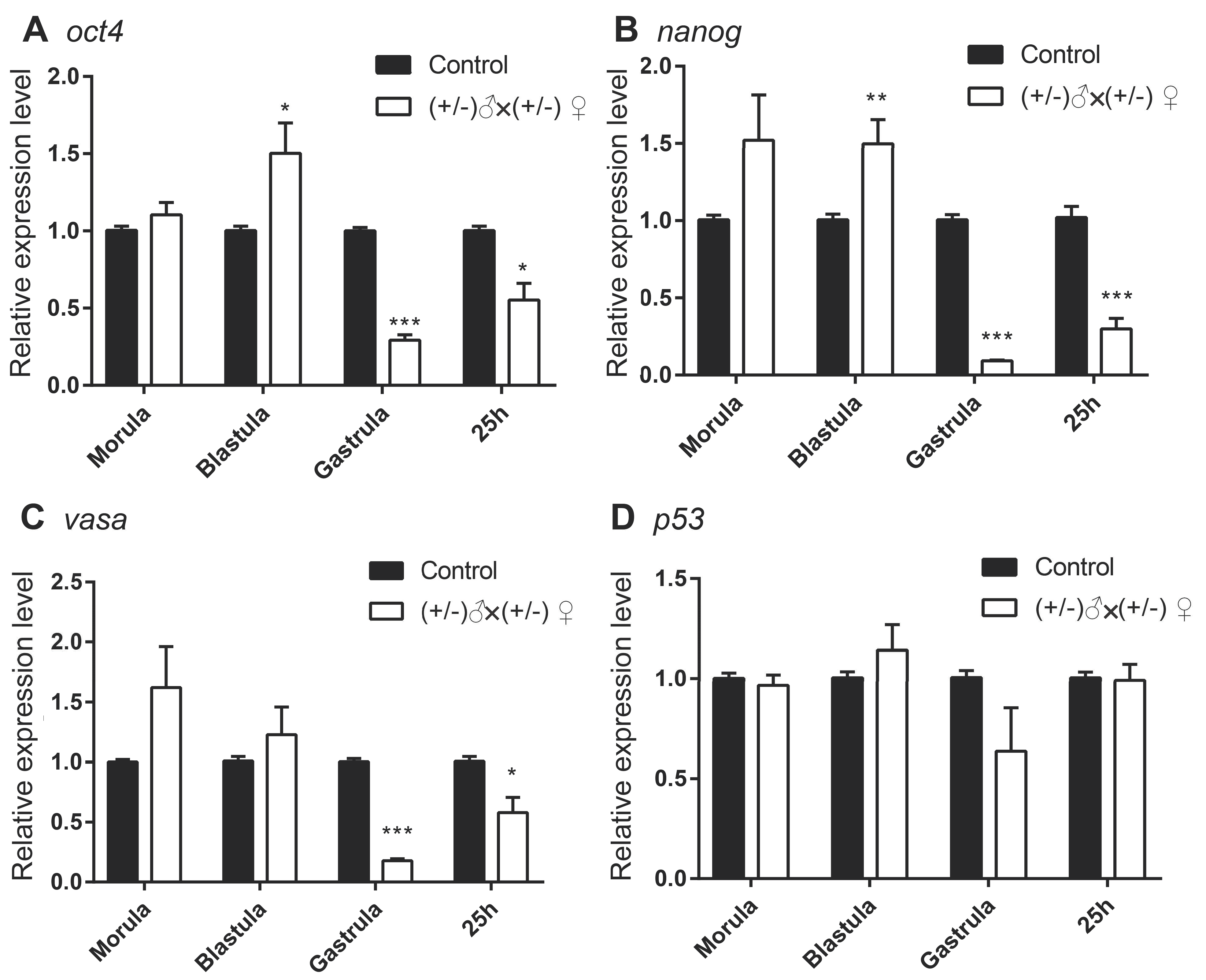
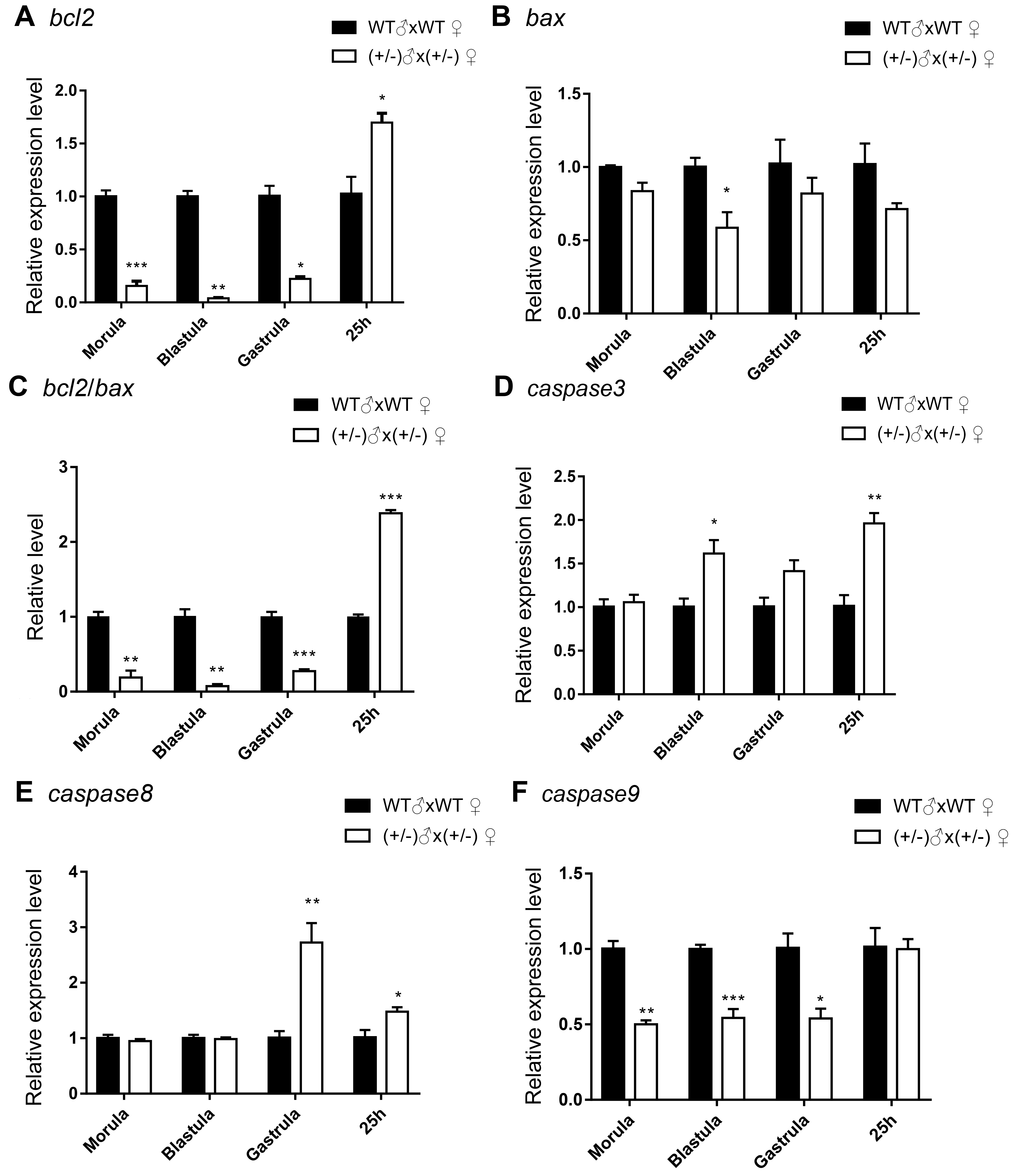
3.2. Gene Expression of the Inbred Offspring of the Heterozygous Mutant during Embryogenesis
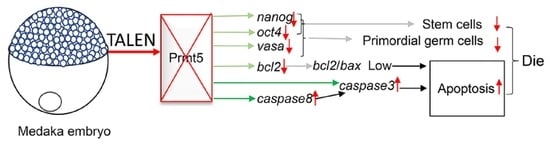
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Rank, G.; Tan, Y.T.; Li, H.; Moritz, R.L.; Simpson, R.J.; Cerruti, L.; Curtis, D.J.; Patel, D.J.; Allis, C.D.; et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 2009, 16, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; Cote, J.; Boulanger, M.C.; Cleroux, P.; Bachand, F.; Autexier, C.; Richard, S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 2002, 159, 957–969. [Google Scholar] [CrossRef]
- Neuenkirchen, N.; Chari, A.; Fischer, U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008, 582, 1997–2003. [Google Scholar] [CrossRef]
- Scoumanne, A.; Zhang, J.; Chen, X. PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res. 2009, 37, 4965–4976. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.; Durant, S.T.; Cho, E.C.; Sheahan, S.; Edelmann, M.; Kessler, B.; La Thangue, N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008, 10, 1431–1439. [Google Scholar] [CrossRef]
- Yang, M.; Sun, J.; Sun, X.; Shen, Q.; Gao, Z.; Yang, C. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009, 5, e1000514. [Google Scholar] [CrossRef]
- Chung, J.; Karkhanis, V.; Tae, S.; Yan, F.; Smith, P.; Ayers, L.W.; Agostinelli, C.; Pileri, S.; Denis, G.V.; Baiocchi, R.A.; et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J. Biol. Chem. 2013, 288, 35534–35547. [Google Scholar]
- Nicholas, C.; Yang, J.; Peters, S.B.; Bill, M.A.; Baiocchi, R.A.; Yan, F.; Sïf, S.; Tae, S.; Gaudio, E.; Wu, X.; et al. PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1). PLoS ONE 2013, 8, e74710. [Google Scholar] [CrossRef]
- Richard, S.; Morel, M.; Cléroux, P. Arginine methylation regulates IL-2 gene expression: A role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 2005, 388, 379–386. [Google Scholar] [CrossRef]
- Krause, C.D.; Yang, Z.H.; Kim, Y.S.; Lee, J.H.; Cook, J.R.; Pestka, S. Protein arginine methyltransferases: Evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 2007, 113, 50–87. [Google Scholar] [CrossRef] [PubMed]
- Tee, W.W.; Pardo, M.; Theunissen, T.W.; Yu, L.; Choudhary, J.S.; Hajkova, P.; Surani, M.A. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010, 24, 2772–2777. [Google Scholar] [CrossRef] [PubMed]
