Abstract
Koi herpesvirus (KHV) is a highly contagious virus that causes high mortality in koi and common carp, leading to a reduction in production worldwide. Recent diagnostic tests based on molecular methods alone (nucleic acid amplification) and indirect immunoassay methods (antibody detection) can be confirmed over KHV infections or prior exposure and latent infections. Unfortunately, there is no established method to detect KHV virus particles, especially when virus titers are low. Therefore, we propose an alternative, direct immunoassay method for viral detection using a single-chain variable fragment (scFv), a specific region of IgG antibodies that binds specifically to KHV particles. The results of functional analyses indicated that four putative scFv candidates, C5, F8, F6, and E4, were specific to KHV, but only F6 and C5 had a high binding affinity. The binding characteristics were confirmed by indirect competitive and sandwich enzyme-linked immunosorbent assays, which indicated that F6 and C5 have a broad penetration area to the binding region and share a similar epitope with commercial KHV monoclonal antibodies. These characteristics were further confirmed by their interactions with purified KHV coat protein by indirect ELISA and Western blot analyses. In conclusion, the F6 and C5 scFvs have adequate binding affinity to KHV particles to permit their use in immunoassays.
1. Introduction
Fisheries production largely occurs via inland aquaculture, and approximately 92.5% of farmed food fish production worldwide was dominated by finfish farming in 20201. Cyprinus carpio (Common carp) is a freshwater fish that accounted for 8% of the global production of finfish in 2016, and it is considered to be the 3rd most produced fish globally, with its production expected to increase further by 2030 [1]. Cyprinus rubrofuscus (Koi carp) is an ornamental fish that is used in common carp breeding. It has been bred outside its country of origin, Japan, since the 1980s and is part of the fish trade worldwide [2]. In 1998, a highly contagious herpesvirus, cyprinid herpesvirus 3, was isolated in the USA [3]. The virus caused mass mortality in both common and koi carp fish, resulting in clinical signs of sunken eyes, gill necrosis, and skin rash [4,5]. Since then, koi herpesvirus (KHV, also known as Cyprinid herpesvirus 3, CyHV-3) disease outbreaks have been reported among common carp worldwide, particularly in Europe, South Africa [4], and some parts of Asia [6,7,8].
KHVD has been listed as a notifiable disease by the World Organization for Animal Health and as a harmful pathogen that may be present in ballast water ships by the International Maritime Organization [9]. KHV can spread rapidly via horizontal transmission from latently infected survivors [10] or even via other vector-borne carriers [11,12] that might persist in wild populations [13].
Since the initial discovery of KHV, many studies have been conducted to learn how to control the disease. Other studies on viral characterization and diagnostic methods have also been conducted. There are two established methods for detecting viruses: nucleic acid testing with various protocols and techniques [14,15,16] and serological-based testing [17,18]. According to Bergmann et al., (2010), real-time polymerase chain reaction (PCR) and the proposed semi-nested PCR have a higher sensitivity than other testing methods. However, this method is limited due to sampling times at high viral titers (>5 dpi), while mucus swabs produced the best results at early infection times (<5 dpi) [19,20]. A valid serological method was developed to detect serum anti-KHV antibodies in infected fish using an enzyme-linked immunosorbent assay (ELISA). This indirect method is effective for detecting viral infections and can provide confirmation when there are no clinical symptoms (latent infection).
An alternative diagnostic method using a recombinant antibody was proposed to detect the presence of virions. Due to their affinity for antigens, antibodies have good binding activity against viruses and can be used for prevention or treatment [21]. Furthermore, their binding to target antigens and the excellent expression of recombinant antibodies in an E. coli system suggests their use as a diagnostic tool via protein–protein interactions [22]. A single-chain variable fragment (scFv) is smaller than a full-sized monoclonal antibody (mAb) [23], but it can more efficiently span diverse phage display libraries and be subjected to multipurpose engineering [24]; thus, an scFv may attach to the virion and block viral replication. Based on this information, this study aimed to develop an immunoassay to detect the KHV virus using a recombinant scFv.
2. Methods
2.1. Subcloning of the pCTCON-KHV Coat Protein
A synthetic KHV capsid triplex subunit 2 gene sequence (modified from GenBank accession number: QAU54957.1 at N286Q) (Macrogen, Seoul, South Korea) was cloned using AflII and NotI (Takara Bio Inc., Tokyo, Japan) into a pCTCON vector that was modified with multiple cloning-site-annealed oligo fragments (Table 1) and transformed into Escherichia coli. The correct clone was confirmed by PCR (Table 1), restriction analysis using SacI (Takara, Tokyo, Japan), and sequencing analysis.

Table 1.
Primer sequence for pCTCON-KHV cloning and confirmation.
2.2. Yeast Transformation and Yeast Surface Display Expression
The Saccharomyces cerevisiae strain EBY100 at an OD600 of 1.3–1.5 was harvested, washed, and resuspended in a LiOAc/DTT buffer (0.1 M lithium acetate, 10 mM dithiothreitol) and then incubated at 28 °C for 30 min with shaking at 200 rpm. The competent cells were resuspended in 1 M sorbitol and electroporated with a pCTCON-KHV plasmid using Gene Xcell Pulser (Bio-Rad, Hercules, CA, USA), then recovered with a YPDS buffer (YPD media, 1 M sorbitol, 1:1) before plating on an SD-Trp plate (2.67% Minimal SD Base, 0.074% DO Supplement-Trp, 2% agar).
