Transcriptomic Analysis Provides Insights into Microplastic and Heavy Metal Challenges in the Line Seahorse (Hippocampus erectus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Design
2.3. Total RNA Extraction from Liver
2.4. cDNA Synthesis
2.5. Transcription Library and Sequencing
2.6. Transcriptome Assembly and Annotation
2.7. Transcriptome Differential Expression Analysis
2.8. GO and KEGG Analysis
2.9. Real-Time Fluorescent Quantitative PCR Analysis
2.10. Statistical Analysis
3. Results
3.1. Sequencing Results
3.2. Transcriptome Assembly Results
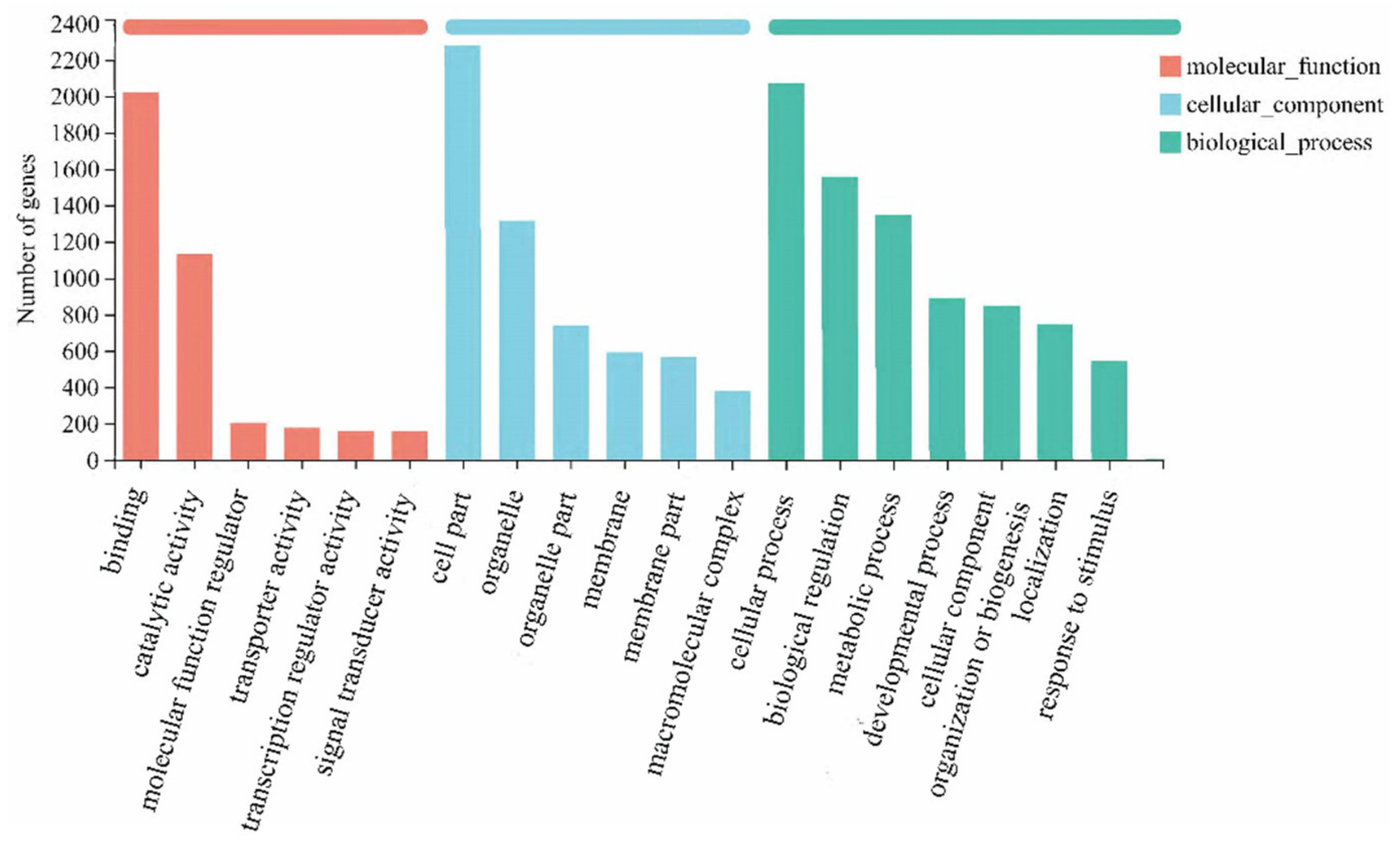
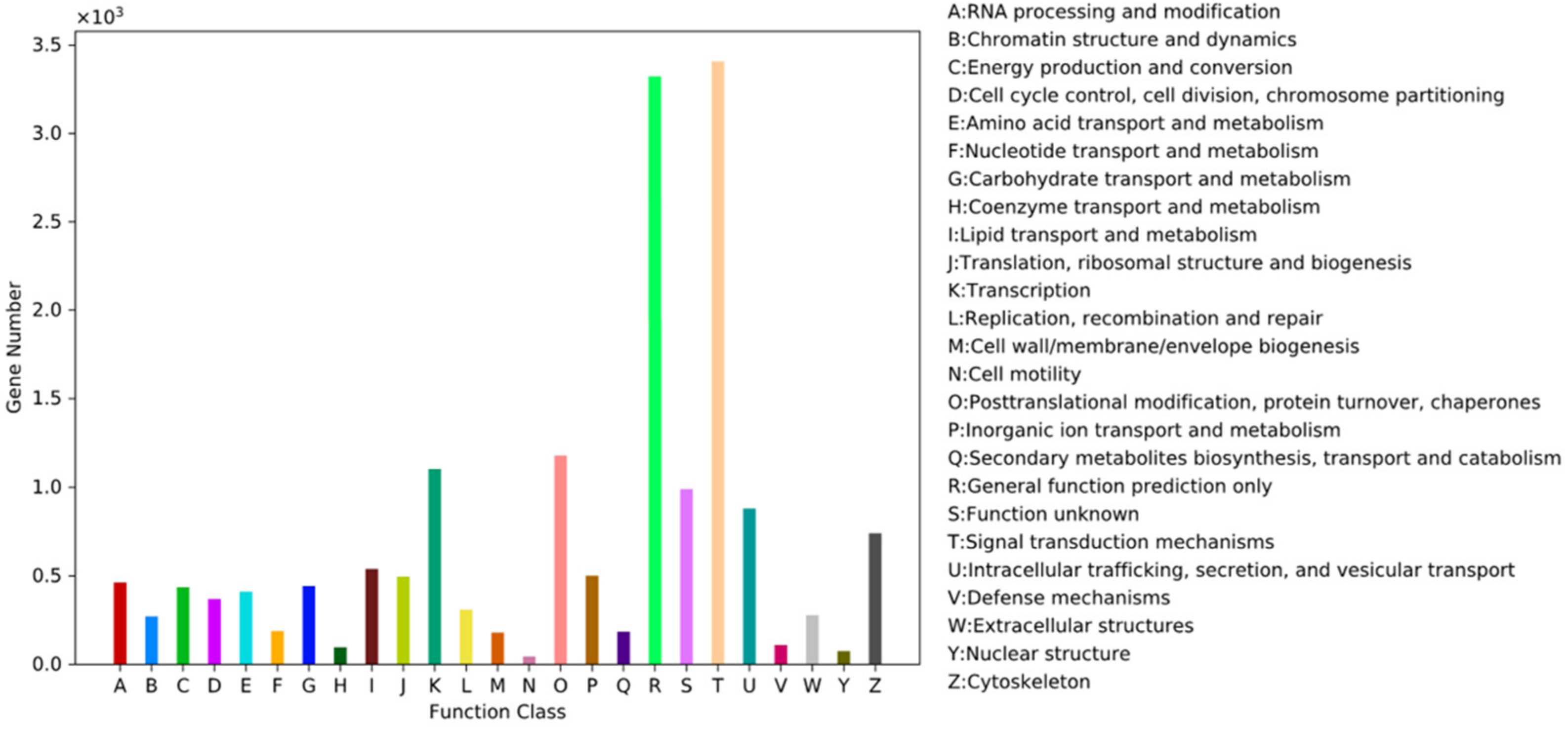
3.3. GO and KOG Statistics
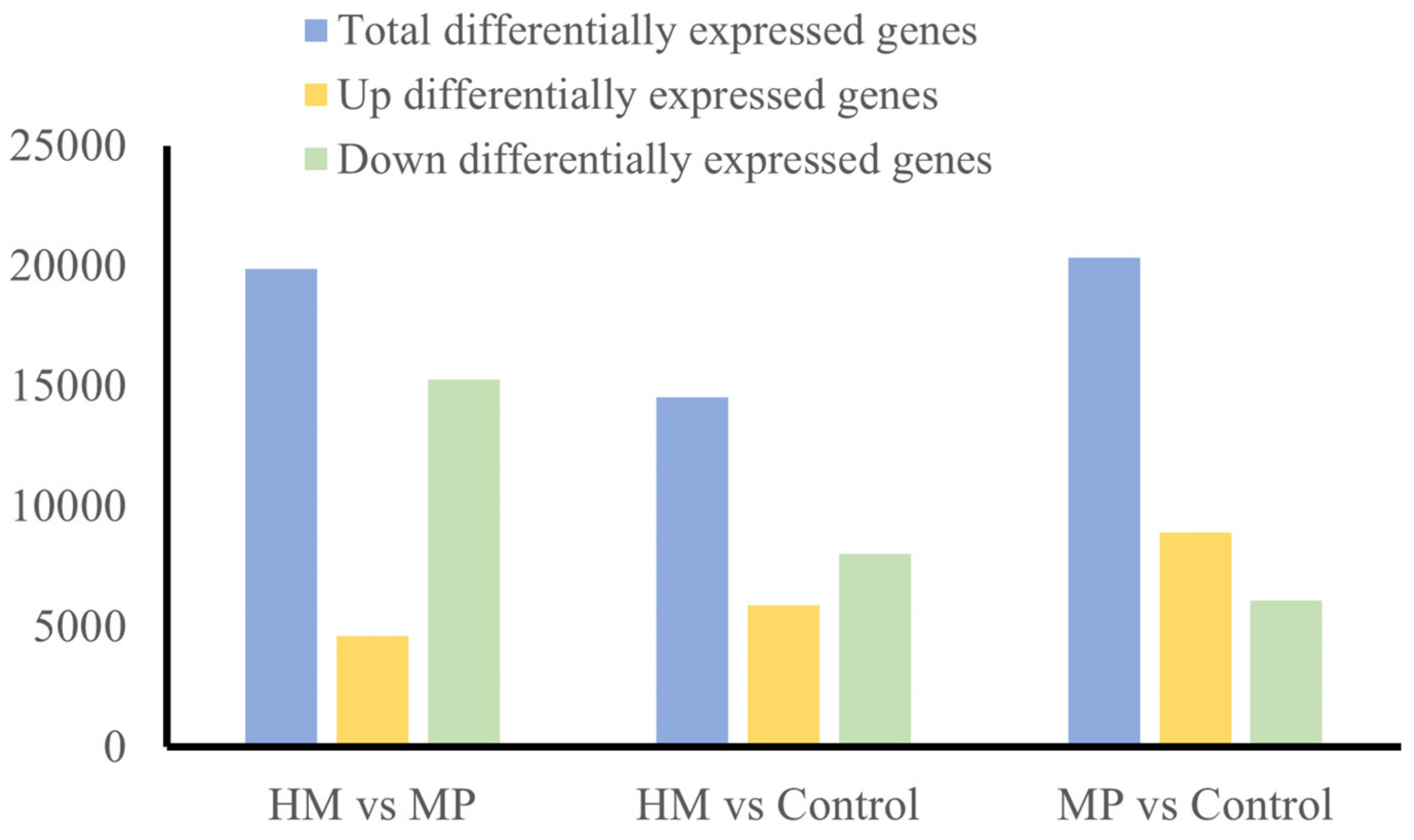
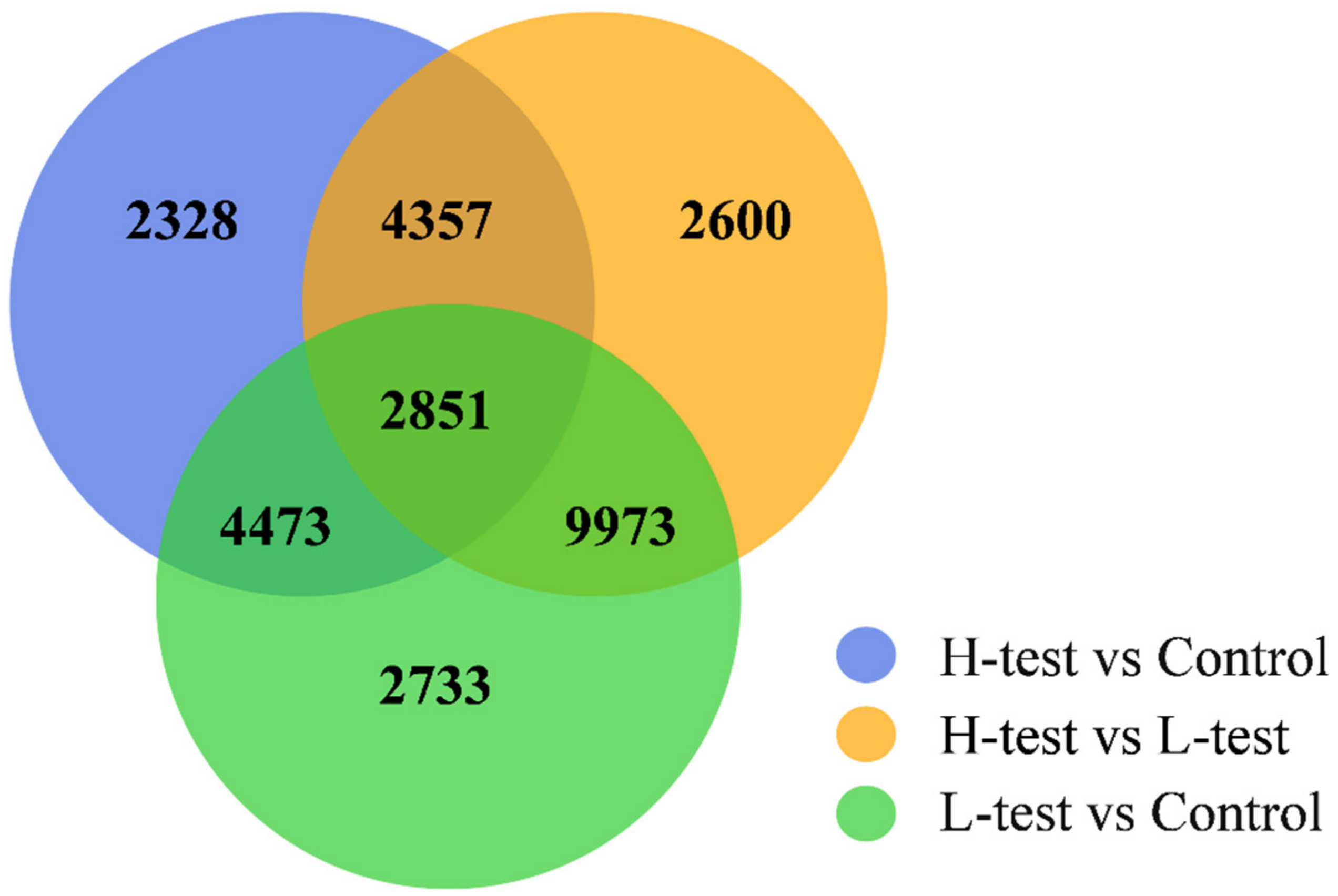
3.4. DEGs
3.5. KEGG Enrichment Analysis
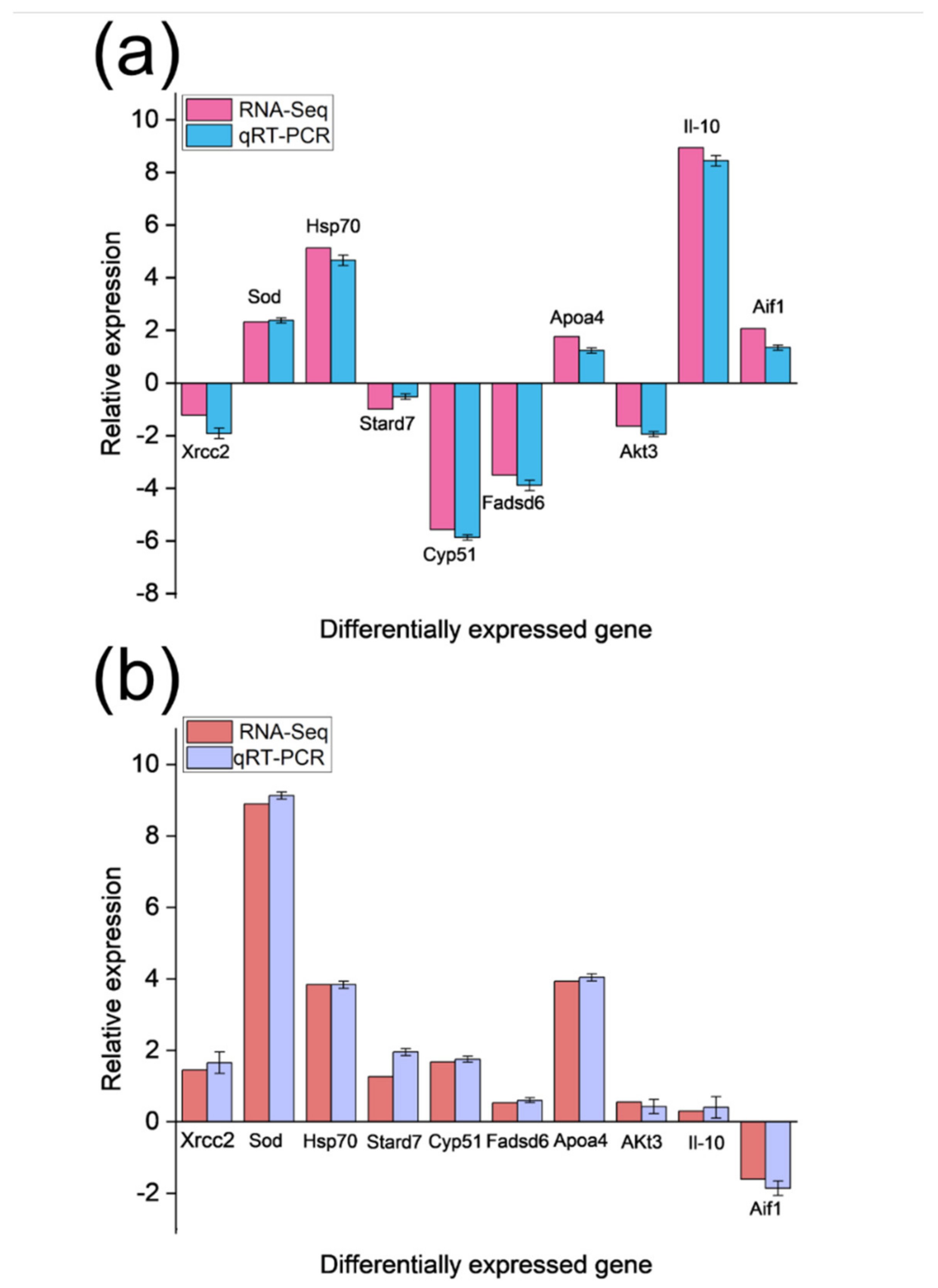
3.6. Real-Time Fluorescent Quantitative PCR Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Huang, H.; Liu, C.; Xia, Y.; Ye, C.; Luo, Z.; Cai, C.; Wang, C.; Lyu, L.; Bi, H.; et al. Waterproof and Breathable Graphene-Based Electronic Fabric for Wearable Sensors. Adv. Mater. Technol. 2022, 7, 2200149. [Google Scholar] [CrossRef]
- Welden, N.A.; Lusher, A.L. Impacts of changing ocean circulation on the distribution of marine microplastic litter. Integr. Environ. Assess. Manag. 2017, 13, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Cózar, A.; Echevarría, F.; González-Gordillo, J.I.; Irigoien, X.; Úbeda, B.; Hernández-León, S.; Palma, Á.T.; Navarro, S.; García-De-Lomas, J.; Ruiz, A.; et al. Plastic debris in the open ocean. Proc. Natl. Acad. Sci. USA 2014, 111, 10239–10244. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, L.; Slat, B.; Ferrari, F.; Sainte-Rose, B.; Aitken, J.; Marthouse, R.; Hajbane, S.; Cunsolo, S.; Schwarz, A.; Levivier, A.; et al. Evidence that the Great Pacific Garbage Patch is rapidly accumulating plastic. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: A review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef] [PubMed]
- Setälä, O.; Fleming-Lehtinen, V.; Lehtiniemi, M. Ingestion and transfer of microplastics in the planktonic food web. Environ. Pollut. 2014, 185, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Li, Y.; Wang, J. Occurrence and ecological impact of microplastics in aquaculture ecosystems. Chemosphere 2021, 274, 129989. [Google Scholar] [CrossRef]