- Nagamatsu, G.; Kosaka, T.; Kawasumi, M.; Kinoshita, T.; Takubo, K.; Akiyama, H.; Sudo, T.; Kobayashi, T.; Oya, M.; Suda, T. A germ cell-specific gene, Prmt5, works in somatic cell reprogramming. J. Biol. Chem. 2011, 286, 10641–10648. [Google Scholar] [CrossRef] [PubMed]
- Dacwag, C.S.; Ohkawa, Y.; Pal, S.; Sif, S.; Imbalzano, A.N. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol. 2007, 27, 384–394. [Google Scholar] [CrossRef]
- Mallappa, C.; Hu, Y.J.; Shamulailatpam, P.; Tae, S.; Sif, S.; Imbalzano, A.N. The expression of myogenic microRNAs indirectly requires protein arginine methyltransferase (Prmt)5 but directly requires Prmt4. Nucleic Acids Res. 2011, 39, 1243–1255. [Google Scholar] [CrossRef]
- Kirino, Y.; Kim, N.; de Planell-Saguer, M.; Khandros, E.; Chiorean, S.; Klein, P.S.; Rigoutsos, I.; Jongens, T.A.; Mourelatos, Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009, 11, 652–658. [Google Scholar] [CrossRef]
- Gonsalvez, G.B.; Rajendra, T.K.; Tian, L.; Matera, A.G. The Sm-protein methyltransferase, Dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006, 16, 1077–1089. [Google Scholar] [CrossRef]
- Anne, J.; Ollo, R.; Ephrussi, A.; Mechler, B.M. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 2007, 134, 137–146. [Google Scholar] [CrossRef]
- Li, Z.; Yu, J.; Hosohama, L.; Nee, K.; Gkountela, S.; Chaudhari, S.; Cass, A.A.; Xiao, X.; Clark, A.T. The Sm protein methyltransferase PRMT5 is not required for primordial germ cell specification in mice. EMBO J. 2015, 34, 748–758. [Google Scholar] [CrossRef]
- Kim, S.; Günesdogan, U.; Zylicz, J.J.; Hackett, J.A.; Cougot, D.; Bao, S.; Lee, C.; Dietmann, S.; Allen, G.E.; Sengupta, R.; et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 2014, 56, 564–579. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, T.; Li, Q.; Liu, C.; Han, F.; Chen, M.; Zhang, L.; Cui, X.; Qin, Y.; Bao, S.; et al. Prmt5 is required for germ cell survival during spermatogenesis in mice. Sci. Rep. 2015, 5, 11031. [Google Scholar] [CrossRef]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.; Bannister, A.J.; Kouzarides, T.; Surani, M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef]
