Abstract
Computer vision has been applied to fish recognition for at least three decades. With the inception of deep learning techniques in the early 2010s, the use of digital images grew strongly, and this trend is likely to continue. As the number of articles published grows, it becomes harder to keep track of the current state of the art and to determine the best course of action for new studies. In this context, this article characterizes the current state of the art by identifying the main studies on the subject and briefly describing their approach. In contrast with most previous reviews related to technology applied to fish recognition, monitoring, and management, rather than providing a detailed overview of the techniques being proposed, this work focuses heavily on the main challenges and research gaps that still remain. Emphasis is given to prevalent weaknesses that prevent more widespread use of this type of technology in practical operations under real-world conditions. Some possible solutions and potential directions for future research are suggested, as an effort to bring the techniques developed in the academy closer to meeting the requirements found in practice.
1. Introduction
Monitoring the many different aspects related to fish, including their life cycle, impacts of human activities, and effects of commercial exploration, is important both for optimizing the fishing industry and for ecology and conservation purposes. There are many activities related to fish monitoring, but these can be roughly divided into four main groups. The first, recognition, has as its main goal to detect and count the number of individuals in a given environment. The second, measurement, aims at non-invasively estimating the dimensions and weight of fish. The third, tracking, aims at following individuals or shoals over time, which is usually done to either aid in the counting process or to determine the behavior of the fish as a response to the environment or to some source of stress. The fourth, classification, aims to identify the species or other factors in order to obtain a better characterization of a given area. Monitoring activities are still predominantly carried out visually, either “in loco” or employing images or videos. There are many limitations associated with visual surveys, including high costs [1,2], low throughput [1,3], subjectivity [2], etc. In addition, human observers have been shown to be less consistent than computer measurements due to individual biases, and the presence of humans and their equipment often causes animals to display avoidance behavior and make data collection unreliable [4]. Imaging devices also enable the collection of data in dangerous areas that could pose risks to a human operator [5].
Given the limitations associated with visual surveys, it is no surprise that new methods capable of automating at least part of the process have been increasingly investigated over the last three decades [6]. Different types of devices have been used for gathering the necessary information from the fish, including acoustic sensors [7], sonars [8], and a wide variety of imaging sensors [9]. In this work, only studies employing conventional RGB (red/green/blue) sensors were considered, as these are by far the most widely used due to their relatively low price and wide availability.
Digital images and videos have been explored for some time, but with the rapid development of new artificial intelligence (AI) algorithms and models and with the increase in computational resources, this type of approach has become the preferred choice in most circumstances [10]. Deep learning models have been particularly successful in dealing with difficult detection, tracking, and classification problems [5]. Despite the significant progress achieved recently, very few methods found in the literature are suitable for use under real conditions. One of the reasons for this is that although the objective of most studies is to eventually arrive at some method for monitoring the fish underwater under real, uncontrolled conditions, a substantial part of the research is carried out using images captured either under controlled underwater conditions (for example, in tanks with artificial illumination) or out of the water. This makes it much easier to generate images in which fish are clearly visible and have good contrast with the background, which is usually not the case in the real world. On the other hand, research carried out under more realistic conditions often has trouble achieving good accuracies [10]. In any case, it is important to recognize that image-based methods may not be suitable to tackle all facets of fish monitoring [5], which underlies the importance of understanding the problem in depth before pursuing a solution that might ultimately be unfeasible. The combination of digital images and videos with artificial intelligence algorithms has been very fruitful in many different areas [5], and significant progress has also been achieved in fish monitoring and management. With hundreds of different strategies already proposed in the literature, it is difficult to keep track of which aspects of the problem have already been successfully tackled and which research gaps still linger. This is particularly relevant considering that the impact of those advancements in practice is still limited. There are many reviews in the literature that offer a detailed technical characterization of the state of the art on the use of digital images/videos for the detection and characterization of fish in different environments (Table 1). Although there is some overlap between this and previous reviews, here the focus is heavily directed at current limitations and what has to be done in order to close the gap between academic research and practical needs. In order to avoid too much redundancy with other reviews and keep the text relatively short, technical details about the articles cited in the next sections are not mentioned unless relevant to the problem or challenge being discussed.

Table 1.
Review and survey articles dealing with computer vision applied to fish recognition, monitoring, and management.
2. Definitions and Acronyms
Some terms deemed of particular importance in the context of this work are defined in this section. Most of the definitions are adapted from [15,32]. A list of acronyms used in this article with their respective meanings is given in Table 2.

Table 2.
Acronyms used in this review.
Artificial intelligence: a computational data-driven approach capable of performing tasks that normally require human intelligence to independently detect, track, or classify fish.
Data annotation: the process of adding metadata to a dataset, such as indicating where the fish are located in the image. This is typically performed manually by human specialists making use of image analysis software.
Data fusion: the process in which different types of data are combined in order to provide results that could not be achieved using single data sources (e.g., combining images and meteorological data to provide accurate detection of a specific fish species).
Deep learning: a special case of machine learning that utilizes artificial neural networks with many layers of processing to implicitly extract features from the data and recognize patterns of interest. Deep learning is appropriate for large datasets with complex features and where there are unknown relationships within the data.
Feature: measurement of a specific property of a data sample. It can be a color, texture, shape, reflectance intensity, index values, or spatial information.
Image augmentation: the process of applying different image processing techniques to alter existing images in order to create more data for training the model.
Imaging: the use of sensors capable of capturing images in a certain range of the electromagnetic spectrum. Imaging sensors include RGB (red/green/blue), multispectral, hyperspectral, and thermal images.
Machine learning: the application of artificial intelligence (AI) algorithms that have the ability to learn characteristics of fish via extraction of features from a large dataset. Machine learning models are often based on knowledge obtained from annotated training data. Once the model is developed, it can be used to predict the desired output on test data or unknown images.
Mathematical morphology: this type of technique, usually applied to binary images, can simplify image data, eliminating irrelevant structures while retaining the basic shapes of the objects. All morphological operations are based on two basic operators, dilation and erosion.
Model: a representation of what a machine learning program has learned from the data.
Overfitting: when a model closely predicts the training data but fails to fit testing data.
Segmentation: the process of partitioning a digital image containing the objects of interest (fish) into multiple segments of similarity or classes (based on sets of pixels with common characteristics of hue, saturation, and intensity) either automatically or manually. In the latter case, the human-powered task is also called image annotation in the context of training AI algorithms. When performed at the pixel level, the process is referred to as semantic segmentation.
Semi-supervised learning: a combination of supervised and unsupervised learning in which a small portion of the data is used for a first supervised training, and the remainder of the process is carried out with unlabeled data.
Supervised learning: a machine learning model based on a known labeled training dataset that is able to predict a class label (classification) or numeric value (regression) for new unknown data.
Unsupervised learning: machine learning that finds patterns in unlabeled data.
Weakly supervised learning: noisy, limited, or imprecise sources are used as references for labeling large amounts of training data, thus reducing the need for hand-labeled data.