- Athey, S.N.; Albotra, S.D.; Gordon, C.A.; Monteleone, B.; Seaton, P.; Andrady, A.L.; Taylor, A.R.; Brander, S.M. Trophic transfer of microplastics in an estuarine food chain and the effects of a sorbed legacy pollutant. Limnol. Oceanogr. Lett. 2020, 5, 154–162. [Google Scholar] [CrossRef]
- Guzzetti, E.; Sureda, A.; Tejada, S.; Faggio, C. Microplastic in marine organism: Environmental and toxicological effects. Environ. Toxicol. Pharmacol. 2018, 64, 164–171. [Google Scholar] [CrossRef]
- Da Costa Araújo, A.P.; De Andrade Vieira, J.E.; Malafaia, G. Toxicity and trophic transfer of polyethylene microplastics from Poecilia reticulata to Danio rerio. Sci. Total Environ. 2020, 742, 140217. [Google Scholar] [CrossRef]
- Jiang, W.; Fang, J.; Du, M.; Gao, Y.; Fang, J.; Jiang, Z. Microplastics influence physiological processes, growth and reproduction in the Manila clam, Ruditapes philippinarum. Environ. Pollut. 2021, 293, 118502. [Google Scholar] [CrossRef] [PubMed]
- Benson, N.U.; Agboola, O.D.; Fred-Ahmadu, O.H.; De-la-Torre, G.E.; Oluwalana, A.; Williams, A.B. Micro (nano) plastics prevalence, food web interactions and toxicity assessment in aquatic organisms: A review. Front. Mar. Sci. 2022, 291, 851281. [Google Scholar] [CrossRef]
- Caruso, G. Microplastics as vectors of contaminants. Mar. Pollut. Bull. 2019, 146, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, D.; Duarte, B.; Paiva, F.; Caçador, I.; Canning-Clode, J. Microplastics as vector for heavy metal contamination from the marine environment. Estuar. Coast. Shelf Sci. 2016, 178, 189–195. [Google Scholar] [CrossRef]
- Liu, S.; Huang, J.; Zhang, W.; Shi, L.; Yi, K.; Yu, H.; Zhang, C.; Li, S.; Li, J. Microplastics as a vehicle of heavy metals in aquatic environments: A review of adsorption factors, mechanisms, and biological effects. J. Environ. Manag. 2021, 302, 113995. [Google Scholar] [CrossRef]
- Kinigopoulou, V.; Pashalidis, I.; Kalderis, D.; Anastopoulos, I. Microplastics as carriers of inorganic and organic contaminants in the environment: A review of recent progress. J. Mol. Liq. 2022, 350, 118580. [Google Scholar] [CrossRef]
- Patrício-Rodrigues, J.; Duarte, A.C.; Santos-Echeandía, J.; Rocha-Santos, T. Significance of interactions between microplastics and POPs in the marine environment: A critical overview. TrAc Trends Anal. Chem. 2019, 111, 252–260. [Google Scholar] [CrossRef]
- El Bahgy, H.E.; Elabd, H.; Elkorashey, R.M. Heavy metals bioaccumulation in marine cultured fish and its probabilistic health hazard. Environ. Sci. Pollut. Res. 2021, 28, 41431–41438. [Google Scholar] [CrossRef]