- Vagin, V.V.; Wohlschlegel, J.; Qu, J.; Jonsson, Z.; Huang, X.; Chuma, S.; Girard, A.; Sachidanandam, R.; Hannon, G.J.; Aravin, A.A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009, 23, 1749–1762. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.M.; Li, C. Identification and phylogenetic analyses of the protein arginine methyltransferase gene family in fish and ascidians. Gene 2004, 340, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cao, M.; Yang, Y.; Nagahama, Y.; Zhao, H. Expression pattern of prmt5 in adult fish and embryos of medaka, Oryzias latipes. Fish Physiol. Biochem. 2009, 35, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Li, C. Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 2012, 279, 932–945. [Google Scholar] [CrossRef]
- Batut, J.; Duboé, C.; Vandel, L. The methyltransferases PRMT4/CARM1 and PRMT5 control differentially myogenesis in zebrafish. PLoS ONE 2011, 6, e25427. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, D.; Liu, X.; Yu, G.; Cai, X.; Xu, C.; Rong, F.; Ouyang, G.; Wang, J.; Xiao, W. Zebrafish prmt5 arginine methyltransferase is essential for germ cell development. Development 2019, 146, dev179572. [Google Scholar] [CrossRef]
- Cheng, N.; Guo, M.; Chang, P.; Zhang, X.; Zhang, R.; Qi, C.; Zhong, X.; Zhou, Q.; Zhao, H. Expression of mep50 in adult and embryos of medaka fish (Oryzias latipes). Fish Physiol. Biochem. 2016, 42, 1053–1061. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, X.; Al Hafiz, M.A.; Liang, X.; Yao, Q.; Guo, M.; Xu, G.; Zhong, X.; Zhou, Q.; Zhao, H. The proteins interacting with Prmt5 in medaka (Oryzias latipes) identified by yeast two-hybridization. Protein Pept. Lett. 2020, 27, 971–978. [Google Scholar] [CrossRef]
- Huang, P.; Xiao, A.; Zhou, M.; Zhu, Z.; Lin, S.; Zhang, B. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 2011, 29, 699–700. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Xiao, A.; Tong, X.; Zu, Y.; Wang, Z.; Zhang, B. TALEN construction via “Unit Assembly” method and targeted genome modifications in zebrafish. Methods 2014, 69, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhang, X.; Cheng, N.; Duan, J.; Wang, J.; Nagahama, Y.; Zhong, X.; Zhou, Q.; Wang, Y. Identification and expression profiles of prdm1 in medaka Oryzias latipes. Mol. Biol. Rep. 2014, 41, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Shinomiya, A.; Tanaka, M.; Kobayashi, T.; Nagahama, Y.; Hamaguchi, S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev. Growth Differ. 2000, 42, 317–326. [Google Scholar] [CrossRef]