Yeast surface display was performed according to the protocol described by Angelini et al. [25] with modifications. A single colony was inoculated in SD-Trp medium at 28 °C until an OD600 of 4–5 was obtained, and the medium was then changed to an SG-CAA medium (pH 6.0; 2% w/v dextrose, 0.67% w/v yeast nitrogen base, 0.5% w/v casamino acids, and 100 mM phosphate buffer) and then harvested and resuspended in PBSCM (phosphate-buffered saline (PBS), pH 7.4, 1 mM CaCl2, 0.5 mM MgSO4). The relative amount of expressed protein was determined using an ELISA with anti-cMyc monoclonal (1:500, Santa Cruz Biotechnology, Dallas, TX, USA) and anti-mouse HRP-linked antibodies (1:1000; Cell Signaling Technology, Danvers, MA, USA).
2.3. Biopanning
An ScFv Tomlinson J phage library stock was revived and purified using a PEG/NaCl buffer (20% (w/v) PEG 6000, 2.5 M sodium chloride, 4:1 ratio (v/v)) and resuspended in a PBS containing 20% glycerol. An initial 109 CFU/mL scFv library in a blocking buffer was sequentially screened from blank, negative samples (red seabream iridovirus (RSIV) coat protein-displaying yeast) to positive samples (KHV coat protein-displaying yeast) with an initial OD600 of 10. The phage scFvs were eluted with 100 mM triethylamine and neutralized with 1 M Tris-HCl, pH 7.4. The number of yeast cells expressing antigen was reduced until the fourth round.
2.4. Phage ELISA
KHV-specific phage scFv clones were prepared from randomly selected individual XL1-blue colonies. A coated KHV yeast plate and an RSIV yeast plate (as control) at an OD600 of 2.5 were blocked using a blocking buffer (3% bovine serum albumin (BSA) in TBS-T). A mixture of 6% BSA in TBS-T and each phage clone supernatant (1:1) was added to each plate in equal amounts, then conjugated with anti-M13 HRP-linked antibodies (Sino Biological, Beijing, China). The absorbance ratio at 405 nm (Tecan, Mannedorf, Switzerland) was calculated, and the putative clones were selected.
2.5. Sequence Analysis and Mutagenesis of Selected KHV-Specific Phage scFvs
The scFv plasmid was extracted and amplified with the LMB3 forward primer 5′-CAGGAAACAGCTATGAC-3′ and the pHENseq primer 5’-CTATGCGGCCCCATTCA-3’ using 2x Taq Mastermix (Bioneer, Daejeon, South Korea). The amplified DNA was sequenced, and complementarity-determining regions (CDRs) were identified using IgBlast (https://www.ncbi.nlm.nih.gov/igblast, accessed on 12 November 2022) [26,27]. All scFv amplicons were subcloned into the pGEMT Easy® (Promega, Madison, WI, USA) vector, and mutagenesis was performed using a Q5® Site-Directed Mutagenesis kit (NEB, Ipswich, MA, USA) according to the manufacturer’s procedures. The correct mutants were confirmed via sequencing by Macrogen.
2.6. Putative scFv Protein Expression and Purification
The putative scFv mutant genes were subcloned using NcoI and NotI (Takara Bio) into a pIg20-3D8scFv plasmid (provided by Dr. Gunsup Lee, Novelgen, Suwon, South Korea) modified with MCS-6×His-Tag oligo primers (Table 2) and expressed in E. coli strain BL21 DE3 (pLysE). An expression test was conducted with 0.1 mM and 1 mM IPTG at 25 °C. The medium fraction was combined with the extracted periplasmic fraction with TES buffer + PMSF (50 mM Tris-HCl, pH 7.2, 0.6 mM EDTA, 20% w/v sucrose, 1 mM PMSF) and filtered through a 0.22 µm vacuum filter system (CORNING, Corning, NY, USA). The filtrate solution was purified with a protein L affinity column (CaptoL™) (GE LifeScience, Uppsala, Sweden), then concentrated with a Vivaspin 15 turbo 10 K MWCO (Sartorius, Goettingen, Germany) filter and resuspended in 1 mL PBS, pH 7.4. Total protein was determined using the Bradford assay (Bio-Rad), and Western blot analysis was performed using anti-6×His-tag monoclonal (1:1000; Abcam, Cambridge, MA, USA) and anti-rabbit HRP-linked antibodies (1:1000; Cell Signaling Technology).

Table 2.
Annealed oligo sequences for pIg20_MCS modification.
2.7. KHV Propagation, Purification, and Viral Titer
Approximately 1 mL of KHV stock (provided by Dr. Jung from the Institute of Animal Medicine, Gyeongsang National University, South Korea) was used to infect a monolayer of CCB cell line (common carp brain; purchased from the Friedrich Loeffler Institute, Germany) in fresh MEM complete media (4 mM L-glutamine, 10 mM HEPES, 10% FBS, nonessential amino acids) for 7 days at 26 °C in a 4% CO2 incubator. The virion was purified according to Bergmann et al. [18]. The active virions were determined using a plaque assay with 2 × 105 cells/mL and 10-fold dilutions (starting from 10−1) and then overlaid with semisolid media (1× MEM complete media with 1% agarose) for 7 days. Viral titers were calculated by serial dilution.