3. Literature Review
The search for articles was carried out in September 2022 on Scopus and Google Scholar, as both encompass virtually all relevant bibliographic databases. The terms used in the search were “fish” and “digital images”. The terms were kept deliberately general in order to reduce the likelihood of relevant work being missed. The downside of this strategy is that the filtering process was labor-intensive and time-consuming. Initially, all journal articles that employed images to tackle any aspect related to fish were considered. Conference articles were excluded as a rule, but a few of those that had highly relevant content were ultimately included. Articles were further filtered by keeping only those devoted either to live fish monitoring or to the analysis of fish shortly after catch (for example, for screening or auditing purposes). Articles aiming at estimating the quality and freshness of fish products were thus removed. Manuscripts published in journals widely considered predatory were also removed. Studies published prior to 2010 were only included when deemed historically relevant in the context of fish monitoring and management. A few articles that were originally missed were subsequently included by careful inspection of the reference lists of some selected review articles (Table 1).
The following four subsections discuss many issues that are specific to each type of application: recognition, tracking, measurement, and classification (Figure 1). However, there are some issues that are common to all of them, such as the difficulties caused by underwater conditions. In those cases, the discussion will be either concentrated in the subsection in which that specific issue has more impact, or in multiple subsections if there are some specific consequences attached to those particular applications.
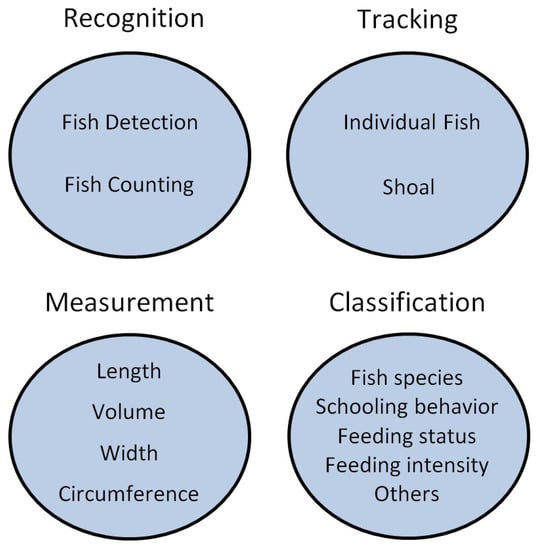
Figure 1.
Categories of applications considered in this work. Applications that involve the detection and analysis of objects other than fish (e.g., food pellets) were not considered. This categorization is one among several possible options and was chosen as the best fit for the purposes of this review.
3.1. Recognition
One of the most basic tasks in fisheries, aquaculture, and ecological monitoring is the detection and counting of fish and other relevant species. This can be done underwater in order to determine the population in a given area [33], over conveyor belts during the process of buying and selling and tank transference [34], or out of the water to determine, for example, the number of fish of different species captured during a catch. Estimating fish populations is important to avoid over-fishing and keep production sustainable [35,36], as well as for wildlife conservation and animal ecology purposes [33]. Automatically counting fish can also be useful for the inspection and enforcement of regulations [37]. Fish detection is often the first step of more complex tasks such as behavior analysis, detection of anomalous events [38], and species classification [39].
Table 3 shows the studies dedicated to this type of task. The column “context” in this and in the following tables categorizes the main application based on their scope, using the classification suggested by Shafait et al. [6], with some adaptations. The term “accuracy” in Table 3, Table 4, Table 5 and Table 6 is used in a wide sense, as several different performance evaluation metrics can be found in the literature. Thus, the numbers in the last column of those tables may represent not only the actual accuracy but also other metrics, which are specified in a legend at the bottom of the tables. Because of this and the fact that most studies use different datasets, direct comparison between different methods is not possible unless new tests using the exact same setups are carried out [19]. A description of several different evaluation metrics can be found in [22].

Table 3.
List of articles dealing with detecting and counting fish.
Lighting variations caused by turbidity, light attenuation at lower depths, and waves on the surface may cause severe detection problems, leading to error [5,10,36,41,52]. Possible solutions for this include the use of larger sensors with better light sensitivity (which usually cost more), or employing artificial lighting, which under natural conditions may not be feasible and can also attract or repel fish, skewing the observations [2]. Additionally, adding artificial lighting often makes the problem worse due to the backscattering of light in front of the camera. Other authors have used infrared sensors together with the RGB cameras to increase the information collected under deep water conditions [42]. Post-processing the images using denoising and enhancement techniques is another option that can at least partially address the issue of poor quality images [41,57], but it is worth pointing out that this type of technique tends to be computationally expensive [52]. Finally, some authors explore the results obtained under more favorable conditions to improve the analysis of more difficult images or video frames [55].
The background of images may contain objects such as cage structures, lice skirts, biofouling organisms, coral, seaweed, etc., which greatly increases the difficulty of the detection task, especially if some of those objects mimic the visual characteristics of the fish of interest [10]. Banno et al. [2] reported a considerable number of false positives due to complex backgrounds, but added that those errors could be easily removed manually. The buildup of fouling on the camera’s lenses was also pointed out by Marini et al. [54] as a potential source of error that should be prevented either by regular maintenance or by using protective gear.
One of the main sources of errors in underwater fish detection and counting is the occlusion by other fish or objects, especially when several individuals are present simultaneously [54]. Some of the methods proposed in the literature were designed specifically to address this problem [5,40,50], but the success under uncontrolled conditions has been limited [2]. Partial success has been achieved by Labao and Naval [36], who devised a cascade structure that automatically performs corrections on the initial estimates by including the contextual information around the objects of interest. Another possible solution is the use of sophisticated tracking strategies applied to video recordings, but even in this case occlusions can lead to low accuracy (see Section 3.3). Structures and objects present in the environment can also cause occlusions, especially considering that fish frequently seek shelter and try to hide whenever they feel threatened. Potential sources of occlusion need to be identified and taken into account if the objective is to reliably estimate the fish population in a given area from digital images taken underwater [33].
Underwater detection, tracking, measurement, and classification of fish requires dealing with the fact that individuals will cross the camera’s line of sight at different distances [58]. This poses several challenges. First, fish outside the range of the camera’s depth of field will appear out of focus and the consequent loss of information can lead to error. Second, fish located too far from the camera will be represented by only a few pixels, which may not be enough for the task at hand [36], thus increasing the number of false negatives [54]. Third, fish that pass too close to the camera may not appear in their entirety in any given image/frame, again limiting the information available. Coro and Walsh [42] explored color distributions in the object to compensate for the lack of resolvability of fish located too close to the camera.
One way to deal with the difficulties mentioned so far is by focusing the detection on certain distinctive body structures rather than the whole body. Costa et al. [44] dealt with problems caused by body movement, bending, and touching specimens by focusing the detection on the eyes, which more unambiguously represented each individual than their whole bodies. Qian et al. [59] focused on the fish heads in order to better track individuals in a fish tank.
The varying quality of underwater images poses challenges not only to automated methods but also to human experts responsible for annotating the data [4]. Especially in the case of low-quality images, annotation errors can be frequent and, as a result, the model ends up being trained with inconsistent data [60]. Banno et al. [2] have shown that the difference in counting results yielded by two different people can surpass 20%, and even repeated counts carried out by the same person can be inconsistent. Annotation becomes even more challenging and prone to subjectivity-related inconsistency with more complex detection tasks, such as pose estimation [51]. With the intrinsic subjectivity of the annotation process, inconsistencies are mostly unavoidable, but their negative effects can be mitigated by using multiple experts and applying a majority rule to assign the definite labels [32]. The downside of this strategy is that manual annotation tends to be expensive and time-consuming, so the best strategy will ultimately depend on how reliable the annotated data needs to be.
