Disentangling the Drivers of the Sampling Bias of Freshwater Fish across Europe
Abstract
1. Introduction
2. Methods
2.1. Study Area and Spatial Records
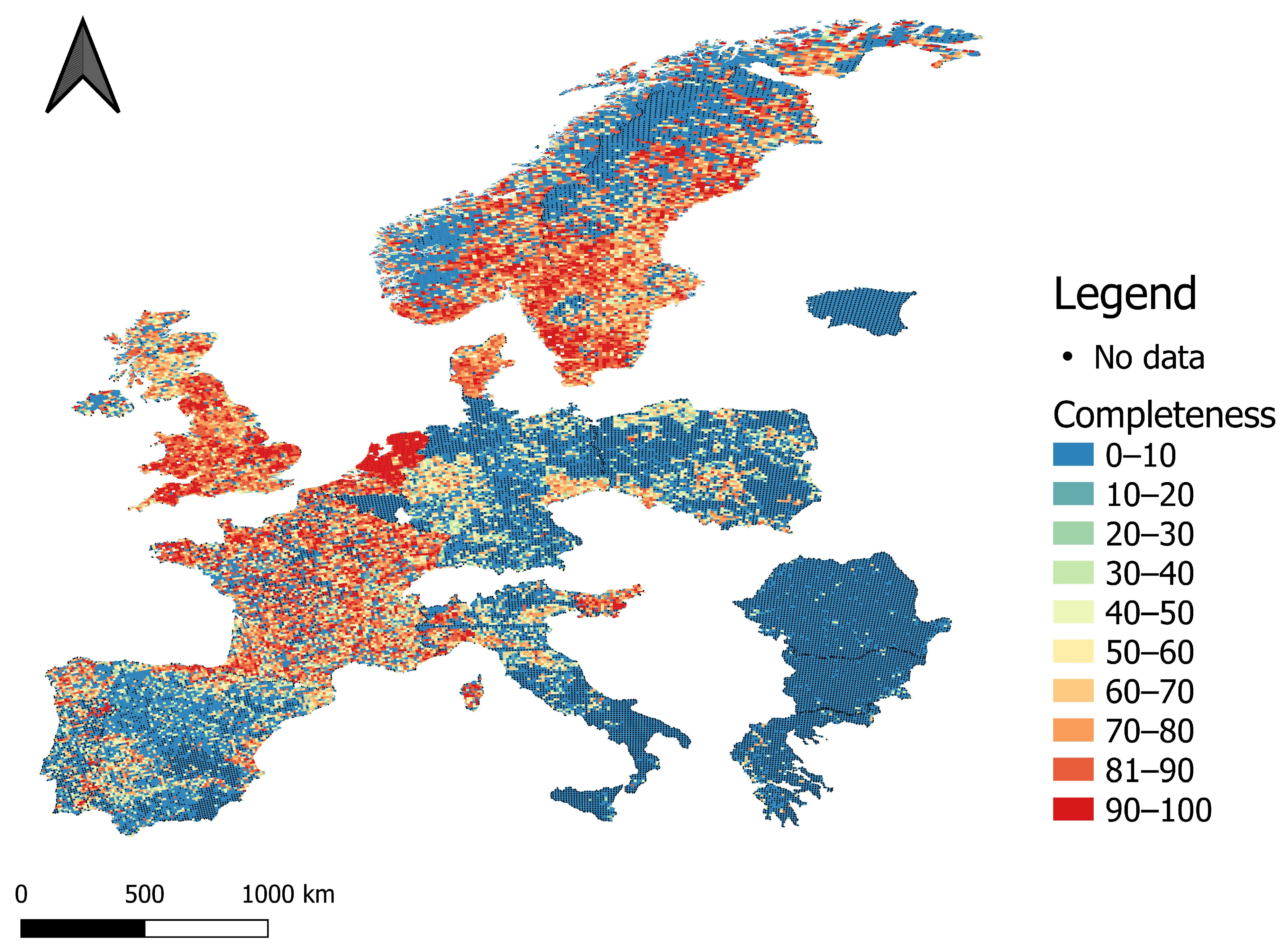
2.2. Completeness Calculation
2.3. Predictors and Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Maldonado, C.; Molina, C.I.; Zizka, A.; Persson, C.; Taylor, C.M.; Albán, J.; Chilquillo, E.; Rønsted, N.; Antonelli, A. Estimating species diversity and distribution in the era of Big Data: To what extent can we trust public databases? Glob. Ecol. Biogeogr. 2015, 24, 973–984. [Google Scholar] [CrossRef] [PubMed]
- NBNAtlas. National Biodiversity Atlas (NBN). Available online: https://www.nbnatlas.org (accessed on 15 June 2021).