2.8. Analysis of the Binding Specificity of Putative scFv Proteins
Protein targets consisted of a blank (BSA), a positive sample (KHV virion), and a negative sample (viral hemorrhagic septicemia virus (VHSV)) virion at 10 ng/µL in coating buffer (0.1 M NaHCO3, pH 8.6) and were conjugated with scFv proteins at a concentration of 2 ng/µL. Anti-6×His-tag monoclonal (1:1000, Abcam) and anti-rabbit HRP-linked antibodies (1:1000; Cell Signaling Technology) were used to detect the scFv.
2.9. Sensitivity Analysis of Putative scFv Proteins
In one group, the KHV virion (10 ng/µL in coating buffer) was conjugated with scFv proteins at 1–10 ng/µL. In another group, a series of KHV virions (5–25 ng/µL) was conjugated with scFv proteins at 4 ng/µL. Anti-6×His-tag monoclonal (1:1000, Abcam) and anti-rabbit HRP-linked antibodies (1:1000; Cell Signaling Technology) were used to detect the scFv proteins.
2.10. Epitope Mapping of Putative scFv Proteins
Competitive ELISAs with KHV virions (10 ng/µL) were performed using various mixtures: anti-KHV mAb (1:100, Aquatic Diagnostics Ltd., Oban, Scotland, UK) and 2 ng/µL C5 scFv (first group); anti-KHV mAb (1:100) and 2 ng/µL F6 scFv (second group); and F6 and C5 scFv protein at 2 ng/µL each (third group), while the scFv protein was conjugated with anti-6×His-tag rabbit monoclonal (1:1000, Abcam). Competitive binding was evaluated using a mixture of different anti-Fc origin antibodies. Sandwich ELISAs of KHV virions (10 ng/µL) were performed using three different combination capture-detection groups containing C5 scFv (2 ng/µL), F6 scFv (2 ng/µL), and anti-KHV mAb (1:100). The detection antibody was conjugated with the proper anti-Fc origin antibodies. Epitope mapping analysis was performed as described by Zhang et al., (2019) [28]. The additivity index was calculated using the following equation:
where A1+2, A1, and A2 represent the OD450 values of the combined scFv and each scFv tested, respectively. The two scFvs recognize different antigenic epitopes only when the AI value is greater than 50%.
3. Results
3.1. Subcloning of KHV Coat Protein and Antigen Expression by Yeast Surface Display
KHV is a herpesvirus with four common morphological parts: a core composed of linear dsDNA, an icosahedral capsid, a tegument, and an envelope consisting of lipids with embedded viral coat proteins (spike glycoprotein) [29]. The capsid triplex subunit 2 (ORF72) is the choice antigen target in KHV as it is a highly conserved region that is present in all Alloherpesviridae [30] and thus is also a diagnostic target [31]. The KHV capsid triplex subunit 2 synthetic gene was modified at N286Q to prevent glycosylation by yeast cells. The gene was successfully cloned into the pCTCON plasmid and was confirmed by targeted PCR (Figure 1a), restriction analysis with SacI digestion (Figure 1b), and sequencing.
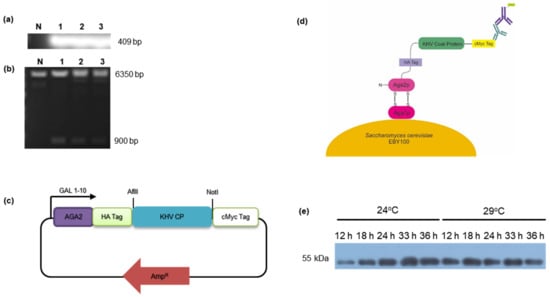
Figure 1.
(a) Targeted PCR detection of the KHV capsid subunit 2 gene with a size of approximately 400 bp. Lane N is the no-template control, and lanes 1–3 are the amplified KHV fragments of the coat protein gene. (b) Digested fragments of the pCTCON-KHV gene with sizes of 6350 bp and 900 bp (lanes 1–3) were digested using SacI. pCTCON-RSIV. (c) Schematic diagram of the yeast surface display system. KHV coat protein expressed with multiple tags (HA and c-Myc) and C-terminus glycoprotein (Aga2p) that covalently bind with Aga1p on the surface membrane. The c-Myc tag was used to detect antigen expression. (d) Schematic diagram of the pCTCON-KHV plasmid containing KHV coat protein (CP) (Genebank Acc. Number: QAU54957.1 with N286Q modification) used for an antigen-expressed yeast surface display. (e) Yeast surface display of KHV CP expression optimization assay at 24 °C and 29 °C and 12–36 h induction time intervals, detected using anti-cMyc (9E10) and anti-mouse-HRP antibodies.
Yeast surface display has emerged as an alternative strategy to evaluate protein antigen expression. This technique can maintain an antigen’s relative structural conformation to allow it to be recognized by an antibody, and it is relatively stable, even when it is not purified. In addition, antigen expression levels can be quantified by recognizing the C-terminal c-Myc tag that is encoded in the plasmid vector (Figure 1c,d) with a fluorescently labeled antibody.
The yeast surface display results are shown in the Western blot data (Figure 1e). Along with the RSIV coat protein surface display, protein expression reached saturation after 18 h of induction at 29 °C. This finding indicates that the yeast cells reached the stationary growth phase, and maximum expression was obtained. In the same manner, a comparable size band was also obtained at 24 °C at 33 h post-induction, suggesting that protein expression by the growing cells was delayed due to the lower temperature. In general, a lower temperature is preferred for protein expression because it improves protein folding and expression levels by reducing the cell growth rate and allowing proper folding [32,33]. In summary, these findings demonstrate that the optimal conditions for protein surface display expression were 33 h prior to induction in SG-CAA medium at 24 °C.