With so many factors affecting the characteristics of the images, especially when captured under uncontrolled conditions, it is necessary to prepare the models to deal with such a variety. In other words, the dataset used to train the models needs to represent the variety of conditions and variations expected to be found in practice. In turn, this often means that thousands of images need to be captured and properly annotated, which explains why virtually all image datasets used in the reported studies have some kind of limitation that decreases the generality of the models trained [38,47,51] and, as a result, limits their potential for practical use [2,4]. This is arguably the main challenge preventing more widespread use of image-based techniques for fish monitoring and management. Given the importance of this issue, it is revisited from slightly different angles both in Section 3.4 and Section 4.
3.2. Measurement
Non-invasively estimating the size and weight of fish is very useful both for ecological and economic purposes. Biomass estimation in particular can provide clues about the feeding process, possible health problems, and potential production in fisheries. It can also reveal important details about the condition of wild species populations in vulnerable areas. In addition, fish length is one of the key variables needed for both taking short-term management decisions and modeling stock trends [1], and automating the measurement process can reduce costs and produce more consistent data [61,62]. Automatic measurement of body traits can also be useful after catch to quickly provide information about the characteristics of the fish batch, which can, for example, be done during transportation on the conveyor belts [34].
Bravata et al. [63] enumerated several shortcomings of manual measurements. In particular, conventional length and weight data collection requires the physical handling of fish, which is time-consuming for personnel and stressful for the fish. Additionally, measurements are commonly taken in the field, where conditions can be suboptimal for ensuring precision and accuracy. This highlights the need for a more objective and systematic way to ensure accurate measurements. Table 4 shows the studies dedicated to measurement tasks.

Table 4.
List of articles dealing with measuring fish.
Table 4.
List of articles dealing with measuring fish.
| Reference | Target | Context | Species | Main Technique | Accuracy |
|---|---|---|---|---|---|
| Al-Jubouri et al. [64] | Length | Underwater (controlled) | Zebrafish | Mathematical model | 0.99 1 |
| Alshdaifat et al. [65] | Body segmentation | Underwater (Fish4Knowledge) | Clownfish | Faster R-CNN, RPN, modified FCN | 0.95 7 |
| Álvarez Ellacuría et al. [1] | Length | Out of water | European hake | Mask R-CNN | 0.92 6 |
| Baloch et al. [66] | Length of body parts | All | Several fish species | Mathematical morphology, rules | 0.87 1 |
| Bravata et al. [63] | Length, weight, circumference | Out of water | 22 species | CNN | 0.73–0.94 1 |
| Fernandes et al. [67] | Length, weight | Out of water | Nile tilapia | SegNet-based model | 0.95–0.96 5 |
| Garcia et al. [68] | Length | Underwater (controlled) | 7 species | Mask R-CNN | 0.58–0.90 5 |
| Jeong et al. [34] | Length, width | Out of water (conveyor belt) | Flatfish | Mathematical morphology | 0.99 1 |
| Konovalov et al. [69] | Weight | Out of water | Asian Seabass | FCN-8s (CNN) | 0.98 8 |
| Monkman et al. [70] | Length | Out of water | European sea bass | NasNet, ResNet-101, MobileNet | 0.93 5 |
| Muñoz-Benavent et al. [61] | Length | Underwater (uncontrolled) | Bluefin tuna | Mathematical model | 0.93–0.97 1 |
| Palmer et al. [62] | Length | Out of water | Dolphinfish | Mask R-CNN | 0.86 1 |
| Rasmussen et al. [71] | Length (larvae) | Petri dishes | 6 species | Mathematical morphology | 0.97 1 |
| Ravanbakhsh et al. [72] | Body segmentation | Underwater (controlled) | Bluefin tuna | PCA, Haar classifier | 0.90–1.00 1 |
| Rico-Díaz et al. [73] | Length | Underwater (uncontrolled) | 3 species | Hough algorithm, ANN | 0.74 1 |
| Shafait et al. [74] | Length | Underwater (uncontrolled) | Southern Bluefin Tuna | Template matching | 0.90–0.99 1 |
| Tseng et al. [75] | Length | Out of water | 3 species | CNN | 0.96 1 |
| White et al. [76] | Length | Out of water | 7 species | Mathematical model | 1.00 1 |
| Yao et al. [77] | Body segmentation | Out of water | Back crucian carp, common carp | K-means | N/A |
| Yu et al. [78] | Length, width, others | Out of water | Silverfish | Mask R-CNN | 0.97–0.99 1 |
| Yu et al. [79] | Length, width, area | Out of water | Silverfish | Improved U-net | 0.97–0.99 1 |
| Zhang et al. [80] | Weight | Out of water | Crucian carp | BPNN | 0.90 8 |
| Zhang et al. [81] | Body segmentation | Underwater (uncontrolled) | Several | DPANet | 0.85–0.91 1 |
| Zhou et al. [82] | Body segmentation | Out of water | 9 species | Atrous pyramid GAN | 0.96–0.98 5 |
Legend: 1 Accuracy; 5 IoU; 6 Pearson correlation; 7 AP; 8 R2.
Fish are not rigid objects and models must learn how to adapt to changes in posture, position, and scale [1]. High accuracies have been achieved with dead fish in an out-of-water context using techniques based on the deep learning concept [1,56,75], although even in those cases errors can occur due to unfavorable fish poses [70]. Measuring fish underwater has proven to be a much more challenging task, with high accuracies being achieved only under tightly controlled or unrealistic conditions [64,65,72], and even in this case, some kind of manual input is sometimes needed [71]. Despite the difficulties, some progress has been achieved under more challenging conditions [66], with body bending models showing promise when paired with stereo vision systems [61]. Other authors have employed a semi-automatic approach, in which the human user needs to provide some information for the system to perform the measurement accurately [74].
Partial or complete body occlusion is a problem that affects all aspects of image-based fish monitoring and management, but it is particularly troublesome in the context of fish measurement [68,75]. Although statistical methods can partially compensate for the lost information under certain conditions [1], usually errors caused by occlusions are unavoidable [66], even if a semi-automatic approach is employed [74].
Some studies dealt with the problem of measuring different fish body parts for a better characterization of the specimens [66]. One difficulty with this approach is that the limits between different body parts are usually not clear even for experienced evaluators, making the problem relatively ill-defined. This is something intrinsic to the problem, which means that some level of uncertainty will likely always be present.
One aspect of body measurement that is sometimes ignored is that converting from pixels to a standard measurement unit such as centimeters is far from trivial [1]. First, it is necessary to know the exact distance between the fish and the camera in order to estimate the dimensions of each pixel, but such a distance changes through the body contours, so in practice, each pixel has a different conversion factor associated. The task is further complicated by the fact that pixels are not circles, but squares. Thus, the diagonal will be more than 40% longer than any line parallel to the square’s sides. These facts make it nearly impossible to obtain an exact conversion, but properly defined statistical corrections can lead to highly accurate estimates [1]. Proper corrections are also critical to compensate for lens distortion, especially considering the growing use of robust and waterproof action cameras which tend to have significant radial distortion [70].
Most models are trained to have maximum accuracy as the target, which normally means properly balancing false positives and false negatives. However, there are some applications for which one or another type of error can be much more damaging. In the context of measurement, fish need to be first detected and then properly measured. If spurious objects are detected as fish, their measurements will be completely wrong, which in practice may cause problems such as lowering prices paid for the fisherman or skewing inspection efforts [60].