- Chandler, M.; See, L.; Copas, K.; Bonde, A.M.; López, B.C.; Danielsen, F.; Legind, J.K.; Masinde, S.; Miller-Rushing, A.J.; Newman, G. Contribution of citizen science towards international biodiversity monitoring. Biol. Conserv. 2017, 213, 280–294. [Google Scholar] [CrossRef]
- García-Navas, V.; Rodríguez-Rey, M. The evolution of climatic niches and its role in shaping diversity patterns in diprotodontid marsupials. J. Mamm. Evol. 2019, 26, 479–492. [Google Scholar] [CrossRef]
- Di Marco, M.; Ferrier, S.; Harwood, T.D.; Hoskins, A.J.; Watson, J.E.M. Wilderness areas halve the extinction risk of terrestrial biodiversity. Nature 2019, 573, 582–585. [Google Scholar] [CrossRef]
- Pacifici, M.; Rondinini, C.; Rhodes, J.R.; Burbidge, A.A.; Cristiano, A.; Watson, J.E.; Woinarski, J.C.; Di Marco, M. Global correlates of range contractions and expansions in terrestrial mammals. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Thomas, C.D.; Cameron, A.; Green, R.E.; Bakkenes, M.; Beaumont, L.J.; Collingham, Y.C.; Erasmus, B.F.N.; de Siqueira, M.F.; Grainger, A.; Hannah, L.; et al. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [CrossRef]
- Hortal, J.; Bello, F.d.; Diniz-Filho, J.A.F.; Lewinsohn, T.M.; Lobo, J.M.; Ladle, R.J. Seven Shortfalls that Beset Large-Scale Knowledge of Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 523–549. [Google Scholar] [CrossRef]
- Lomolino, M.V. Conservation biogeography. In Frontiers of Biogeography: New Directions in the Geography of Nature; Lomolino, M.V., Heaney, L.R., Eds.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2004; Available online: https://www.researchgate.net/profile/Mark-Lomolino/publication/285850561_Conservation_biogeography/links/584c7c3c08aeb989251f778d/Conservation-biogeography.pdf (accessed on 15 June 2021).
- Proença, V.; Martin, L.J.; Pereira, H.M.; Fernandez, M.; McRae, L.; Belnap, J.; Böhm, M.; Brummitt, N.; García-Moreno, J.; Gregory, R.D. Global biodiversity monitoring: From data sources to essential biodiversity variables. Biol. Conserv. 2017, 213, 256–263. [Google Scholar] [CrossRef]
- Bowler, D.E.; Callaghan, C.T.; Bhandari, N.; Henle, K.; Benjamin Barth, M.; Koppitz, C.; Klenke, R.; Winter, M.; Jansen, F.; Bruelheide, H.; et al. Temporal trends in the spatial bias of species occurrence records. Ecography 2022, 2022, e06219. [Google Scholar] [CrossRef]
- Heberling, J.M.; Miller, J.T.; Noesgaard, D.; Weingart, S.B.; Schigel, D. Data integration enables global biodiversity synthesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018093118. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: What it does, and why inputs and settings matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Meyer, C.; Kreft, H.; Guralnick, R.; Jetz, W. Global priorities for an effective information basis of biodiversity distributions. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Troudet, J.; Grandcolas, P.; Blin, A.; Vignes-Lebbe, R.; Legendre, F. Taxonomic bias in biodiversity data and societal preferences. Sci. Rep. 2017, 7, 1–14. [Google Scholar]
- Yang, W.; Ma, K.; Kreft, H. Geographical sampling bias in a large distributional database and its effects on species richness–environment models. J. Biogeogr. 2013, 40, 1415–1426. [Google Scholar] [CrossRef]
- Leitão, P.J.; Moreira, F.; Osborne, P.E. Effects of geographical data sampling bias on habitat models of species distributions: A case study with steppe birds in southern Portugal. Int. J. Geogr. Inf. Sci. 2011, 25, 439–454. [Google Scholar] [CrossRef]
- Beever, E.A.; Ray, C.; Wilkening, J.L.; Brussard, P.F.; Mote, P.W. Contemporary climate change alters the pace and drivers of extinction. Glob. Change Biol. 2011, 17, 2054–2070. [Google Scholar] [CrossRef]