- Zamora-Ledezma, C.; Negrete-Bolagay, D.; Figueroa, F.; Zamora-Ledezma, E.; Ni, M.; Alexis, F.; Guerrero, V.H. Heavy metal water pollution: A fresh look about hazards, novel and conventional remediation methods. Environ. Technol. Innov. 2021, 22, 101504. [Google Scholar] [CrossRef]
- Steinhausen, S.L.; Agyeman, N.; Turrero, P.; Ardura, A.; Garcia-Vazquez, E. Heavy metals in fish nearby electronic waste may threaten consumer’s health. Examples from Accra, Ghana. Mar. Pollut. Bull. 2022, 175, 113162. [Google Scholar] [CrossRef]
- Renu, K.; Chakraborty, R.; Myakala, H.; Koti, R.; Famurewa, A.C.; Madhyastha, H.; Vellingiri, B.; George, A.; Gopalakrishnan, A.V. Molecular mechanism of heavy metals (Lead, Chromium, Arsenic, Mercury, Nickel and Cadmium)-induced hepatotoxicity–A review. Chemosphere 2021, 271, 129735. [Google Scholar] [CrossRef] [PubMed]
- Edem, C.A.; Vincent, O.; Grace, I.; Rebecca, E.; Joseph, E. Distribution of heavy metals in bones, gills, livers and muscles of (Tilapia) Oreochromis niloticus from Henshaw Town Beach market in Calabar Nigeria. Pak. J. Nutr. 2009, 8, 1209–1211. [Google Scholar]
- Duran, A.; Tuzen, M.; Soylak, M. Assessment of trace metal concentrations in muscle tissue of certain commercially available fish species from Kayseri, Turkey. Environ. Monit. Assess. 2014, 186, 4619–4628. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.A.; Dehler, C.E.; Król, E. Transcriptomic responses in the fish intestine. Dev. Comp. Immunol. 2016, 64, 103–117. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, Y.; Xiong, D.; Zhang, J. RNA-seq revealed the signatures of immunity and metabolism in the Litopenaeus vannamei intestine in response to dietary succinate. Fish Shellfish Immunol. 2019, 95, 16–24. [Google Scholar] [CrossRef]
- Qiu, W.; Liu, S.; Yang, F.; Dong, P.; Yang, M.; Wong, M.; Zheng, C. Metabolism disruption analysis of zebrafish larvae in response to BPA and BPA analogs based on RNA-Seq technique. Ecotoxicol. Environ. Saf. 2019, 174, 181–188. [Google Scholar] [CrossRef]
- Li, Y.; Tsim, K.W.-K.; Wang, W.-X. Copper promoting oyster larval growth and settlement: Molecular insights from RNA-seq. Sci. Total Environ. 2021, 784, 147159. [Google Scholar] [CrossRef]
- Mohsen, M.; Sun, L.; Lin, C.; Huo, D.; Yang, H. Mechanism underlying the toxicity of the microplastic fibre transfer in the sea cucumber Apostichopus japonicus. J. Hazard. Mater. 2021, 416, 125858. [Google Scholar] [CrossRef]