- Camp, E.; Sánchez-Sánchez, A.V.; García-España, A.; Desalle, R.; Odqvist, L.; Enrique O’Connor, J.; Mullor, J.L. Nanog regulates proliferation during early fish development. Stem Cells 2009, 27, 2081–2091. [Google Scholar] [CrossRef]
- Wang, D.; Manali, D.; Wang, T.; Bhat, N.; Hong, N.; Li, Z.; Wang, L.; Yan, Y.; Liu, R.; Hong, Y. Identification of pluripotency genes in the fish medaka. Int. J. Biol. Sci. 2011, 7, 440–451. [Google Scholar] [CrossRef]
- Shen, J.; Yokota, S.; Yokoi, H.; Suzuki, T. Diethylnitrosamine-induced expression of germline-specific genes and pluripotency factors, including vasa and oct4, in medaka somatic cells. Biochem. Biophys. Res. Commun. 2016, 478, 858–863. [Google Scholar] [CrossRef]
- Krause, M.K.; Rhodes, L.D.; Van Beneden, R.J. Cloning of the p53 tumor suppressor gene from the Japanese medaka (Oryzias latipes) and evaluation of mutational hotspots in MNNG-exposed fish. Gene 1997, 189, 101–106. [Google Scholar] [CrossRef]
- Barjhoux, I.; Gonzalez, P.; Baudrimont, M.; Cachot, J. Molecular and phenotypic responses of Japanese medaka (Oryzias latipes) early life stages to environmental concentrations of cadmium in sediment. Environ. Sci. Pollut. Res. Int. 2016, 23, 17969–17981. [Google Scholar] [CrossRef]
- Iijima, N.; Yokoyama, T. Apoptosis in the medaka embryo in the early developmental stage. Acta Histochem. Cytochem. 2007, 40, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sakamaki, K.; Nozaki, M.; Kominami, K.; Satou, Y. The evolutionary conservation of the core components necessary for the extrinsic apoptotic signaling pathway, in medaka fish. BMC Genom. 2007, 8, 141. [Google Scholar] [CrossRef]
- Lee, J.W.; Choi, Y.C.; Kim, R.; Lee, S.K. Multiwall carbon nanotube-induced apoptosis and antioxidant gene expression in the gills, liver, and intestine of Oryzias latipes. BioMed Res. Int. 2015, 2015, 485343. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Niu, B.; Zhu, H.; He, X.; Bai, C.; Li, G.; Hua, J. PRMT5 enhances generation of induced pluripotent stem cells from dairy goat embryonic fibroblasts via down-regulation of p53. Cell Prolif. 2015, 48, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Chavez, S.L.; Pera, R.A.R. Generation of human induced pluripotent stem cells using epigenetic regulators reveals a germ cell-like identity in partially reprogrammed colonies. PLoS ONE 2013, 8, e82838. [Google Scholar] [CrossRef] [PubMed]