- Gallien, L.; Douzet, R.; Pratte, S.; Zimmermann, N.E.; Thuiller, W. Invasive species distribution models—How violating the equilibrium assumption can create new insights. Glob. Ecol. Biogeogr. 2012, 21, 1126–1136. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Rome, Q.; Villemant, C.; Courchamp, F. Can species distribution models really predict the expansion of invasive species? PLoS ONE 2018, 13, e0193085. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species Distribution Models: Ecological Explanation and Prediction across Space and Time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Araujo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Kadmon, R.; Farber, O.; Danin, A. Effect of roadside bias on the accuracy of predictive maps produced by bioclimatic models. Ecol. Appl. 2004, 14, 401–413. [Google Scholar] [CrossRef]
- Hortal, J.; Lobo, J.M.; Jiménez-Valverde, A. Limitations of biodiversity databases: Case study on seed-plant diversity in Tenerife, Canary Islands. Conserv. Biol. 2007, 21, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Ranc, N.; Santini, L.; Rondinini, C.; Boitani, L.; Poitevin, F.; Angerbjörn, A.; Maiorano, L. Performance tradeoffs in target-group bias correction for species distribution models. Ecography 2017, 40, 1076–1087. [Google Scholar] [CrossRef]
- Maasri, A.; Jähnig, S.C.; Adamescu, M.C.; Adrian, R.; Baigun, C.; Baird, D.J.; Batista-Morales, A.; Bonada, N.; Brown, L.E.; Cai, Q.; et al. A global agenda for advancing freshwater biodiversity research. Ecol. Lett. 2022, 25, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Romo, H.; García-Barros, E.; Lobo, J.M. Identifying recorder-induced geographic bias in an Iberian butterfly database. Ecography 2006, 29, 873–885. [Google Scholar] [CrossRef]
- Barbosa, A.M.; Pautasso, M.; Figueiredo, D. Species–people correlations and the need to account for survey effort in biodiversity analyses. Divers. Distrib. 2013, 19, 1188–1197. [Google Scholar] [CrossRef]
- Martinez, J.; Wool, D. Sampling bias in roadsides: The case of galling aphids on Pistacia trees. Biodivers. Conserv. 2006, 15, 2109–2121. [Google Scholar] [CrossRef]
- Barends, J.M.; Pietersen, D.; Zambatis, G.; Tye, D.R.; Maritz, B. Sampling bias in reptile occurrence data for the Kruger National Park. Koedoe: Afr. Prot. Area Conserv. Sci. 2020, 62, 1–9. [Google Scholar] [CrossRef]
- Moua, Y.; Roux, E.; Seyler, F.; Briolant, S. Correcting the effect of sampling bias in species distribution modeling—A new method in the case of a low number of presence data. Ecol. Inform. 2020, 57, 101086. [Google Scholar] [CrossRef]
- Inman, R.; Franklin, J.; Esque, T.; Nussear, K. Comparing sample bias correction methods for species distribution modeling using virtual species. Ecosphere 2021, 12, e03422. [Google Scholar] [CrossRef]
- Boyd, R.J.; Powney, G.; Carvell, C.; Pescott, O. occAssess: An R package for assessing potential biases in species occurrence data. Ecol. Evol. 2021, 11, 16177–16187. [Google Scholar] [CrossRef] [PubMed]
- Zizka, A.; Antonelli, A.; Silvestro, D. sampbias, a method for quantifying geographic sampling biases in species distribution data. Ecography 2021, 44, 25–32. [Google Scholar] [CrossRef]
- La Sorte, F.A.; Somveille, M. Survey completeness of a global citizen-science database of bird occurrence. Ecography 2020, 43, 34–43. [Google Scholar] [CrossRef]
- Freitas, T.M.d.S.; Stropp, J.; Calegari, B.B.; Calatayud, J.; De Marco, P., Jr.; Montag, L.F.d.A.; Hortal, J. Quantifying shortfalls in the knowledge on Neotropical Auchenipteridae fishes. Fish Fish. 2021, 22, 87–104. [Google Scholar] [CrossRef]