Research on the use of computer vision techniques for measuring fish is still in its infancy. Because many of the studies aim at proving a solid proof of concept instead of generating models ready to be used in practice, the datasets used in such studies are usually limited in terms of both the number of samples and variability [67,72,82]. As the state of the art evolves, more comprehensive databases will be needed (see Section 4). One negative consequence of dataset limitations is that overfitting occurs frequently [63]. Overfitting is a phenomenon in which the model adapts very well to the data used for training but lacks generality to deal with new data, leading to low accuracies. There are a few measures that can be taken to avoid overfitting, such as early training stop and image augmentation applied to the training subset, but the best way to deal with the problem is to increase the number and variability of the training dataset [4,5].
One major reason for the lack of truly representative datasets in the case of fish segmentation and measuring is that the point-level annotations needed in this case are significantly more difficult to acquire than image-level annotations. If the fish population is large, a more efficient approach would be to indicate that the image contains at least one fish, and then let the model locate all the individuals in the image [47], thus effectively automating part of the annotation process. More research effort is needed to improve accuracy in order for this type of approach to become viable.
3.3. Tracking
Many studies dedicated to the detection, counting, measurement, and classification of fish use individual images to reach their goal. However, videos or multiple still images are frequently used in underwater applications. This implies that each fish will likely appear in multiple frames/images, some of which will certainly be more suitable for image analysis. Thus, considering multiple recognition candidates for the same fish seems a reasonable strategy [6,39]. This approach implicitly requires that individual fish be tracked. Fish tracking is also a fundamental step in determining the behavior of individuals or shoals [59,83,84], which in turn is used to detect problems such as diseases [85], lack of oxygenation [86], the presence of ammonia [87] and other pollutants [88], feeding status [58,89], changes in the environment [86], welfare status [90,91], etc. The detection of undesirable behaviors, such as a rise in the willingness to escape from aquaculture tanks, is another important application that has been explored [92,93]. Table 5 shows the studies dedicated to this type of task.
The term “tracking” is adopted here in a broad sense, as it includes not only studies dedicated to determining the trajectory of fish over time but also those focusing on the activity and behavior of fish over time, in which case the exact trajectory may not be as relevant as other cues extracted from videos or sequences of images [84].

Table 5.
List of articles dealing with tracking fish.
Table 5.
List of articles dealing with tracking fish.
| Reference | Target | Context | Species | Main Technique | Accuracy |
|---|---|---|---|---|---|
| Abe et al. [35] | Individual | Underwater (uncontrolled) | Bluefin tuna | SegNet | 0.72 2 |
| Anas et al. [85] | Individual | Underwater (controlled) | Goldfish, tilapia | YOLO, NB, kNN, RF | 0.8–0.9 1 |
| Atienza-Vanacloig et al. [60] | Individual | Underwater (uncontrolled) | Bluefin tuna | Deformable adaptive 2D model | 0.9 1 |
| Boom et al. [94] | Individual | Underwater (Fish4Knowledge) | Several | GMM, APMM, ViBe, Adaboost, SVM | 0.80–0.93 1 |
| Cheng et al. [95] | Individual | Underwater (controlled) | N/A | CNN | 0.93–0.97 2 |
| Delcourt et al. [96] | Individual | Underwater (controlled) | Tilapia | Mathematical morphology | 0.83–0.99 1 |
| Ditria et al. [58] | Individual | Underwater | Luderick | ResNet50 | 0.92 2 |
| Duarte et al. [90] | Individual | Underwater (controlled) | Senegalese sole | Mathematical model | 0.8–0.9 8 |
| Han et al. [86] | Shoal | Underwater (controlled) | Zebrafish | CNN | 0.71–0.82 1 |
| Huang et al. [97] | Individual | Underwater (uncontrolled) | N/A | Kalman filter, SSD, YOLOv2 | 0.94–0.96 1 |
| Li et al. [98] | Individual | Underwater (controlled) | N/A | CMFTNet | 0.66 1 |
| Liu et al. [99] | Individual | Underwater (controlled) | Zebrafish | Mathematical models | 0.95 1 |
| Papadakis et al. [92] | Individual | Underwater (controlled) | Gilthead sea bream | LABView (3rd party software) | N/A |
| Papadakis et al. [93] | Individual | Underwater (controlled) | Sea bass, see bream | Mathematical model | N/A |
| Pérez-Escudero et al. [100] | Individual | Underwater (controlled) | Zebrafish, medaka | Mathematical morphology | 0.99 1 |
| Pinkiewicz et al. [91] | Individual | Underwater (uncontrolled) | Atlantic salmon | Kalman filter | 0.99 1 |
| Qian et al. [59] | Individual | Underwater (controlled) | Zebrafish | Kalman filter, feature matching | 0.96–0.99 1 |
| Qian et al. [83] | Individual | Underwater (controlled) | Zebrafish | Mathematical model | 0.97–0.98 1 |
| Saberioon and Cisar [101] | Individual | Underwater (controlled) | Nile tilapia | Mathematical morphology | 0.97–0.98 1 |
| Sadoul et al. [102] | Shoal | Underwater (controlled) | Rainbow trout | Mathematical model | 0.94 1 |
| Sun et al. [103] | Shoal | Underwater (controlled) | Crucian | K-means | 0.93 1 |
| Teles et al. [104] | Individual | Underwater (controlled) | Zebrafish | PNN, SOM | 0.94 1 |
| Wageeh et al. [105] | Individual | Underwater (controlled) | Goldfish | MSR-YOLO | N/A |
| Wang et al. [106] | Individual | Underwater (controlled) | Zebrafish | CNN | 0.94–0.99 2 |
| Wang et al. [84] | Individual | Underwater (controlled) | Spotted knifejaw | FlowNet2, 3D CNN | 0.95 1 |
| Xia et al. [107] | Individual | Underwater (controlled) | Zebrafish | Mathematical model | 0.98–1.00 1 |
| Xu et al. [87] | Individual | Underwater (controlled) | Goldfish | Faster R-CNN, YOLO-V3 | 0.95–0.98 1 |
| Zhao et al. [88] | Individual | Underwater (controlled) | Red snapper | Thresholding, Kalman filter | 0.98 1 |
Legend: 1 Accuracy; 2 F-score; 8 R2.
There are many challenges that need to be overcome for proper fish tracking. Arguably, the most difficult one is to keep track of large populations containing many visually similar individuals. This is particularly challenging if the intention is to track individual fish instead of whole shoals [35,96]. Occlusions can be particularly insidious because as fish merge and separate, their identities can be swapped, and tracking fails [13]. In order to deal with a problem as complex as this, some authors have employed deep learning techniques such as semantic segmentation [35], which can implicitly extract features from the images which enable more accurate tracking. Other authors adopted a sophisticated multi-step approach designed specifically to deal with this kind of challenge [94]. However, when too little individual information is available, which is usually the case in densely packed shoals with a high rate of occlusions [60], camera-based individual tracking becomes nearly unfeasible. For this reason, some authors have adopted strategies that try to track the shoal as a whole, rather than following individual fish [86,102].
Another challenge is the fact that it is more difficult to detect and track fish as they move farther away from the camera [35]. There are two main reasons for this. First, the farther away the fish are from the camera, the smaller the number of pixels available to characterize the animal. Second, some level of turbidity will almost always be present, so visibility can decrease rapidly with distance. In addition, real underwater fish images are generally of poor quality due to limited range, non-uniform lighting, low contrast, color attenuation, and blurring [60]. These problems can be mitigated using image enhancement and noise reduction techniques such as Retinex-based and bilateral trigonometric filters [35,85], but not completely overcome. A possible way to deal with this issue is to employ multiple cameras bringing an extended field of view, which can be very useful not only to counteract visibility issues but also to meet the requirements of shoal tracking and monitoring [86]. However, the additional equipment may cause costs to rise to unacceptable levels and make it more complex to manage the system and to track across multiple cameras.