- Hernández-Pérez, J.; Naderi, F.; Chivite, M.; Soengas, J.L.; Míguez, J.M.; López-Patiño, M.A. Influence of Stress on Liver Circadian Physiology. A Study in Rainbow Trout, Oncorhynchus mykiss, as Fish Model. Front. Physiol. 2019, 10, 611. [Google Scholar] [CrossRef]
- Liu, Y.; Jia, X.; Zhu, H.; Zhang, Q.; He, Y.; Shen, Y.; Xu, X.; Li, J. The effects of exposure to microplastics on grass carp (Ctenopharyngodon idella) at the physiological, biochemical, and transcriptomic levels. Chemosphere 2021, 286, 131831. [Google Scholar] [CrossRef]
- Vieira, K.S.; Neto, J.A.B.; Crapez, M.A.C.; Gaylarde, C.; Pierri, B.d.; Saldaña-Serrano, M.; Bainy, A.C.D.; Nogueira, D.J.; Fonseca, E.M. Occurrence of microplastics and heavy metals accumulation in native oysters Crassostrea Gasar in the Paranaguá estuarine system, Brazil. Mar. Pollut. Bull. 2021, 166, 112225. [Google Scholar] [CrossRef] [PubMed]
- Tse-Lynn, L.; Alexander, T.; Lindsay, A.; Ratanawalee, P. Species in wildlife trade: Socio-economic factors influence seahorse relative abundance in Thailand. Biol. Conserv. 2016, 201, 301–308. [Google Scholar]
- Groten, J.P.; Feron, V.J.; Sühnel, J. Toxicology of simple and complex mixtures. Trends Pharmacol. Sci. 2001, 22, 316–322. [Google Scholar] [CrossRef]
- Prato, E.; Biandolino, F. Combined toxicity of mercury, copper and cadmium on embryogenesis and early larval stages of the Mytilus galloprovincialis. Environ. Technol. 2007, 28, 915–920. [Google Scholar] [CrossRef]
- Jinhui, S.; Sudong, X.; Yan, N.; Xia, P.; Jiahao, Q.; Yongjian, X. Effects of microplastics and attached heavy metals on growth, immunity, and heavy metal accumulation in the yellow seahorse, Hippocampus kuda Bleeker. Mar. Pollut. Bull. 2019, 149, 110510. [Google Scholar] [CrossRef]
- Brown, R.A.M.; Epis, M.R.; Horsham, J.L.; Kabir, T.D.; Richardson, K.L.; Leedman, P.J. Total RNA extraction from tissues for microRNA and target gene expression analysis: Not all kits are created equal. BMC Biotechnol. 2018, 18, 1–11. [Google Scholar] [CrossRef]
- Zhao, S. Assessment of the impact of using a reference transcriptome in mapping short RNA-Seq reads. PLoS ONE 2014, 9, e101374. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Alvarez, R.V.; Vidal, N.M.; Garzón-Martínez, G.A.; Barrero, L.S.; Landsman, D.; Mariño-Ramírez, L. Workflow and web application for annotating NCBI BioProject transcriptome data. Database 2017, 2017, bax008. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Kima, H.; Lee, B.; Han, J.; Lee, Y.H.; Min, G.; Kim, S.; Lee, J. De novo assembly and annotation of the Antarctic copepod (Tigriopus kingsejongensis) transcriptome. Mar. Genom. 2016, 28, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Peixoto, D.; Torreblanca, A.; Pereira, S.; Vieira, M.N.; Varó, I. Effect of short-term exposure to fluorescent red polymer microspheres on Artemia franciscana nauplii and juveniles. Environ. Sci. Pollut. Res. 2021, 29, 6080–6092. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Xue, L. Liver transcriptome sequencing and de novo annotation of the large yellow croaker (Larimichthy crocea) under heat and cold stress. Mar. Genom. 2016, 25, 95–102. [Google Scholar] [CrossRef]
- Wang, S.; Xie, S.; Wang, Z.; Zhang, C.; Pan, Z.; Sun, D.; Xu, G.; Zou, J. Single and Combined Effects of Microplastics and Cadmium on the Cadmium Accumulation and Biochemical and Immunity of Channa argus. Biol. Trace Element Res. 2021, 200, 3377–3387. [Google Scholar] [CrossRef]
- Yuan, W.; Zhou, Y.; Chen, Y.; Liu, X.; Wang, J. Toxicological effects of microplastics and heavy metals on the Daphnia magna. Sci. Total Environ. 2020, 746, 141254. [Google Scholar] [CrossRef]
- Boopathy, L.R.A.; Jacob-Tomas, S.; Alecki, C.; Vera, M. Mechanisms tailoring the expression of heat shock proteins to proteostasis challenges. J. Biol. Chem. 2022, 198, 101796. [Google Scholar] [CrossRef]
- Dubrez, L.; Causse, S.; Bonan, N.B.; Dumétier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef]
- Farrell, P.; Nelson, K. Trophic level transfer of microplastic: Mytilus edulis (L.) to Carcinus maenas (L.). Environ. Pollut. 2013, 177, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Luís, L.G.; Ferreira, P.; Fonte, E.; Oliveira, M.; Guilhermino, L. Does the presence of microplastics influence the acute toxicity of chromium (VI) to early juveniles of the common goby (Pomatoschistus microps)? A study with juveniles from two wild estuarine populations. Aquat. Toxicol. 2015, 164, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Mates, J. Effects of antioxidant enzymes in the molecular control of reactive oxygen species toxicology. Toxicology 2000, 153, 83–104. [Google Scholar] [CrossRef]
- Schülke, S.; Dreidax, D.; Malik, A.; Burmester, T.; Nevo, E.; Band, M.; Avivi, A.; Hankeln, T. Living with stress: Regulation of antioxidant defense genes in the subterranean, hypoxia-tolerant mole rat, Spalax. Gene 2012, 500, 199–206. [Google Scholar] [CrossRef]