- Troia, M.J.; McManamay, R.A. Filling in the GAPS: Evaluating completeness and coverage of open-access biodiversity databases in the United States. Ecol. Evol. 2016, 6, 4654–4669. [Google Scholar] [CrossRef]
- Reid, A.J.; Carlson, A.K.; Creed, I.F.; Eliason, E.J.; Gell, P.A.; Johnson, P.T.; Kidd, K.A.; MacCormack, T.J.; Olden, J.D.; Ormerod, S.J. Emerging threats and persistent conservation challenges for freshwater biodiversity. Biol. Rev. 2019, 94, 849–873. [Google Scholar] [CrossRef]
- WWF. Living Planet Report 2020-Bending the Curve of Biodiversity Loss: A Deep Dive into Freshwater; Almond, R.E., Grooten, M., Peterson, T., Eds.; WWF: Gland, Switzerland, 2020. [Google Scholar]
- Limburg, K.E.; Hughes, R.M.; Jackson, D.C.; Czech, B. Human Population Increase, Economic Growth, and Fish Conservation: Collision Course or Savvy Stewardship? Fisheries 2011, 36, 27–35. [Google Scholar] [CrossRef]
- Shelton, J.M.; Weyl, O.L.F.; Chakona, A.; Ellender, B.R.; Esler, K.J.; Impson, N.D.; Jordaan, M.S.; Marr, S.M.; Ngobela, T.; Paxton, B.R.; et al. Vulnerability of Cape Fold Ecoregion freshwater fishes to climate change and other human impacts. Aquat. Conserv. Mar. Freshw. Ecosyst. 2018, 28, 68–77. [Google Scholar] [CrossRef]
- Sánchez-Fernández, D.; Lobo, J.M.; Abellán, P.; Ribera, I.; Millán, A. Bias in freshwater biodiversity sampling: The case of Iberian water beetles. Divers. Distrib. 2008, 14, 754–762. [Google Scholar] [CrossRef]
- Pelayo-Villamil, P.; Guisande, C.; Manjarrés-Hernández, A.; Jiménez, L.F.; Granado-Lorencio, C.; García-Roselló, E.; González-Dacosta, J.; Heine, J.; González-Vilas, L.; Lobo, J.M. Completeness of national freshwater fish species inventories around the world. Biodivers. Conserv. 2018, 27, 3807–3817. [Google Scholar] [CrossRef]
- Ballesteros-Mejia, L.; Kitching, I.J.; Jetz, W.; Nagel, P.; Beck, J. Mapping the biodiversity of tropical insects: Species richness and inventory completeness of African sphingid moths. Glob. Ecol. Biogeogr. 2013, 22, 586–595. [Google Scholar] [CrossRef]
- Narváez-Gómez, J.P.; Guedes, T.B.; Lohmann, L.G. Recovering the drivers of sampling bias in Bignonieae (Bignoniaceae) and identifying priority areas for new survey efforts. Biodivers. Conserv. 2021, 30, 2319–2339. [Google Scholar] [CrossRef]
- Sánchez-Fernández, D.; Yela, J.L.; Acosta, R.; Bonada, N.; García-Barros, E.; Guisande, C.; Heine, J.; Millán, A.; Munguira, M.L.; Romo, H.; et al. Are patterns of sampling effort and completeness of inventories congruent? A test using databases for five insect taxa in the Iberian Peninsula. Insect Conserv. Divers. 2022, 15, 406–415. [Google Scholar] [CrossRef]
- de Almeida, T.C.; Tessarolo, G.; Nabout, J.C.; Teresa, F.B. Non-stationary drivers on fish sampling efforts in Brazilian freshwaters. Divers. Distrib. 2021, 27, 1224–1234. [Google Scholar] [CrossRef]
- Water Framework Directive. Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 establishing a framework for Community action in the field of water policy. Off. J. Eur. Communities 2000, 22, 2000. [Google Scholar]
- GBIF. GBIF Home Page. Available online: https://www.gbif.org (accessed on 15 June 2021).
- SMNH. Ichthyology Database; Swedish Museum of Natural History: Stockholm, Sweden, 2021. [Google Scholar]
- Keith, P.; Persat, H.; Feunteun, É.; Allardi, J. Les Poissons d’eau douce de France; Biotope: Paris, France, 2011. [Google Scholar]
- SIBIC. Carta Piscícola Española. Publicación electrónica. Available online: https://www.cartapiscicola.es/ (accessed on 26 May 2021).
- Marčeta, B.; Pliberšek, J. (Eds.) BiosWeb; Fisheries Research Institute of Slovenia: Ljubljana, Slovenia.
- Froese, R.; Pauly, D. FishBase. Available online: https://www.www.fishbase.org (accessed on 15 June 2021).