- Gkountela, S.; Li, Z.; Chin, C.J.; Lee, S.A.; Clark, A.T. PRMT5 is required for human embryonic stem cell proliferation but not pluripotency. Stem Cell Rev. Rep. 2014, 10, 230–239. [Google Scholar] [CrossRef]
- Hong, Y.; Winkler, C.; Liu, T.; Chai, G.; Schartl, M. Activation of the mouse Oct4 promoter in medaka embryonic stem cells and its use for ablation of spontaneous differentiation. Mech. Dev. 2004, 121, 933–943. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, A.V.; Camp, E.; García-España, A.; Leal-Tassias, A.; Mullor, J.L. Medaka Oct4 is expressed during early embryo development, and in primordial germ cells and adult gonads. Dev. Dyn. 2010, 239, 672–679. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, A.V.; Camp, E.; Leal-Tassias, A.; Atkinson, S.P.; Armstrong, L.; Díaz-Llopis, M.; Mullor, J.L. Nanog regulates primordial germ cell migration through Cxcr4b. Stem Cells 2010, 28, 1457–1764. [Google Scholar] [CrossRef]
- Liu, R.; Li, M.; Li, Z.; Hong, N.; Xu, H.; Hong, Y. Medaka Oct4 is essential for pluripotency in blastula formation and ES cell derivation. Stem Cell Rev. Rep. 2015, 11, 11–23. [Google Scholar] [CrossRef]
- Sun, B.; Gui, L.; Liu, R.; Hong, Y.; Li, M. Medaka oct4 is essential for gastrulation, central nervous system development and angiogenesis. Gene 2020, 733, 144270. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Vogel, G.; Yu, Z.; Almazan, G.; Richard, S. Type II arginine methyltransferase PRMT5 regulates gene expression of inhibitors of differentiation/DNA binding Id2 and Id4 during glial cell differentiation. J. Biol. Chem. 2011, 286, 44424–44432. [Google Scholar] [CrossRef] [PubMed]
- Norrie, J.L.; Li, Q.; Co, S.; Huang, B.L.; Ding, D.; Uy, J.C.; Ji, Z.; Mackem, S.; Bedford, M.T.; Galli, A.; et al. PRMT5 is essential for the maintenance of chondrogenic progenitor cells in the limb bud. Development 2016, 143, 4608–4619. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, J.; Liu, Z.; Gray, R.S.; Vokes, S.A. PRMT5 is necessary to form distinct cartilage identities in the knee and long bone. Dev. Biol. 2019, 456, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Günther, S.; Looso, M.; Künne, C.; Krüger, M.; Kim, J.; Zhou, Y.; Braun, T. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat. Commun. 2015, 6, 7140. [Google Scholar] [CrossRef]