- Karimian, A.; Mir, S.M.; Parsian, H.; Refieyan, S.; Mirza-Aghazadeh-Attari, M.; Yousefi, B.; Majidinia, M. Crosstalk between Phosphoinositide 3-kinase/Akt signaling pathway with DNA damage response and oxidative stress in cancer. J. Cell. Biochem. 2018, 120, 10248–10272. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, X.; Wang, G.; Lin, S.; Zeng, X.; Wang, Y.; Zhang, Z. PI3K-AKT signaling pathway is involved in hypoxia/thermal-induced immunosuppression of small abalone Haliotis diversicolor. Fish Shellfish Immunol. 2016, 59, 492–508. [Google Scholar] [CrossRef]
- Bottiglione, F.; Dee, C.T.; Lea, R.; Zeef, L.A.H.; Badrock, A.P.; Wane, M.; Bugeon, L.; Dallman, M.J.; Allen, J.E.; Hurlstone, A.F.L. Zebrafish IL-4–like Cytokines and IL-10 Suppress Inflammation but Only IL-10 Is Essential for Gill Homeostasis. J. Immunol. 2020, 205, 994–1008. [Google Scholar] [CrossRef]
- Baumann, L.; Schmidt-Posthaus, H.; Segner, H.; Wolf, J.C. Comment on “uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver”. Environ. Sci. Technol. 2016, 50, 12521–12522. [Google Scholar] [CrossRef]
- Ren, Y.; Ding, D.; Pan, B.; Bu, W. The TLR13-MyD88-NF-κB signalling pathway of Cyclina sinensis plays vital roles in innate immune responses. Fish Shellfish Immun. 2017, 70, 720–730. [Google Scholar] [CrossRef]
- Nasser, F.; Lynch, I. Secreted protein eco-corona mediates uptake and impacts of polystyrene nanoparticles on Daphnia magna. J. Proteom. 2016, 137, 45–51. [Google Scholar] [CrossRef]
- Li, D.; Yao, H.; Du, L.; Zeng, X.; Xiao, Q. Thallium(I and III) exposure leads to liver damage and disorders of fatty acid metabolism in mice. Chemosphere 2022, 307, 135618. [Google Scholar] [CrossRef] [PubMed]
- Campani, T.; Baini, M.; Giannetti, M.; Cancelli, F.; Mancusi, C.; Serena, F.; Marsili, L.; Casini, S.; Fossi, M.C. Presence of plastic debris in loggerhead turtle stranded along the Tuscany coasts of the Pelagos Sanctuary for Mediterranean Marine Mammals (Italy). Mar. Pollut. Bull. 2013, 74, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.L. Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1771, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Debeljak, N.; Horvat, S.; Vouk, K.; Lee, M.; Rozman, D. Characterization of the Mouse Lanosterol 14α-Demethylase (CYP51), a New Member of the Evolutionarily Most Conserved Cytochrome P450 Family. Arch. Biochem. Biophys. 2000, 379, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.; Clemente-Blanco, A. Cell Cycle and DNA Repair Regulation in the Damage Response: Protein Phosphatases Take Over the Reins. Int. J. Mol. Sci. 2020, 21, 446. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Primer Sequences (5′–3′) | Amplification Efficiencies |