- Menegotto, A.; Rangel, T.F. Mapping knowledge gaps in marine diversity reveals a latitudinal gradient of missing species richness. Nat. Commun. 2018, 9, 4713. [Google Scholar] [CrossRef]
- Papeş, M.; Havel, J.E.; Vander Zanden, M.J. Using maximum entropy to predict the potential distribution of an invasive freshwater snail. Freshw. Biol. 2016, 61, 457–471. [Google Scholar] [CrossRef]
- Jenkins, C.N.; Pimm, S.L.; Joppa, L.N. Global patterns of terrestrial vertebrate diversity and conservation. Proc. Natl. Acad. Sci. USA 2013, 110, E2602–E2610. [Google Scholar] [CrossRef]
- Lobo, J.M.; Hortal, J.; Yela, J.L.; Millán, A.; Sánchez-Fernández, D.; García-Roselló, E.; González-Dacosta, J.; Heine, J.; González-Vilas, L.; Guisande, C. KnowBR: An application to map the geographical variation of survey effort and identify well-surveyed areas from biodiversity databases. Ecol. Indic. 2018, 91, 241–248. [Google Scholar] [CrossRef]
- Ugland, K.I.; Gray, J.S.; Ellingsen, K.E. The species–accumulation curve and estimation of species richness. J. Anim. Ecol. 2003, 72, 888–897. [Google Scholar] [CrossRef]
- Sánchez-Fernández, D.; Fox, R.; Dennis, R.L.H.; Lobo, J.M. How complete are insect inventories? An assessment of the british butterfly database highlighting the influence of dynamic distribution shifts on sampling completeness. Biodivers. Conserv. 2021, 30, 889–902. [Google Scholar] [CrossRef]
- Stropp, J.; Umbelino, B.; Correia, R.; Campos-Silva, J.; Ladle, R.; Malhado, A. The ghosts of forests past and future: Deforestation and botanical sampling in the Brazilian Amazon. Ecography 2020, 43, 979–989. [Google Scholar] [CrossRef]
- Sporbert, M.; Bruelheide, H.; Seidler, G.; Keil, P.; Jandt, U.; Austrheim, G.; Biurrun, I.; Campos, J.A.; Čarni, A.; Chytrý, M. Assessing sampling coverage of species distribution in biodiversity databases. J. Veg. Sci. 2019, 30, 620–632. [Google Scholar] [CrossRef]
- Kummu, M.; Taka, M.; Guillaume, J.H. Gridded global datasets for gross domestic product and Human Development Index over 1990–2015. Sci. Data 2018, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Venter, O.; Sanderson, E.W.; Magrach, A.; Allan, J.R.; Beher, J.; Jones, K.R.; Possingham, H.P.; Laurance, W.F.; Wood, P.; Fekete, B.M. Global terrestrial Human Footprint maps for 1993 and 2009. Sci. Data 2016, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tessarolo, G.; Rangel, T.F.; Araújo, M.B.; Hortal, J. Uncertainty associated with survey design in Species Distribution Models. Divers. Distrib. 2014, 20, 1258–1269. [Google Scholar] [CrossRef]
- Shirey, V.; Belitz, M.W.; Barve, V.; Guralnick, R. A complete inventory of North American butterfly occurrence data: Narrowing data gaps, but increasing bias. Ecography 2021, 44, 537–547. [Google Scholar] [CrossRef]
- Linke, S.; Lehner, B.; Ouellet Dallaire, C.; Ariwi, J.; Grill, G.; Anand, M.; Beames, P.; Burchard-Levine, V.; Maxwell, S.; Moidu, H.; et al. Global hydro-environmental sub-basin and river reach characteristics at high spatial resolution. Sci. Data 2019, 6, 283. [Google Scholar] [CrossRef]
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 170122. [Google Scholar] [CrossRef]
- QGIS. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2020. Available online: http://qgis.osgeo.org (accessed on 12 March 2021).
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- R CoreTeam. Package “Stats.”. R Stats Package 2018. Available online: https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html (accessed on 14 September 2022).
- Barton, K. Package ‘MuMIn’. cran. r-project. org/web/packages. 2015. Available online: MuMIn/MuMIn.pdf. (accessed on 8 November 2021).