- Webb, L.M.; Sengupta, S.; Edell, C.; Piedra-Quintero, Z.L.; Amici, S.A.; Miranda, J.N.; Bevins, M.; Kennemer, A.; Laliotis, G.; Tsichlis, P.N.; et al. Protein arginine methyltransferase 5 promotes cholesterol biosynthesis-mediated Th17 responses and autoimmunity. J. Clin. Investig. 2020, 130, 1683–1698. [Google Scholar] [CrossRef]
- Litzler, L.C.; Zahn, A.; Meli, A.P.; Hébert, S.; Patenaude, A.M.; Methot, S.P.; Sprumont, A.; Bois, T.; Kitamura, D.; Costantino, S.; et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat. Commun. 2019, 10, 22. [Google Scholar] [CrossRef]
- Tanaka, Y.; Nagai, Y.; Okumura, M.; Greene, M.I.; Kambayashi, T. PRMT5 is required for T cell survival and proliferation by maintaining cytokine signaling. Front. Immunol. 2020, 11, 621. [Google Scholar] [CrossRef]
- Korsmeyer, S.J.; Shutter, J.R.; Veis, D.J.; Merry, D.E.; Oltvai, Z.N. Bcl-2/Bax: A rheostat that regulates an anti-oxidant pathway and cell death. Semin. Cancer Biol. 1993, 4, 327–332. [Google Scholar]
- Korsmeyer, S.J. Regulators of cell death. Trends Genet. 1995, 11, 101–105. [Google Scholar] [CrossRef]
- Galluzzi, L.; López-Soto, A.; Kumar, S.; Kroemer, G. Caspases connect cell-death signaling to organismal homeostasis. Immunity 2016, 44, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.J.; Han, L.H.; Cong, R.S.; Liang, J. Caspase family proteases and apoptosis. Acta Biochim. Biophys. Sin. (Shanghai) 2005, 37, 719–727. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| prmt5 | prmt5F | ACCTGTAGCTGTTAATTTGATCTGC |
| prmt5R | GAATCCAAAAGCAGAGCATCCT | |
| β-actin | β-actinF | CACACCTTCTACAATGAGCTG |
| β-actinR | CCAGATCTGCTGGAAGGTGG | |
| oct4 | oct4-qF | TCTTTGGCGTAAACTCGTCTCA |
| oct4-qR | CTTGCGTAAAACCCAAAGTGAT | |
| nanog | nanog-qF | TACTCCAAACGCCCCGAAAG |
| nanog-qR | GTGTCCTTCTGATGCCTCCTAA | |
| vasa | vasa-qF | GCTCATCAACCAGATTTACCA |
| vasa-qR | ATCTCCCTCATCTGGTAGCCG | |
| p53 | p53-qF | TCTACAAGAAGACGGAGCACG |
| p53-qR | ACTGTAACACTCTGCCTTTTGGTAT | |
| bcl2 | bcl2-qF | TCGACAGTTTTCCCCTGCAA |
| bcl2-qR | GAAACCCCCTGAACGGAACT | |
| bax | bax-qF | GCGATCAAGGTAGCGAAAAAT |
| bax-qR | CAGGTTTCTCCTGACCCGTT | |
| caspase3 | caspase3-qF | AACAAGACGCCGACCCTTAC |