3.2. Screening of Anti-KHV-Specific scFv by Biopanning
The relative protein expression levels of antigen-displaying yeast cells were linearly correlated with the number of yeast cells (Supplementary Figure S1a,b). Therefore, the decreased number of yeast cells in every panning round exhibited reduced antigen levels. A lower concentration of the target antigen selectively enriched the high-affinity libraries in each round of panning. As the number of rounds increased, the scFv-expressing phage tended to have increased specificity and positive binding affinity, as shown by the fold enrichment at the end of panning (Table 3). After the fourth round of panning, the results of affinity evaluation obtained with the four clones C5, E4, F6, and F8 demonstrated ratios greater than 2.0 (Figure 2a); this value is considered to be a threshold value for determining the high specificity of KHV antigen affinity compared to the RSIV antigen.

Table 3.
Summary of biopanning round selection. Yield = Phage eluted/Phage added.
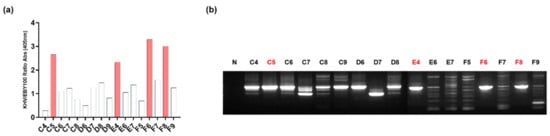
Figure 2.
(a) The binding affinity of monoclonal phage display scFv was determined after the 4th round of panning. Approximately 17 clones appeared and were selected using an ELISA. The C5, E4, F6, and F8 clones have higher affinity ratios than the other clones. Data are presented as a comparison of absorbance values of KHV antigen to RSIV antigen. (b) The scFv fragments of each clone were amplified by PCR using universal primer, pHEN_seq, and LMB3. The amplicons are separated using 1% gel electrophoresis containing ethidium bromide and are compared to the no-template control (lane N). The full-length scFv fragments of C5, E4, F6, and F8 at a size of 1000 bp were collected by gel purification for further analysis.
3.3. ScFv Sequence Analysis and CDR Determination
To obtain the scFv sequence, the plasmid of each clone was amplified by PCR and visualized using 1% agarose gel electrophoresis (Figure 2b). The C5, E4, F6, and F8 fragments with a size of approximately 1000 bp were purified using a DNA sequencing service. The putative scFv clones originated from a naïve human library (Tomlinson protocol) to allow the CDR region to be determined using the IgBlast online site. Surprisingly, the C5, F8, and E4 sequences had one missing nucleotide base in the VH region; thus, the frame was shifted, and a stop codon appeared. In addition, in the F6 sequence, the amber stop codon (TAG) appeared within the sequence. To correct the sequences, mutagenesis was performed either by substitution (F6 clone) or insertion (C5, F8, and E4 clones) (Supplementary Figure S2). All of the clones were similar to some human germlines. The heavy chain locus consisted of the variable regions of either IGHV3-23*01 or IGHV3-23*05. These indicate the identity value variable among the clones, which built FR1 until half of CDR3 was recombined. The other half was recombined with a small part of IGHD2-21*01 as the diversity region and IGHJ4*02 as the joining region, consecutively. In the light chain locus, all of the putative scFv clones had a canonical sequence to the IGKV1-39*01 germline, a kappa-type chain, although some different amino acids were present in the CDR2 and CDR3 regions and were joined by IGKJ1*01. All of the sequences had various germline recombination, with the most variable motifs located at both CDR2 and CDR3 loci (shown in bold, Table 4).

Table 4.
CDR determination from four scFv amino acid sequences of the variable heavy (VH) region and variable kappa (Vκ) region using IgBlast.
3.4. Protein Expression and Characterization of Anti-KHV scFv
For the expression of scFv, an expression vector was constructed by subcloning four scFv sequences (C5, E4, F6, and F8) into a modified pIg20 vector encoded with a 6×HisTag in the 3′ terminus (Figure 3a) and expressed in BL21 (DE3) pLysE. The expression of scFv was determined at various IPTG concentrations (0.1 and 1 mM) and confirmed by Western blot analysis (Figure 3b). After large-scale production, the scFv protein was purified by affinity chromatography with Protein L, which binds specifically to the kappa light chain region in scFv [34] and is confirmed by Western blot analysis and Coomassie staining (Figure 3c,d). Functional analyses were performed to characterize the binding interactions. To determine the specificity of scFv binding, each protein was quantitated by ELISA against the KHV virion (positive sample) and compared to the VHSV virion (negative sample) and BSA (control) (Figure 4a). According to individual p-value analyses between positive and negative samples compared to the control group, all scFvs bound specifically to KHV. On the other hand, the binding interaction of scFv protein was measured to evaluate the binding affinity to KHV. A series of antibodies were applied to fixed antigen and vice versa. F6 and C5 showed a higher binding affinity to KHV than the others at low antigen concentrations (Figure 4b) or at low antibody concentrations (Figure 4c). These results suggest that F6 and C5 have higher affinity against KHV, with a limit of detection of 5 ng/µL (versus 15 ng/µL for E4 and F8) and a minimum scFv concentration of 4 ng/µL.
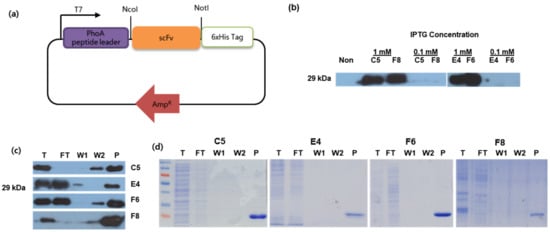
Figure 3.