Due to body bending while free swimming, the same individual can be observed with very different shapes and fish size and orientation can vary [60]. If not taken into account, this can cause intermittency in the tracking process [59,83]. A solution that is frequently employed in situations such as this is to use deformable models capable of mirroring the actual fish poses [60,91]. Some studies explore the posture patterns of the fish to draw conclusions about their behavior and for early detection of potential problems [107].
Tracking is usually carried out using videos captured with a relatively high frame rate, so when occlusions occur, tracking may resume as soon as the individual reappears a few frames later. However, there are instances in which plants and algae (both moving and static), rocks, or other fish hide a target for too long a time for the tracker to be able to properly resume tracking. In cases such as this, it may be possible to apply statistical techniques (e.g., covariance-based models) to refine tracking decisions [94], but tracking failures are likely to happen from time to time, especially if many fish are being tracked simultaneously [59,101]. If the occlusion is only partial, there are approaches based on deep learning techniques that have achieved some degree of success in avoiding tracking errors [98]. Another solution that has been explored is a multi-view setup in which at least two cameras with different orientations are used simultaneously for tracking [99]. Exploring only the body parts that have more distinctive features, such as the head [106], is another way that has been tested to counterbalance the difficulties involved in tracking large groups of individuals. Under tightly controlled conditions, some studies have been successful in identifying the right individuals and resuming tracking even days after the first detection [100].
As in the case of fish measurement, the majority of studies related to fish tracking are performed using images captured in tanks with at least partially controlled conditions. In addition, many of the methods proposed in the literature require that the data be recorded in shallow tanks with depths of no more than a few centimeters [101]. While these constraints are acceptable in prospective studies, they often are too restrictive for practical use. Thus, further progress depends on investigating new algorithms more adapted to the conditions expected to occur in the real world.
One limitation of many fish tracking studies is that the trajectories are followed in a 2D plane, while real movement occurs in a tridimensional space, thus limiting the conclusions that can be drawn from the data [80,101]. In order to deal with this limitation, some authors have been investigating 3D models more suitable for fish tracking [84,87,95,97,99]. Many of those efforts rely on stereo-vision strategies that require accurate calibration of multiple cameras or unrealistic assumptions about the data acquired, making them unsuitable for real-time tracking [101]. This has led some authors to explore single sensors with the ability to acquire depth information, such as Microsoft’s Kinect, although in this case, the maximum distance for detectability can be limited [101].
3.4. Classification
When multiple species are present, simply counting the number of individuals may not be enough to draw reliable conclusions. In cases such as this, it is necessary to identify the species of each detected individual [108], especially if the objective is to obtain a detailed survey of the fish resources available in a given area [109]. Species classification is also useful for the detection of unwanted or invasive species, so control measures can be adopted [110]. Identification of fish species can be useful for after-catch inspection purposes, as many countries have a list of protected species that should not be fished and vessels may have quotas that should not be exceeded. Additionally, keeping track of fish harvests in a cheaper and more effective way is important to building sustainable and profitable fisheries, a goal that relies heavily on the correct identification and enumeration of different species [111]. Manual classification of fish in images and videos normally requires highly trained specialists and is a demanding, time-consuming, and expensive task, so efficient automatic methods are in high demand [108]. Table 6 shows the studies dedicated to fish classification. All values in the last column are actual accuracies, so in this case, the legend specifying the performance metrics is not shown.

Table 6.
List of articles dealing with the classification of fish.
Table 6.
List of articles dealing with the classification of fish.
| Reference | Target | Context | Species | Main Technique | Accuracy |
|---|---|---|---|---|---|
| Ahmed et al. [112] | Diseased/healthy | Out of water | Salmon | SVM | 0.91–0.94 |
| Allken et al. [113] | Species | Underwater (controlled) | 3 fish species | CNN | 0.94 |
| Alsmadi et al. [114] | Broad classes | Out of water | Several fish species | Memetic algorithm | 0.82–0.90 |
| Alsmadi [115] | Broad classes | Out of water | Several fish species | Hybrid Tabu search, genetic algorithm | 0.82–0.87 |
| Banan et al. [116] | Species | Out of water | Carp (4 species) | CNN | 1.00 |
| Banerjee et al. [117] | Species | Out of water | Carp (3 species) | Deep convolutional autoencoder | 0.97 |
| Boom et al. [94] | Species | Underwater (Fish4Knowledge) | Several | GMM, APMM, ViBe, Adaboost, SVM | 0.80–0.93 |
| Chuang et al. [118] | Species | Underwater (Fish4Knowledge) | Several fish species | Hierarchical partial classifier (SVM) | 0.92–0.97 |
| Coro and Walsh [42] | Size categories | Underwater (uncontrolled) | Tuna, sharks, mantas | YOLOv3 | 0.65–0.75 |
| Hernández-Serna and Jiménez-Segura [119] | Species | Out of water | Several fish species | MLPNN | 0.88–0.92 |
| Hsiao et al. [120] | Species | Underwater (uncontrolled) | Several fish species | SRC-MP | 0.82–0.96 |
| Hu et al. [121] | Species | Out of water | 6 fish species | Multi-class SVM | 0.98 |
| Huang et al. [39] | Species | Underwater (uncontrolled) | 15 fish species | Hierarchical tree, GMM | 0.97 |
| Iqbal et al. [122] | Species | All | 6 fish species | Reduced AlexNet (CNN) | 0.9 |
| Iqbal et al. [89] | Feeding status | Underwater (controlled) | Black scrapers | CNN | 0.98 |
| Ismail et al. [123] | Species | All | 18 species | AlexNet, GoogleNet, ResNet50 | 0.99 |
| Jalal et al. [124] | Species | Underwater (Fish4Knowledge) | Several fish species | YOLO-based model, GMM | 0.8–0.95 |
| Joo et al. [125] | Species | Underwater (controlled) | Cichlids (12 species) | SVM, RF | 0.67–0.78 |
| Ju and Xue [126] | Species | All | Several fish species | AlexNet (CNN) | 0.91–0.97 |
| Knausgård et al. [127] | Species | Underwater (Fish4Knowledge) | 23 fish species | YOLOv3, CNN | 0.84–0.99 |
| Kutlu et al. [128] | Species | Out of water | 25 fish species | kNN | 0.99 |
| Li et al. [129] | Fish face recognition | Underwater (controlled) | Golden crucian carp | Self-SE module, FFRNet | 0.9 |
| Liu et al. [130] | Feeding activity | Underwater (controlled) | Atlantic salmon | Mathematical model | 0.92 |
| Lu et al. [131] | Species | Out of water | 6 species | Modified VGG-16 (CNN) | 0.96 |
| Måløy et al. [132] | Feeding status | Underwater (uncontrolled) | Salmon | DSRN | 0.8 |
| Mana and Sasipraba [133] | Species | Underwater (Fish4Knowledge) | Corkwing, pollack, coalfish | Mask R-CNN, ODKELM | 0.94–0.96 |
| Mathur et al. [134] | Species | Underwater (Fish4Knowledge) | 23 species | ResNet50 (CNN) | 0.98 |
| Meng et al. [135] | Species | Underwater (uncontrolled) | Guppy, snakehead, medaka, neontetora | GoogleNet, AlexNet (CNN) | 0.85–0.87 |
| Ovalle et al. [111] | Species | Out of water | 14 species | Mask R-CNN, MobileNet-V1 | 0.75–0.98 |