|---|---|---|
| XRCC2 | F: CCGCCAAAGTGTTCGTGA R: TGATCCAAATGCGCCATG | 99 |
| Sod | F: TCACATACTTCACGGGTTTCG R: AGGGAAATGTTCAAGGTACTGC | 102 |
| Hsp70 | F: GTCGGTGAAATAACAGGGAACA R: CTCTGGGTCTACAGGTATTAAGGTG | 95 |
| Stard7 | F: CGTTTGGCTCCTTTTGTGC R: GCGGGTTTGTCTACTCCTCTG | 97 |
| Cyp51a1 | F: CGCCTGGTTGGGTTTCTT R: TCAAACAACGCTCCAACAGG | 102 |
| Fadsd6 | F: TGGTGCTTGTCATACTACCTTCG R: CTTGATGCTTCTCGTGGTCG | 104 |
| Apoa4 | F: CGTCTTCGTTTTGGCTGTTT R: TCTTTTCCGAGATCCGACTGT | 103 |
| Akt3 | F: GCGACGGAGAAGTTGTTGAG R: GTTGTCAAGGAAGGATGGGTC | 96 |
| Il-10 | F: GGAGGACACGAGGGACTTGA R: GCCTTTGTTTTGCATCTGACTG | 100 |
| Aif1 | F: ACGCCATCAATGAGGCTTTT R: TCTTGGCAAGTCCCAGTTTCT | 102 |
| B2m | F: TACACCCACCAGCCAGGAAA R: GGACTCGACGACATCGAACATC | 100 |
| Pathway ID | Upregulated DEGs | Downregulated DEGs | Q Value | Metabolic Pathway Annotation |
|---|---|---|---|---|
| map04974 | 23 | 45 | 4.9 × 10−3 | Protein digestion and absorption |
| map00100 | 3 | 13 | 5.9 × 10−3 | Steroid biosynthesis |
| map04512 | 22 | 44 | 1.23 × 10−2 | ECM-receptor interaction |
| map04923 | 9 | 31 | 7.38 × 10−2 | Regulation of lipolysis in adipocytes |
| map04975 | 20 | 5 | 1.947 × 10−1 | Fat digestion and absorption |
| map00561 | 17 | 19 | 1.947 × 10−1 | Glycerolipid metabolism |
| map04672 | 7 | 8 | 1.947 × 10−1 | Intestinal immune network for IgA Production |
| map05134 | 21 | 4 | 1.947 × 10−1 | Legionellosis |
| map05145 | 31 | 23 | 1.947 × 10−1 | Toxoplasmosis |
| map04978 | 12 | 9 | 2.153 × 10−1 | Mineral absorption |
| map04151 | 62 | 81 | 2.153 × 10−1 | PI3K-Akt signaling pathway |
| map04930 | 6 | 24 | 2.153 × 10−1 | Type II diabetes mellitus |
| map00604 | 6 | 5 | 2.153 × 10−1 | Glycosphingolipid biosynthesis—ganglio series |
| map00830 | 11 | 8 | 2.153 × 10−1 | Retinol metabolism |
| map04721 | 17 | 19 | 2.153 × 10−1 | Synaptic vesicle cycle |
| map02010 | 18 | 16 | 2.802 × 10−1 | ABC transporters |
| map00520 | 23 | 5 | 2.802 × 10−1 | Amino sugar and nucleotide sugar metabolism |
| map05414 | 27 | 24 | 3.534 × 10−1 | Dilated cardiomyopathy |
| map04612 | 19 | 1 | 6.349 × 10−1 | Antigen processing and presentation |