(a) Schematic diagram of the pIg20-HisTag vector encoded with an scFv sequence by insertion between the NcoI and NotI sites. This recombinant plasmid was expressed as protein in BL21 pLysE strain bacteria. (b) Optimum expression conditions were obtained with 1 mM IPTG induction at 25 °C, which differed significantly from those at 0.1 mM IPTG. Large-scale expression was performed under optimum conditions and the product was purified using CaptoL™ column chromatography with normal gravity. Purification was confirmed by Western blot analysis (c) and by SDS-PAGE and Coomassie staining (d) with eluate samples from T = Total (filtered media + periplasmic fraction), FT = flow through, W1 = wash 1 elute (binding buffer), W2 = wash 2 elute (citrate/glycine buffer, pH 3.5), P = purified protein (elution buffer). There are no visible impurities in lane P, suggesting that the scFv recombinant protein was purified successfully.
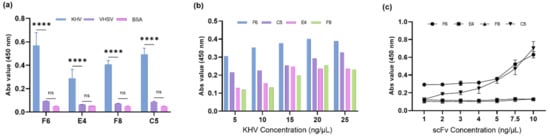
Figure 4.
(a) Quantitative analysis of F6, E4, F8, and C5 scFv proteins against KHV virions (positive sample), VHSV virions (negative sample), and BSA (control) using an ELISA and two-way ANOVA to measure binding specificity. Data are presented as the mean ± SEM (**** p < 0.0001; ns, nonsignificant). (b) Sensitivity tests with 400 ng of scFv against serial dilutions of KHV virions demonstrate that F6 and C5 have a higher affinity to virion antigens than the others at a concentration as low as 5 ng/µL. (c) Sensitivity test using a series of scFv concentrations (1–10 ng/µL) against 10 ng/µL KHV virion.
3.5. Epitope Mapping of Putative scFv Proteins
KHV virion was obtained in KHV infected CCB cell line, which showed cytopathic effect (CPE). CPE started to appear at 3 dpi, and complete cell lysis was observed at 7 dpi (Supplementary Figure S3). After propagation, KHV particles were titrated using plaque assay. The titer was 7 × 105 pfu/mL, approximately. To avoid background noise while performing the ELISA, the KHV virion was purified using a sucrose gradient, and the concentration obtained was 250 ng/µL. The amount of purified virion was confirmed by an ELISA and remained detectable at concentrations as low as 5 ng/µL (Supplementary Figure S4). F6 and C5 were selected for further analysis based on previous data and were freshly produced on a large scale (Supplementary Figure S5). This KHV virion was used as an antigen in the competitive ELISA scheme (Figure 5a), which demonstrated the competition between the binding of F6 and C5 to anti-KHV monoclonal antibody (KHV mAb) protein (at a concentration ratio of 1:1), as the absorbance value of the two differed significantly compared to their individual values (Figure 5b). Therefore, scFv and the anti-KHV mAb compete for binding at the same epitope region, whereas F6 and C5 have slightly different effects on the two individual values. The additivity index value shows that the F6 scFv has a ratio below 50%, but this value was higher compared to the other three scFvs. This result indicates that F6 binds to a similar epitope region as C5 but with a higher affinity. In addition, competitive ELISAs with different amounts of one protein in the mixture (the same in total) were performed to determine the additivity effect of one protein using a different method. In the F6/C5 combination group, the addition of one of the proteins tended to increase the total affinity value, whereas there was no significant difference in the other two groups (Figure 5c). This may be because the steric movement of scFv to access the binding site on the antigen surface in the scFv/KHV mAb group is hindered by the KHV mAb.
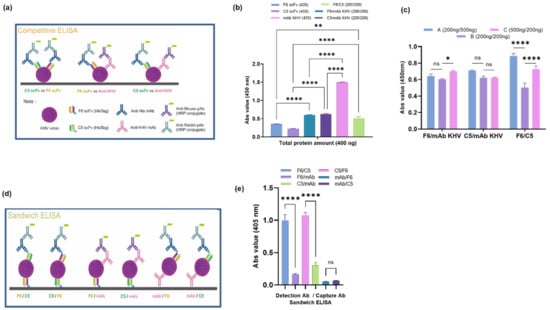
Figure 5.
(a) Diagram of the competitive ELISA. (b) The affinity of F6, C5, and KHV mAb was compared to the same amount of cocktail antibody (400 ng single protein, 200 ng/200 ng combination) to perform epitope mapping of the antibody by competitive ELISA. The correlation between groups was measured statistically (** p = 0.0051; **** p < 0.0001; ns, nonsignificant). The results are shown as an additivity index in Table 5. (c) Comparison of affinity values determined by competitive ELISAs with different amounts of one protein in the total mixture demonstrates different interaction effects (* p = 0.0298; **** p < 0.0001; ns, nonsignificant). (d) Affinity measurements for F6, C5, and KHV mAb were determined by sandwich ELISAs. (e) Quantitative analysis of sandwich ELISAs demonstrates that the F6 and C5 interactions are more favorable for detecting KHV virions than combinations with the commercial KHV mAb. Data are presented as the mean ± SEM.