| Pramunendar et al. [136] | Species | Underwater (Fish4Knowledge) | 23 species | MLPNN | 0.93–0.96 |
| Qin et al. [137] | Species | Underwater (Fish4Knowledge) | 23 species | SVM, CNN | 0.98 |
| Qiu et al. [138] | Species | Underwater (uncontrolled) | Several | Bilinear CNN | 0.72–0.95 |
| Rauf et al. [139] | Species identification | Out of water | 6 species | 32-layer CNN | 0.85–0.96 |
| Rohani et al. [140] | Fish eggs (dead/alive) | Out of water | Rainbow trout | MLPNN, SVM | 0.99 |
| Saberioon et al. [141] | Feeding status | Out of water (anesthetized fish) | Rainbow trout | RF, SVM, LR, kNN | 0.75–0.82 |
| Saitoh et al. [142] | Species | Out of water | 129 species | RF | 0.30–0.87 |
| Salman et al. [143] | Species | Underwater (Fish4Knowledge) | 15 species | CNN | 0.90 |
| dos Santos and Gonçalves [144] | Species | All | 68 species | CNN | 0.87 |
| Shafait et al. [6] | Species | Underwater (Fish4Knowledge) | 10 species | PCA, nearest neighbor classifier | 0.94 |
| Sharmin et al. [145] | Species | Out of water | 6 species | PCA, SVM | 0.94 |
| Siddiqui et al. [146] | Species | Underwater (uncontrolled) | 16 species | CNN, SVM | 0.94 |
| Smadi et al. [147] | Species | Out of water | 8 species | CNN | 0.98 |
| Spampinato et al. [148] | Species | Underwater (Fish4Knowledge) | 10 species | SIFT, LTP, SVM | 0.85–0.99 |
| Štifanić et al. [149] | Species | Underwater (Fish4Knowledge) | 4 species | CNN | 0.99 |
| Storbeck and Daan [150] | Species | Out of water (conveyor belt) | 6 species | MLPNN | 0.95 |
| Tharwat et al. [151] | Species | Out of water | 4 species | LDA, AdaBoost | 0.96 |
| Ubina et al. [152] | Feeding intensity | Water tank | N/A | 3D CNN | 0.95 |
| Villon et al. [108] | Species | Underwater (uncontrolled) | 20 species | CNN | 0.95 |
| White et al. [76] | Species | Out of water | 7 species | Canonical discriminant analysis | 1.00 |
| Wishkerman et al. [153] | Pigmentation patterns | Out of water | Senegalese sole | GLCM, PCA, LDA | >0.9 |
| Xu et al. [154] | Species | Out of water | 6 species | SE-ResNet152 | 0.91–0.98 |
| Zhang et al. [110] | Species | Out of water | 8 species | AdaBoost | 0.99 |
| Zhang et al. [109] | Species | Underwater (uncontrolled) | 9 species | ResNet50 (CNN) | 0.85–0.90 |
| Zhou et al. [155] | Feeding intensity | Underwater (controlled) | Tilapia | LeNet5 (CNN) | 0.9 |
| Zion et al. [156] | Species | Underwater (controlled) | 3 species | Mathematical model | 0.91–1.00 |
| Zion et al. [157] | Species | Underwater (controlled) | 3 species | Mathematical model | 0.89–1.00 |
The majority of the studies dedicated to fish classification aim at species recognition, but there are a few exceptions. Alsmadi et al. [114] and Alsmadi [115] tried to classify fish into four broad groups: garden, food, predatory, and poisonous. Coro and Walsh [42] proposed a system for counting large fish, so a necessary intermediate step was to classify individuals according to their size. Iqbal et al. [89] classified fish according to their feeding status (normal or underfed), Liu et al. [130] and Måløy et al. [132] tried to classify feeding and non-feeding periods by analyzing consecutive video frames, and Ubina et al. [152] and Zhou et al. [155] categorized feeding intensity according to four classes. Saberioon et al. [141] tested a number of machine learning models to determine the type of diet ingested by rainbow trout (fish or plants). Li et al. [129] aimed at recognizing individual fish from distinctive visual cues in their faces.
Most techniques for classification under unconstrained conditions assume that the fish have already been detected and properly delineated [6], with only a few exceptions [42,146]. This happens because both the detection and the classification tasks are difficult under uncontrolled conditions, and detection errors propagate to the classification case, leading to low accuracies. This is a limitation that needs to be addressed in order to improve the usefulness of the classifiers proposed in the literature.
Species classification using digital images relies on visual cues that need to be clearly visible in order to provide enough information for the classifier to properly perform the task. Under uncontrolled conditions, visibility can be limited and lighting conditions vary unpredictably, making it harder for the models to extract the information needed [94,108,111,118,124,132,143,146], for example, by changing the animal textural features or their contrast with the background [54]. To make matters more complicated, fish can be partially occluded and their orientation with respect to the camera may not be favorable [42,111,113,130,143,146]. Depending on the severity of these effects, the amount of information that can be extracted from the images will not be enough to resolve the species, leading to error [148]. For this reason, authors have proposed some techniques to enhance the images and make them more suitable for information extraction [14,158]. Artificial intelligence techniques, and deep learning in particular, are well suited for extracting information from less than ideal data [108,111], as long as they have been trained to deal with those more challenging conditions [146]. This underlies the importance to train AI models with data that truly represent the entire variability associated with the problem being addressed. With so many variables being at play under real uncontrolled conditions, such a comprehensive image dataset should include a large number of images captured under a wide range of conditions. This is far from trivial, especially considering the intrinsic difficulty of capturing underwater images. One solution that has been increasingly explored is the generation of realistic synthetic images using techniques such as transfer learning, augmentation, and generative adversarial networks (GANs), or simply by pasting images of real fish into synthetic backgrounds [113]. This procedure can certainly increase the robustness of the models under limited real data availability, but in many cases, real data are still essential to meet the requirements of real-world applications [4].
The distinction between species with similar characteristics is another major challenge with no simple solution, especially in an uncontrolled underwater context [113,124,143,144]. The body color of imaged fish, which is one of the major cues used for distinguishing between species, is strongly affected by the fact that different light wavelengths are absorbed at different rates as depth changes [36,52,132], increasing the color variability, and, as a consequence, the color variation between species becomes less evident [131,148]. Color variations that often exist within the same species make the problem even more challenging. In this context, the subtler the differences between species, the better must be the quality of the images used for classification, which has led some authors to use only ideal images (out-of-water images with high contrast between fish and background) in their studies [116,117,121,125]. In an extreme case of a study dealing with similar species, Joo et al. [125] tried to classify 12 species of the Cichlidae family that populate Lake Malawi, reporting that the relatively poor results were due mostly to the model’s inability to capture distinctive traits using the setup adopted in the experiments, the genetic similarity between the species, and potential cross-breeding between species mixing their traits to some degree. In any case, all methods trying to discriminate between relatively similar species employ some kind of machine learning model [35], and the degree of success seems to be directly related to the representativeness of the dataset used for training, reinforcing the importance of creating good quality datasets as discussed in the previous paragraph.