| map04216 | 18 | 4 | 6.349 × 10−1 | Ferroptosis |
| Pathway ID | Upregulated DEGs | Downregulated DEGs | Q Value | Metabolic Pathway Annotation |
|---|---|---|---|---|
| map03440 | 39 | 1 | 2.7 × 10−3 | Homologous recombination |
| map03030 | 34 | 0 | 9.2 × 10−3 | DNA replication |
| map00590 | 27 | 6 | 2.594 × 10−1 | Arachidonic acid metabolism |
| map04110 | 92 | 2 | 2.594 × 10−1 | Cell cycle |
| map03460 | 40 | 4 | 2.594 × 10−1 | Fanconi anemia pathway |
| map04724 | 80 | 32 | 2.594 × 10−1 | Glutamatergic synapse |
| map00480 | 33 | 3 | 2.594 × 10−1 | Glutathione metabolism |
| map00240 | 62 | 9 | 2.594 × 10−1 | Pyrimidine metabolism |
| map04977 | 25 | 4 | 2.594 × 10−1 | Vitamin digestion and absorption |
| map03420 | 32 | 0 | 2.685 × 10−1 | Nucleotide excision repair |
| map00520 | 35 | 4 | 5.093 × 10−1 | Amino sugar and nucleotide sugar metabolism |
| map03410 | 25 | 1 | 5.093 × 10−1 | Base excision repair |
| map00010 | 44 | 6 | 5.093 × 10−1 | Glycolysis/Gluconeogenesis |
| map00340 | 11 | 7 | 5.093 × 10−1 | Histidine metabolism |
| map03430 | 15 | 0 | 5.093 × 10−1 | Mismatch repair |
| map04114 | 80 | 12 | 5.093 × 10−1 | Oocyte meiosis |
| map03050 | 28 | 0 | 5.093 × 10−1 | Proteasome |
| map00230 | 113 | 14 | 5.093 × 10−1 | Purine metabolism |
| map04742 | 24 | 12 | 5.093 × 10−1 | Taste transduction |
| map05414 | 59 | 15 | 5.416 × 10−1 | Dilated cardiomyopathy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Shang, D.; Yang, Y.; Cui, P.; Sun, J. Transcriptomic Analysis Provides Insights into Microplastic and Heavy Metal Challenges in the Line Seahorse (Hippocampus erectus). Fishes 2022, 7, 338. https://doi.org/10.3390/fishes7060338
Liu Y, Shang D, Yang Y, Cui P, Sun J. Transcriptomic Analysis Provides Insights into Microplastic and Heavy Metal Challenges in the Line Seahorse (Hippocampus erectus). Fishes. 2022; 7(6):338. https://doi.org/10.3390/fishes7060338
Chicago/Turabian StyleLiu, Ying, Dongwei Shang, Yanjing Yang, Pei Cui, and Jinhui Sun. 2022. "Transcriptomic Analysis Provides Insights into Microplastic and Heavy Metal Challenges in the Line Seahorse (Hippocampus erectus)" Fishes 7, no. 6: 338. https://doi.org/10.3390/fishes7060338
APA StyleLiu, Y., Shang, D., Yang, Y., Cui, P., & Sun, J. (2022). Transcriptomic Analysis Provides Insights into Microplastic and Heavy Metal Challenges in the Line Seahorse (Hippocampus erectus). Fishes, 7(6), 338. https://doi.org/10.3390/fishes7060338