Sandwich ELISAs are frequently used as a diagnostic tool for detecting macromolecule antigens. This method is used to capture and detect antibodies (Figure 5d), which bind to different epitopes. From three different proteins, six distinct groups were used as capture/detection antibody combinations. The F6/C5 scFv sandwich combination (and vice versa) tended to exhibit significantly higher sensitivity than the other groups (Figure 5e). Furthermore, when the KHV mAb was used as the capture antibody, the sensitivity was very low when scFv protein was added. However, when the KHV mAb was used as a detection antibody, the sensitivity improved slightly. This result demonstrates a different binding effect on the viral epitope; the penetrating effect per surface area of scFv is better than that of KHV mAb due to the protein’s size. Hence, F6 and C5 anti-KHV scFv exhibited good binding affinity to the KHV virion.
4. Discussion
Many diagnostic methods have been reported the detection of KHV. The method used in the current study will hopefully overcome the limitations of the indirect immunoassay method during the early infection period, where the serum antibody titer is likely to be low. Competitive ELISAs and sandwich ELISAs are potential models that may be used for diagnostic purposes, and each has its own advantages and disadvantages. Sandwich ELISAs have high sensitivity, specificity, and flexibility, but it may be difficult to optimize the antibodies used due to the cross-reactivity between capture and detection antibodies. Competitive ELISAs are more robust and consistent procedures than sandwich ELISAs but have limitations like those of other types of ELISAs [35].
The KHV antigen is structurally “close” to the RSIV antigen; both antigens represent the capsid sequences, and they may share an evolutionary similarity [36]. To ensure that the selected scFv is specific to the KHV antigen, we screened a library against the RSIV antigen displayed on the yeast surface immediately after prescreening the yeast S. cerevisiae EBY100 strain. This method enhances specificity and avoids cross-reactivity against relevant structures.
In addition, capsid triplex subunit 2 (VP23) expressed by yeast surface display may be considered a sole protein that has a partially folded conformational state that exists as a “molten globule” [37], a folding intermediate between native states (N states) and unfolding states (U states) [38]. This suggests that its complex form is influenced by the environment and interactions with other molecules. Two copies of VP23 with one copy of subunit 1 assemble to form a heterotrimer that stabilizes the pentamer to build a capsid [39]. The capsid is not exposed to the outside environment when it is already packaged as a free virion, yet it could be presented as an antigen by host cells to introduce it to the immune system.
The reduced phage yield value after the fourth round of panning indicated reduced library diversity, while the specificity was enhanced. The quality of naïve human origin libraries (Tomlinson I and J) was increased due to NNK and NNS-mutated codons, which exert a broad-spectrum ability, but also an excess of inactive clones and simultaneous saturation of the libraries [40].
The different concentrations of expressed proteins per liter after purification indicated the different intrinsic properties of each scFv candidate. This phenomenon may also be affected by protein expression parameters, such as temperature and IPTG concentration. Temperature is known to affect protein stability, which reflects the proper folding of expressed proteins. The melting temperatures (Tm) of peptide sequences and signal peptides are the main factors affecting protein characteristics; a higher melting temperature means that the protein is more stable [41]. The signal peptide plays a role in protein localization into the periplasmic membrane in various pathway mechanisms that depend on the type of signal peptide sequence; however, this might limit its production since protein saturation occurs in the periplasmic spaces. Purification using CaptoL™ selectively binds the kappa chain variable fragment with protein L, which can be altered by a change in pH [42]. When the pH is reduced to 2.2, dissociation occurs via a conformational change of both proteins, causing the scFv-protein L complexes to become unbound.
Functional analysis of putative scFv was performed using ELISAs and Western blots. All scFvs exhibited specific binding to the KHV virion, while the F6 and C5 scFvs had a greater affinity than the other two scFvs. The results of competitive and sandwich ELISAs demonstrated that the scFv protein is a small protein with broad penetration capability that shares binding epitope regions like a commercial KHV mAb. In conclusion, the F6 and C5 scFv candidates exhibited promising binding characteristics against the KHV virion.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7060370/s1, Figure S1: Quantification of yeast surface display expressing antigen level values; Figure S2: Nucleic acid sequence of putative anti-KHV scFv; Figure S3: KHV virion propagation and titer assay; Figure S4: Purified KHV virion quantification; Figure S5: F6 and C5 scFv protein characterization.