Non-lateral fish views or curved body shapes can lead to loss of critical information [118,146]. In cases such as these, the answer provided by the model might not be reliable. Since it is not possible to explore information that is not there, one way to deal with the problem is to evaluate the confidence level associated with each output provided by the model, and then act accordingly. Chuang et al. [118] proposed a hierarchical partial classifier to deal with the uncertainty and to reduce misclassifications by avoiding making guesses with low confidence. Villon et al. [159] proposed a framework that identifies the fish species and associates a “sure” or “unsure” label based on a confidence threshold applied to the output of the deep learning model.
A problem that arises often in classification problems is the imbalance between classes [94,127,131]. It is common for some classes to have many more samples than others, a situation that may impact negatively the training of artificial intelligence models, as these will tend to strongly favor the most numerous classes. This can lead, for example, to rare species being misidentified, which might be particularly important when conducting fish surveys. The most straightforward way to compensate for this problem is to equalize the classes by either augmenting the smaller classes with artificially generated images (image augmentation) or reducing the larger classes by removing part of the samples for training. More sophisticated approaches in which the classes are properly weighted during training also exist, for instance by using deep learning models with class-balanced focal loss functions [154].
As discussed in Section 3.1, annotation of the reference data can be prone to inconsistencies. In the case of classification problems, the data used to train and validate the models are often unambiguous enough to avoid this type of issue. However, classification tasks that are inherently more ambiguous, such as the analysis of feeding activity [130], can pose annotation difficulties that are at least as damaging as those found in detection and counting applications. Because of these challenges, datasets used for classification purposes are often limited both in terms of the number of samples and representativeness. Models trained under these conditions tend to overfit the data and lack robustness when dealing with new data [132]. Overfitting becomes an even greater problem if the number of classes is high [146].
Due to all challenges mentioned above, some classification problems may not be properly solved by fully automatic methods even when state-of-the-art classifiers are employed, as images may simply not carry enough information to resolve all possible ambiguities. One solution adopted by some authors is to employ a semi-automatic approach [121], in which the human user is required to provide some kind of input to refine the classification, for example by manually correcting possible misclassifications.
4. General Remarks
Almost all articles describing techniques designed to work with images captured underwater under uncontrolled conditions mention image quality degradation as a major source of misestimates and misclassifications [12]. Saleh et al. [22] divided these environmental challenges into five categories: (1) the underwater environment is noisy and has large lighting variation, causing the same object to potentially having a wide range of visual characteristics; (2) underwater scenes are highly dynamic and can change quickly; (3) depth and distance perception can be incorrect due to refraction; (4) images are affected by water turbidity, light scattering, shading, and multiple scattering; (5) image data are frequently under-sampled due to low-resolution cameras and power constraints. Due to the importance of these issues, many authors have proposed different types of techniques to enhance the images and make them more suitable for further analysis [158,160,161,162,163], and review articles on the subject have already been published [14]. It is important to consider, however, that these techniques cannot recover information that has been lost due to poor conditions, rather they can only emphasize image features that are obscured and might be useful for the model being used. One way to tackle this unavoidable loss of information is to employ other types of sensors, such as sonars [164] and near-infrared images [165], and then apply data fusion techniques to merge those different types of data into a meaningful answer [16,22].
As discussed throughout Section 3, the variability associated with images and videos captured under real conditions is very high. As a consequence, in order to cover the conditions found in practice and avoid overfitting [23,63,132], a large amount of images or videos needs to be captured and, considering that the vast majority of methods proposed in the literature require some kind of supervised training, the data collected need to be properly labeled [15,108]. Unsupervised and semi-supervised approaches can drastically decrease the amount of data that needs to be actually labeled, but the former rarely is suitable for fish-related applications and the latter still needs to be properly investigated [15], because it is not clear if existing techniques can be successfully adapted to the characteristics of fish images. Weakly supervised methods which use noisy labels to train the model can also be applied, but the training process becomes substantially more challenging [22].
The labeling process is usually labor-intensive and expensive, especially in the case of fine-grained object recognition, which often requires a deep understanding of the specific domain by the human operator [148]. Those facts make building truly comprehensive and representative datasets a task that is largely out of the reach of most (if not all) research groups. An undesirable consequence of this situation is that most methods found in the literature are developed and tested using data that do not properly represent the conditions likely to be found in a potential practical application [54,146]. At the same time, there is a reluctance or inability of some parties to share annotated datasets, which certainly slows the progress in the development and applications of computer vision techniques for monitoring [45]. The situation is slowly changing though, with at least 10 datasets currently being made available for training and testing computer vision models [45]. One of these datasets, known as Fish4Knowledge (http://www.fish4knowledge.eu/) (accessed on 16 November 2022), has been extensively used in many studies as it provides a large number of images of several marine species of interest. As useful and important an initiative as Fish4Knowledge is, it is not without limitations. This dataset only provides underwater images acquired during the daylight in oligotrophic and transparent coral reef waters [54], conditions that can differ greatly from those potentially facing practical technologies. It is also worth pointing out that most datasets, including Fish4Knowledge, do not cover the characteristics found in marine and freshwater aquaculture [52], resulting in an even deeper data gap. Thus, the challenge of building more realistic datasets is yet to be met.
A possible solution to increase both the number of samples and data variety is to involve individuals outside the research community in the efforts to build datasets, using the principles of citizen science [148,166,167]. There are many incentives that can be applied in order to engage people, including the reward mechanisms extensively used in social networks, early or free access to new technologies and applications, and direct access to ichthyologists, among others. This type of approach may not be applicable in the case of problems that require expert knowledge for proper image annotation [148].
Deep learning models are particularly useful for analyzing underwater images due to their superior ability to model complex and highly nonlinear attributes often found in this type of environment [10,15]. Because deep learning models are capable of finely capturing the characteristics of those attributes, they frequently have trouble dealing with data with characteristics that were not present in the original training dataset. Given the difficulties involved in building truly comprehensive datasets, some alternative solutions have been proposed. Two of those solutions, transfer learning and image augmentation, are widely applied.
Although deep learning architectures can be developed from scratch by properly combining different types of layers (fully connected, convolution, LSTM, etc.), there are several standard architectures available that have been thoroughly tested with a wide range of applications. More importantly, almost all available architectures have versions that were pretrained using massive datasets such as ImageNet [168]. Interestingly, those networks can be retrained for a new application simply by “freezing” most of the layers and using the new data to update only those dedicated to the classification itself [15]. This process, called transfer learning, usually greatly reduces the amount of data needed to properly train the deep learning model while producing results that can be even more accurate than those yielded by models trained from scratch [3,138]. For this reason, this technique is widely employed in many areas of research [169].
Image augmentation is the process of applying different image processing techniques to alter existing images in order to create more data for training the model. It is frequently applied when the data available are deemed insufficient for proper model training. This strategy has been used by almost 20% of the articles cited in this review. Image augmentation must be applied only to the training set after the division of the original dataset. Unfortunately, many authors first apply image augmentation, and only afterward the division into training and test sets is carried out [18,46,122,123,126,135,144,155]. As a result, both sets will contain the exact same samples, with only some minute differences, making it very easy for the model to perform well on the test set. This produces heavily biased results that in no way represent the true accuracy of the proposed model. It is important that both authors and reviewers be aware of this fact, in order to avoid the publication of articles with a methodological error as serious as this.