| caspase3-qR | TGTACCGTTACGAGGACCCA | |
| caspase8 | caspase8-qF | TGACCCTACCCTTTCCCAGT |
| caspase8-qR | CACTTTGGGCTAGTGTGCCT | |
| caspase9 | caspase9-qF | AACTTCGTGGTGGAAGTCCG |
| caspase9-qR | AGCTCCCAGCTGATCTGATCTT | |
| RPS18 | s18-qF | GTGTGGTGACCATCATGCAGAA |
| s18-qR | TGGCAAGGACCTGGCTGTATT |
| Generation | Prmt5 (+/−) × (+/−) | Wild Type | p Value | ||
|---|---|---|---|---|---|
| All Stages | To Gastrula | All Stages | To Gastrula | ||
| F2 | 29.9% | 19.9% | 4.2% | - | 2.3 × 10−27 |
| F3 | 24.3% | 18.1% | 5.7% | - | 3.1 × 10−5 |
| F8 | - | 83.3% | - | 6.9% | 1.9 × 10−101 |
| F9 | - | 2.4% | - | 3.5% | 0.31 |
| F10 | - | 2.4% | - | 2.7% | 0.84 |
| F11 | - | 1.9% | - | 2.8% | 0.97 |
| Mating Pattern | The Offspring Tested | (+/−) | (+/+) | (−/−) | p Value |
|---|---|---|---|---|---|
| F2 (+/−) × WT | 42 | 16 | 26 | - | 0.12 |
| F2 (+/−) × F2 (+/−) | 43 | 28 | 15 | 0 | 0.83 |
| F3 (+/−) × F3( +/−) | 43 | 25 | 18 | 0 | 0.24 |
| WT × F8 (+/−) | 33 | 14 | 19 | - | 0.38 |
| F9 (+/−) × F9 (+/−) | 46 | 26 | 20 | 0 | 0.14 |
| F11 (+/−) × F11 (+/−) | 48 | 33 | 15 | 0 | 0.76 |
| Total | 255 | 142 | 113 | - | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, X.; Duan, S.; Yang, Q.; Ma, X.; Li, Z.; Yao, Q.; Wu, K.; Chang, P.; Feng, G.; Hong, W.; et al. Protein Arginine Methyltransferase 5 Is Necessary for Embryonic Development in Medaka Oryzias latipes. Fishes 2023, 8, 19. https://doi.org/10.3390/fishes8010019
Liang X, Duan S, Yang Q, Ma X, Li Z, Yao Q, Wu K, Chang P, Feng G, Hong W, et al. Protein Arginine Methyltransferase 5 Is Necessary for Embryonic Development in Medaka Oryzias latipes. Fishes. 2023; 8(1):19. https://doi.org/10.3390/fishes8010019
Chicago/Turabian StyleLiang, Xiaoting, Shi Duan, Qing Yang, Xiaoqin Ma, Zhenyu Li, Qiting Yao, Kongyue Wu, Pei Chang, Gongqing Feng, Wentao Hong, and et al. 2023. "Protein Arginine Methyltransferase 5 Is Necessary for Embryonic Development in Medaka Oryzias latipes" Fishes 8, no. 1: 19. https://doi.org/10.3390/fishes8010019
APA StyleLiang, X., Duan, S., Yang, Q., Ma, X., Li, Z., Yao, Q., Wu, K., Chang, P., Feng, G., Hong, W., Cao, M., Zhou, Q., Zhong, X., & Zhao, H. (2023). Protein Arginine Methyltransferase 5 Is Necessary for Embryonic Development in Medaka Oryzias latipes. Fishes, 8(1), 19. https://doi.org/10.3390/fishes8010019