Author Contributions
A.D.M.L., H.S. and S.L. designed the experiment and concepts. A.D.M.L. and H.S. performed the experiments, analyzed the data, and drafted the manuscript. T.-J.C., T.-S.J., T.-K.L. and S.L. assisted with writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a grant (NRF-2017M3A9E4072753) from the Bio & Medical Technology Development Program of the National Research Foundation (NRF), which is funded by the Ministry of Science and ICT, and a grant (202104662) from the Korea Institute of Marine Science & Technology Promotion in the Ministry of Oceans and Fisheries (MOF) of the Republic of Korea.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- FAO. The State of World Fisheries and Aquaculture 2018; FAO: Rome, Italy, 2018. [Google Scholar]
- Pokorova, D.; Vesely, T.; Piackova, V.; Reschova, S.; Hulova, J. Current knowledge on koi herpesvirus (KHV): A review. Vet. Med. 2005, 50, 139–148. [Google Scholar] [CrossRef]
- Waltzek, T.; Kelley, G.O.; Stone, D.M.; Way, K.; Hanson, L.; Fukuda, H.; Hirono, I.; Aoki, T.; Davison, A.J.; Hedrick, R.P. Koi herpesvirus represents a third cyprinid herpesvirus (CyHV-3) in the family Herpesviridae. J. Gen. Virol. 2005, 86, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
- Haenen, O.; Way, K.; Bergmann, S.; Ariel, E. The emergence of koi herpesvirus and its significance to European aquaculture. Bull. Eur. Assoc. Fish Pathol. 2004, 24, 293–307. [Google Scholar]
- Bretzinger, A.; Fischer-Scherl, T.; Oumouna, M.; Hoffmann, R.; Truyen, U. Mass mortalities in Koi carp, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish Pathol. 1999, 19, 182–185. [Google Scholar]
- Sunarto, A.; McColl, K.A.; Crane, M.; Sumiati, T.; Hyatt, A.D.; Barnes, A.; Walker, P.J. Isolation and characterization of koi herpesvirus (KHV) from Indonesia: Identification of a new genetic lineage. J. Fish Dis. 2010, 34, 87–101. [Google Scholar] [CrossRef]
- Tu, C.; Weng, M.-C.; Shiau, J.-R.; Lin, S.-Y. Detection of Koi Herpesvirus in Koi Cyprinus carpio in Taiwan. Fish Pathol. 2004, 39, 109–110. [Google Scholar] [CrossRef]
- Gomez, D.K.; Joh, S.J.; Jang, H.; Shin, S.P.; Choresca, C.H.; Han, J.E.; Kim, J.H.; Jun, J.W.; Park, S.C. Detection of koi herpesvirus (KHV) from koi (Cyprinus carpio koi) broodstock in South Korea. Aquaculture 2011, 311, 42–47. [Google Scholar] [CrossRef]
- Cohen, A. Non-Native Bacterial and Viral Pathogens in Ballast Water: Potential for Impacts to ESA-Listed Species under NOAA’s Jurisdiction; Endangered Species Division, Silver Spring; MD Center for Research on Aquatic Bioinvasions (CRAB): Richmond, CA, USA, 2010. [Google Scholar]
- Uchii, K.; Matsui, K.; Iida, T.; Kawabata, Z. Distribution of the introduced cyprinid herpesvirus 3 in a wild population of common carp, Cyprinus carpio L. J. Fish Dis. 2009, 32, 857–864. [Google Scholar] [CrossRef]
- Kielpinski, M.; Kempter, J.; Panicz, R.; Sadowski, J.; Schütze, H.; Ohlemeyer, S.; Bergmann, S.M. Detection of KHV in Freshwater Mussels and Crustaceans from Ponds with KHV History in Common Carp (Cyprinus carpio). Isr. J. Aquac.-Bamidgeh 2010, 62, 20576. [Google Scholar] [CrossRef]
- Boutier, M.; Ronsmans, M.; Rakus, K.; Jazowiecka-Rakus, J.; Vancsok, C.; Morvan, L.; Peñaranda, M.M.D.; Stone, D.M.; Way, K.; van Beurden, S.J.; et al. Cyprinid herpesvirus 3: An archetype of fish alloherpesviruses. Adv. Virus Res. 2015, 93, 161–256. [Google Scholar]
- Uchii, K.; Minamoto, T.; Honjo, M.N.; Kawabata, Z. Seasonal reactivation enables Cyprinid herpesvirus 3 to persist in a wild host population. FEMS Microbiol. Ecol. 2014, 87, 536–542. [Google Scholar] [CrossRef]
- Gilad, O.; Yun, S.; Zagmutt-Vergara, F.; Leutenegger, C.; Bercovier, H.; Hedrick, R. Concentrations of a Koi herpesvirus (KHV) in tissues of experimentally-infected Cyprinus carpio koi as assessed by real-time TaqMan PCR. Dis. Aquat. Org. 2004, 60, 179–187. [Google Scholar] [CrossRef]
- Bercovier, H.; Fishman, Y.; Nahary, R.; Sinai, S.; Zlotkin, A.; Eyngor, M.; Gilad, O.; Eldar, A.; Hedrick, R.P. Cloning of the koi herpesvirus (KHV) gene encoding thymidine kinase and its use for a highly sensitive PCR based diagnosis. BMC Microbiol. 2005, 5, 13. [Google Scholar] [CrossRef]
- Yoshino, M.; Watari, H.; Kojima, T.; Ikedo, M.; Kurita, J. Rapid, sensitive and simple detection method for koi herpesvirus using loop-mediated isothermal amplification. Microbiol. Immunol. 2009, 53, 375–383. [Google Scholar] [CrossRef]
- Adkison, M.A.; Gilad, O.; Hedrick, R.P. An Enzyme Linked Immunosorbent Assay (ELISA) for Detection of Antibodies to the Koi Herpesvirus (KHV) in the Serum of Koi Cyprinus carpio. Fish Pathol. 2005, 40, 53–62. [Google Scholar] [CrossRef]
- Bergmann, S.M.; Wang, Q.; Zeng, W.; Li, Y.; Wang, Y.; Matras, M.; Reichert, M.; Fichtner, D.; Lenk, M.; Morin, T.; et al. Validation of a KHV antibody enzyme-linked immunosorbent assay (ELISA). J. Fish Dis. 2017, 40, 1511–1527. [Google Scholar] [CrossRef]