- Carrizo, S.F.; Lengyel, S.; Kapusi, F.; Szabolcs, M.; Kasperidus, H.D.; Scholz, M.; Markovic, D.; Freyhof, J.; Cid, N.; Cardoso, A.C.; et al. Critical catchments for freshwater biodiversity conservation in Europe: Identification, prioritisation and gap analysis. J. Appl. Ecol. 2017, 54, 1209–1218. [Google Scholar] [CrossRef]
- Manjarrés-Hernández, A.; Guisande, C.; García-Roselló, E.; Heine, J.; Pelayo-Villamil, P.; Pérez-Costas, E.; González-Vilas, L.; González-Dacosta, J.; Duque, S.R.; Granado-Lorencio, C. Predicting the effects of climate change on future freshwater fish diversity at global scale. Nat. Conserv. 2021, 43, 1–24. [Google Scholar] [CrossRef]
- Soberón, M.J.; Llorente, B.J. The use of species accumulation functions for the prediction of species richness. Conserv. Biol. 1993, 7, 480–488. [Google Scholar] [CrossRef]
- Jarić, I.; Roll, U.; Arlinghaus, R.; Belmaker, J.; Chen, Y.; China, V.; Douda, K.; Essl, F.; Jähnig, S.C.; Jeschke, J.M.; et al. Expanding conservation culturomics and iEcology from terrestrial to aquatic realms. PLOS Biol. 2020, 18, e3000935. [Google Scholar] [CrossRef] [PubMed]
- Mair, L.; Ruete, A. Explaining Spatial Variation in the Recording Effort of Citizen Science Data across Multiple Taxa. PLoS ONE 2016, 11, e0147796. [Google Scholar] [CrossRef]
- Crandall, C.A.; Monroe, M.; Dutka-Gianelli, J.; Fitzgerald, B.; Lorenzen, K. How to Bait the Hook: Identifying What Motivates Anglers to Participate in a Volunteer Angler Data Program. Fisheries 2018, 43, 517–526. [Google Scholar] [CrossRef]
- Carraro, L.; Mächler, E.; Wüthrich, R.; Altermatt, F. Environmental DNA allows upscaling spatial patterns of biodiversity in freshwater ecosystems. Nat. Commun. 2020, 11, 3585. [Google Scholar] [CrossRef]
- Muha, T.P.; Rodríguez-Rey, M.; Rolla, M.; Tricarico, E. Using environmental DNA to improve species distribution models for freshwater invaders. Front. Ecol. Evol. 2017, 5, 158. [Google Scholar] [CrossRef]
- Lessa, T.; Dos Santos, J.W.; Correia, R.A.; Ladle, R.J.; Malhado, A.C.M. Known unknowns: Filling the gaps in scientific knowledge production in the Caatinga. PLoS ONE 2019, 14, e0219359. [Google Scholar] [CrossRef]
- Escribano, N.; Galicia, D.; Ariño, A.H. Completeness of Digital Accessible Knowledge (DAK) about terrestrial mammals in the Iberian Peninsula. PLoS ONE 2019, 14, e0213542. [Google Scholar] [CrossRef]
- Titley, M.A.; Butchart, S.H.; Jones, V.R.; Whittingham, M.J.; Willis, S.G. Global inequities and political borders challenge nature conservation under climate change. Proc. Natl. Acad. Sci. USA 2021, 118, e2011204118. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Böller, M.; Erhardt, A.; Schwanghart, W. Spatial bias in the GBIF database and its effect on modeling species' geographic distributions. Ecol. Inform. 2014, 19, 10–15. [Google Scholar] [CrossRef]
- Girardello, M.; Chapman, A.; Dennis, R.; Kaila, L.; Borges, P.A.; Santangeli, A. Gaps in butterfly inventory data: A global analysis. Biol. Conserv. 2019, 236, 289–295. [Google Scholar] [CrossRef]
- Speed, J.D.M.; Bendiksby, M.; Finstad, A.G.; Hassel, K.; Kolstad, A.L.; Prestø, T. Contrasting spatial, temporal and environmental patterns in observation and specimen based species occurrence data. PLoS ONE 2018, 13, e0196417. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Logez, M.; Xu, J.; Tao, S.; Villéger, S.; Brosse, S. Human impacts on global freshwater fish biodiversity. Science 2021, 371, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Schipper, A.M.; Barbarossa, V. Global congruence of riverine fish species richness and human presence. Glob. Ecol. Biogeogr. 2022, 31, 1501–1512. [Google Scholar] [CrossRef]