- Bergmann, S.M.; Riechardt, M.; Fichtner, D.; Lee, P.; Kempter, J. Investigation on the diagnostic sensitivity of molecular tools used for detection of koi herpesvirus. J. Virol. Methods 2010, 163, 229–233. [Google Scholar] [CrossRef]
- Monaghan, S.; Thompson, K.; Adams, A.; Bergmann, S.M. Sensitivity of seven PCR s for early detection of koi herpesvirus in experimentally infected carp, Cyprinus carpio L., by lethal and non-lethal sampling methods. J. Fish Dis. 2015, 38, 303–319. [Google Scholar] [CrossRef]
- Salazar, G.; Zhang, N.; Fu, T.-M.; An, Z. Antibody therapies for the prevention and treatment of viral infections. npj Vaccines 2017, 2, 19. [Google Scholar] [CrossRef]
- Trier, N.H.; Houen, G. Antibodies as Diagnostic Targets and as Reagents for Diagnostics; Multidisciplinary Digital Publishing Institute: Basel, Switzerland, 2020. [Google Scholar]
- Nelson, A.L.; Reichert, J.M. Development trends for therapeutic antibody fragments. Nat. Biotechnol. 2009, 27, 331–337. [Google Scholar] [CrossRef]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv Antibody: Principles and Clinical Application. J. Immunol. Res. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Angelini, A.; Chen, T.F.; de Picciotto, S.; Yang, N.J.; Tzeng, A.; Santos, M.S.; Van Deventer, J.A.; Traxlmayr, M.W.; Wittrup, K.D. Protein engineering and selection using yeast surface display. In Yeast Surface Display; Springer: Berlin/Heidelberg, Germany, 2015; pp. 3–36. [Google Scholar]
- Martin, A.C. Accessing the Kabat Antibody Sequence Database by Computer; Wiley Online Library: New York, NY, USA, 1996. [Google Scholar]
- Dondelinger, M.; Filée, P.; Sauvage, E.; Quinting, B.; Muyldermans, S.; Galleni, M.; Vandevenne, M.S. Understanding the Significance and Implications of Antibody Numbering and Antigen-Binding Surface/Residue Definition. Front. Immunol. 2018, 9, 2278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Chen, Y.; Ke, Y.; Zhang, L.; Zhang, B.; Yang, L.; Zhu, J. Single Chain Fragment Variable (scFv) Antibodies Targeting the Spike Protein of Porcine Epidemic Diarrhea Virus Provide Protection against Viral Infection in Piglets. Viruses 2019, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Mettenleiter, T.C. Intriguing interplay between viral proteins during herpesvirus assembly or: The herpesvirus assembly puzzle. Veter-Microbiol. 2006, 113, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Kurobe, T.; Gatherer, D.; Cunningham, C.; Korf, I.; Fukuda, H.; Hedrick, R.P.; Waltzek, T.B. Comparative Genomics of Carp Herpesviruses. J. Virol. 2013, 87, 2908–2922. [Google Scholar] [CrossRef]
- Tu, C.; Lu, Y.-P.; Hsieh, C.-Y.; Huang, S.-M.; Chang, S.-K.; Chen, M.-M. Production of monoclonal antibody against ORF72 of koi herpesvirus isolated in Taiwan. Folia Microbiol. 2013, 59, 159–165. [Google Scholar] [CrossRef]
- Hong, F.; Meinander, N.Q.; Jönsson, L.J. Fermentation strategies for improved heterologous expression of laccase inPichia pastoris. Biotechnol. Bioeng. 2002, 79, 438–449. [Google Scholar] [CrossRef]
- Camarero, S.; Pardo, I.; Cañas, A.I.; Molina-Espeja, P.; Record, E.; Martínez, A.T.; Martínez, M.J.; Alcalde, M. Engineering Platforms for Directed Evolution of Laccase from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 2012, 78, 1370–1384. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.; Li, Y. Protein L chromatography: A useful tool for monitoring/separating homodimers during the purification of IgG-like asymmetric bispecific antibodies. Protein Expr. Purif. 2020, 175, 105711. [Google Scholar] [CrossRef]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2017, 72, 32–42. [Google Scholar] [CrossRef]
- Caprari, S.; Metzler, S.; Lengauer, T.; Kalinina, O.V. Sequence and Structure Analysis of Distantly-Related Viruses Reveals Extensive Gene Transfer between Viruses and Hosts and among Viruses. Viruses 2015, 7, 5388–5409. [Google Scholar] [CrossRef]
- Kirkitadze, M.D.; Barlow, P.N.; Price, N.C.; Kelly, S.M.; Boutell, C.J.; Rixon, F.J.; McClelland, D.A. The herpes simplex virus triplex protein, VP23, exists as a molten globule. J. Virol. 1998, 72, 10066–10072. [Google Scholar] [CrossRef]
- Kuwajima, K. The Molten Globule, and Two-State vs. Non-Two-State Folding of Globular Proteins. Biomolecules 2020, 10, 407. [Google Scholar] [CrossRef]
- Heming, J.D.; Conway, J.F.; Homa, F.L. Herpesvirus capsid assembly and DNA packaging. Cell Biol. Herpes Viruses 2017, 223, 119–142. [Google Scholar]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef]
- Rees, D.C.; Robertson, A.D. Some thermodynamic implications for the thermostability of proteins. Protein Sci. 2001, 10, 1187–1194. [Google Scholar] [CrossRef]
- Rodrigo, G.; Gruvegård, M.; Van Alstine, J.M. Antibody fragments and their purification by protein L affinity chromatography. Antibodies 2015, 4, 259–277. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).