- Riibak, K.; Bennett, J.A.; Kook, E.; Reier, Ü.; Tamme, R.; Bueno, C.G.; Pärtel, M. Drivers of plant community completeness differ at regional and landscape scales. Agric. Ecosyst. Environ. 2020, 301, 107004. [Google Scholar] [CrossRef]
- Tiago, P.; Ceia-Hasse, A.; Marques, T.A.; Capinha, C.; Pereira, H.M. Spatial distribution of citizen science casuistic observations for different taxonomic groups. Sci. Rep. 2017, 7, 12832. [Google Scholar] [CrossRef]
- Lobo, J.M.; Baselga, A.; Hortal, J.; Jiménez-Valverde, A.; Gómez, J.F. How does the knowledge about the spatial distribution of Iberian dung beetle species accumulate over time? Divers. Distrib. 2007, 13, 772–780. [Google Scholar] [CrossRef]
- Nuñez-Penichet, C.; Cobos, M.E.; Soberón, J.; Gueta, T.; Barve, N.; Barve, V.; Navarro-Sigüenza, A.G.; Peterson, A.T. Selection of sampling sites for biodiversity inventory: Effects of environmental and geographical considerations. Methods Ecol. Evol. 2022, 13, 1595–1607. [Google Scholar] [CrossRef]
- Bohlin, T.; Hamrin, S.; Heggberget, T.G.; Rasmussen, G.; Saltveit, S.J. Electrofishing—theory and practice with special emphasis on salmonids. Hydrobiologia 1989, 173, 9–43. [Google Scholar] [CrossRef]
- Mccabe, J.D.; Anich, N.M.; Brady, R.S.; Zuckerberg, B. Raising the bar for the next generation of biological atlases: Using existing data to inform the design and implementation of atlas monitoring. Ibis 2018, 160, 528–541. [Google Scholar] [CrossRef]
- Xu, H.; Cao, M.; Wu, Y.; Cai, L.; Cao, Y.; Ding, H.; Cui, P.; Wu, J.; Wang, Z.; Le, Z.; et al. Optimized monitoring sites for detection of biodiversity trends in China. Biodivers. Conserv. 2017, 26, 1959–1971. [Google Scholar] [CrossRef]
- Velásquez-Tibatá, J. WhereNext: Biological Survey Recommending System Based on General Dissimilarity Modeling. R Package. Available online: https://github.com/jivelasquezt/WhereNext-Pkg/. (accessed on 28 July 2021).
- D’Antraccoli, M.; Bacaro, G.; Tordoni, E.; Bedini, G.; Peruzzi, L. More species, less effort: Designing and comparing sampling strategies to draft optimised floristic inventories. Perspect. Plant Ecol. Evol. Syst. 2020, 45, 125547. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, Y.; Engler, J.O.; Rödder, D.; Secondi, J. Mapping Species Distributions with MAXENT Using a Geographically Biased Sample of Presence Data: A Performance Assessment of Methods for Correcting Sampling Bias. PLoS ONE 2014, 9, e97122. [Google Scholar] [CrossRef]
- Tessarolo, G.; Ladle, R.J.; Lobo, J.M.; Rangel, T.F.; Hortal, J. Using maps of biogeographical ignorance to reveal the uncertainty in distributional data hidden in species distribution models. Ecography 2021, 44, 1743–1755. [Google Scholar] [CrossRef]
- Oliveira, U.; Paglia, A.P.; Brescovit, A.D.; de Carvalho, C.J.B.; Silva, D.P.; Rezende, D.T.; Leite, F.S.F.; Batista, J.A.N.; Barbosa, J.P.P.P.; Stehmann, J.R.; et al. The strong influence of collection bias on biodiversity knowledge shortfalls of Brazilian terrestrial biodiversity. Divers. Distrib. 2016, 22, 1232–1244. [Google Scholar] [CrossRef]
- Lima, L.B.; De Marco Júnior, P.; Lima-Junior, D.P. Trends and gaps in studies of stream-dwelling fish in Brazil. Hydrobiologia 2021, 848, 3955–3968. [Google Scholar] [CrossRef]
- Pelayo-Villamil, P.; Guisande, C.; Vari, R.P.; Manjarrés-Hernández, A.; García-Roselló, E.; González-Dacosta, J.; Heine, J.; González Vilas, L.; Patti, B.; Quinci, E.M.; et al. Global diversity patterns of freshwater fishes—Potential victims of their own success. Divers. Distrib. 2015, 21, 345–356. [Google Scholar] [CrossRef]
| Variable | Description | Source |
|---|---|---|
| Airport | Euclidean distance to the closest airport | http://worldmap.harvard.edu |
| Cities | Euclidean distance to the closest major city | https://hub.arcgis.com/maps/esri::world-cities-1/ |
| Road | Road density | https://www.hydrosheds.org/page/hydroatlas |
| Population | Population density in 2010 | https://www.hydrosheds.org/page/hydroatlas |
| HDI | Human Development Index in 2015 [65] | https://www.hydrosheds.org/page/hydroatlas |
| HFT | Human Footprint [66] | https://www.hydrosheds.org/page/hydroatlas |
| Bio 1 | Annual Mean Temperature | https://chelsa-climate.org/bioclim/ |
| Bio 2 | Mean Diurnal Range (mean of monthly temp (max temp–min temp)) | https://chelsa-climate.org/bioclim/ |
| Bio 3 | Isothermality (BIO2/BIO7) (* 100) | https://chelsa-climate.org/bioclim/ |
| Bio 4 | Temperature Seasonality (standard deviation *100) | https://chelsa-climate.org/bioclim/ |
| Bio 5 | Max Temperature of Warmest Month | https://chelsa-climate.org/bioclim/ |
| Bio 6 | Min Temperature of Coldest Month | https://chelsa-climate.org/bioclim/ |
| Bio 7 | Temperature Annual Range (BIO5-BIO6) | https://chelsa-climate.org/bioclim/ |
| Bio 8 | Mean Temperature of Wettest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 9 | Mean Temperature of Driest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 10 | Mean Temperature of Warmest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 11 | Mean Temperature of Coldest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 12 | Annual Precipitation | https://chelsa-climate.org/bioclim/ |
| Bio 13 | Precipitation of Wettest Month | https://chelsa-climate.org/bioclim/ |
| Bio 14 | Precipitation of Driest Month | https://chelsa-climate.org/bioclim/ |
| Bio 15 | Precipitation Seasonality (Coefficient of Variation) | https://chelsa-climate.org/bioclim/ |
| Bio 16 | Precipitation of Wettest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 17 | Precipitation of Driest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 18 | Precipitation of Warmest Quarter | https://chelsa-climate.org/bioclim/ |
| Bio 19 | Precipitation of Coldest Quarter | https://chelsa-climate.org/bioclim/ |
| Belgium | Germany | Greece | Italy | Netherlands | Norway | Poland | Portugal | Slovenia | Spain | Sweden | United Kingdom | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Climate PC1 | −2.7456 | −0.1909 | - | - | - | −1.1168 | 0.2427 | - | −1.4714 | - | −0.2945 | −0.4970 |
| Climate PC2 | - | −0.1945 | −0.4574 | - | - | −0.8693 | - | - | −0.8778 | −0.6246 | −0.3006 | −0.2778 |
| Nature Reserves | - | - | 0.2608 | −0.4622 | −0.2873 | −0.1568 | 0.1158 | - | - | - | −0.3434 | 0.1261 |
| Distance to airports | - | −0.3652 | −0.1318 | - | - | - | - | 0.1438 | −0.3290 | - | ||
| Distance to big cities | - | −0.1938 | - | −0.8213 | 0.4747 | −0.1593 | −0.3428 | 0.2814 | - | −0.2493 | - | −0.1686 |
| Population density | - | 0.2222 | - | −0.4364 | - | - | - | - | - | - | 0.0896 | - |
| HDI | 0.7212 | - | - | 0.6697 | - | - | −0.1088 | −0.2542 | 0.6577 | 0.1345 | −0.2426 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Rey, M.; Grenouillet, G. Disentangling the Drivers of the Sampling Bias of Freshwater Fish across Europe. Fishes 2022, 7, 383. https://doi.org/10.3390/fishes7060383
Rodríguez-Rey M, Grenouillet G. Disentangling the Drivers of the Sampling Bias of Freshwater Fish across Europe. Fishes. 2022; 7(6):383. https://doi.org/10.3390/fishes7060383
Chicago/Turabian StyleRodríguez-Rey, Marta, and Gaël Grenouillet. 2022. "Disentangling the Drivers of the Sampling Bias of Freshwater Fish across Europe" Fishes 7, no. 6: 383. https://doi.org/10.3390/fishes7060383
APA StyleRodríguez-Rey, M., & Grenouillet, G. (2022). Disentangling the Drivers of the Sampling Bias of Freshwater Fish across Europe. Fishes, 7(6), 383. https://doi.org/10.3390/fishes7060383