Improper image augmentation is not the only methodological problem associated with the application of deep learning. The deep learning community is very active and has made available many platforms, architectures, and tutorials, making it relatively straightforward to apply deep learning models without much training. However, the correct application of those models has many subtleties and intricacies that, if not properly addressed, can lead to unrealistic results and render the investigation useless. These intricacies include the number of images used for training, how images are preprocessed, how models are built, how models are fine-tuned, how features are extracted, how features are combined to produce final predictions, what metric is used to assess the model performance, among others [22]. Thus, despite all tools available to assist deep learning users, some effort is needed to achieve results that are actually relevant and useful.
Deep learning models are often viewed as computer-intensive techniques that require powerful and expensive hardware to work effectively [19,23]. While it is true that, depending on the size of the training dataset and the number of parameters to be tuned, the training process can take a long time to be completed even if powerful graphics processing units (GPUs) are employed, in general, the final trained models can be executed quickly and effectively even in devices with limited computational power, including smartphones and small computers such as the Raspberry Pi [170]. Therefore, contrary to what is sometimes claimed in the literature [28], in general, deep learning models do not require considerable investment in dedicated hardware from end users.
Besides the technical difficulties, there are some practical challenges that need to be taken into account, especially for underwater imaging. For example, communications between the underwater cameras and storage servers can be troublesome, with loss of data being always a possibility if expensive redundant systems are not employed [54]. Additionally, debris and algae can accumulate on the lenses, causing blurring and reduction of the field of view [52,54,94]. Underwater systems may also consume too much power, thus requiring an expensive infrastructure to work properly [42]. The excessive storage requirements for long-term monitoring have been mentioned by the authors of [107], who argued that it may be necessary to employ low-resolution images/videos in order to reduce the amount of data to be recorded. Another possible solution for the storage problem is to process the data in real time through edge computing, and store/transmit only the information of interest [3]. It is worth considering, however, that unforeseen new techniques and ideas might greatly benefit from the raw data being discarded, so the benefits of having a robust storage infrastructure can frequently outweigh its drawbacks.
The main challenges and issues identified in this review are synthesized in Figure 2.

Figure 2.
Synthesis of the main challenges and issues identified in this review.
5. Concluding Remarks
The use of computer vision for fish recognition, monitoring, and management has been growing steadily as imaging technologies and artificial intelligence techniques evolve (Figure 3). The literature contains several investigations addressing a wide range of applications. The good results reported in the literature often lead to a false sense of “mission accomplished”, when in fact most studies suffer from severe limitations that prevent the proposed techniques to be readily applicable in the real world. In most cases, those limitations are linked to the fact that collecting good-quality image data on fish is very difficult, especially in an underwater setup. Overcoming the challenges discussed in this review will require considerable effort, but this is essential for enabling practical technologies capable of improving the way fish resources are managed and explored.

Figure 3.
Evolution of the number of computer vision articles applied to fish monitoring and management. Numbers were compiled using the search results described in Section 3.
Knowing the challenges in need of suitable solutions and understanding the real stage of maturity of the techniques and related technologies is very important not only for scientists and researchers but also for entrepreneurs willing to explore the potential market, in order to avoid botched products and services. Other economic sectors for which computer vision and artificial intelligence have been explored for a longer period of time have seen many technology-based companies and startups fail due to the introduction of immature technologies to the market. Beyond the loss caused by those failures, flawed products often ruin the perception of potential customers regarding that particular technology, making it more difficult for new entrants to succeed, even if the product is good. Hopefully, this situation can be avoided in the fish sector.
It is difficult to foresee how the research will progress in the future, especially considering how dynamic and fast changing the computer vision and artificial intelligence areas have been so far. However, two trends seem more likely to continue to gain momentum, the use of deep learning techniques and the application of data fusion principles to bring together the information yielded by different types of data sources. As more representative datasets are collected and made available, the gap between academic results and real-world requirements will tend to close.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Álvarez Ellacuría, A.; Palmer, M.; Catalán, I.A.; Lisani, J.L. Image-based, unsupervised estimation of fish size from commercial landings using deep learning. ICES J. Mar. Sci. 2020, 77, 1330–1339. [Google Scholar] [CrossRef]
- Banno, K.; Kaland, H.; Crescitelli, A.M.; Tuene, S.A.; Aas, G.H.; Gansel, L.C. A novel approach for wild fish monitoring at aquaculture sites: Wild fish presence analysis using computer vision. Aquac. Environ. Interact. 2022, 14, 97–112. [Google Scholar] [CrossRef]
- Saleh, A.; Sheaves, M.; Rahimi Azghadi, M. Computer vision and deep learning for fish classification in underwater habitats: A survey. Fish Fish. 2022, 23, 977–999. [Google Scholar] [CrossRef]
- Ditria, E.M.; Sievers, M.; Lopez-Marcano, S.; Jinks, E.L.; Connolly, R.M. Deep learning for automated analysis of fish abundance: The benefits of training across multiple habitats. Environ. Monit. Assess. 2020, 192, 698. [Google Scholar] [CrossRef] [PubMed]
- Ditria, E.M.; Lopez-Marcano, S.; Sievers, M.; Jinks, E.L.; Brown, C.J.; Connolly, R.M. Automating the Analysis of Fish Abundance Using Object Detection: Optimizing Animal Ecology With Deep Learning. Front. Mar. Sci. 2020, 7. [Google Scholar] [CrossRef]
- Shafait, F.; Mian, A.; Shortis, M.; Ghanem, B.; Culverhouse, P.F.; Edgington, D.; Cline, D.; Ravanbakhsh, M.; Seager, J.; Harvey, E.S. Fish identification from videos captured in uncontrolled underwater environments. ICES J. Mar. Sci. 2016, 73, 2737–2746. [Google Scholar] [CrossRef]
- Noda, J.J.; Travieso, C.M.; Sánchez-Rodríguez, D. Automatic Taxonomic Classification of Fish Based on Their Acoustic Signals. Appl. Sci. 2016, 6, 443. [Google Scholar] [CrossRef]
- Helminen, J.; Linnansaari, T. Object and behavior differentiation for improved automated counts of migrating river fish using imaging sonar data. Fish. Res. 2021, 237, 105883. [Google Scholar] [CrossRef]
- Saberioon, M.; Gholizadeh, A.; Cisar, P.; Pautsina, A.; Urban, J. Application of machine vision systems in aquaculture with emphasis on fish: State-of-the-art and key issues. Rev. Aquac. 2017, 9, 369–387. [Google Scholar] [CrossRef]
- Salman, A.; Siddiqui, S.A.; Shafait, F.; Mian, A.; Shortis, M.R.; Khurshid, K.; Ulges, A.; Schwanecke, U. Automatic fish detection in underwater videos by a deep neural network-based hybrid motion learning system. ICES J. Mar. Sci. 2020, 77, 1295–1307. [Google Scholar] [CrossRef]
- Alsmadi, M.K.; Almarashdeh, I. A survey on fish classification techniques. J. King Saud-Univ.—Comput. Inf. Sci. 2022, 34, 1625–1638. [Google Scholar] [CrossRef]
- An, D.; Hao, J.; Wei, Y.; Wang, Y.; Yu, X. Application of computer vision in fish intelligent feeding system—A review. Aquac. Res. 2021, 52, 423–437. [Google Scholar] [CrossRef]
- Delcourt, J.; Denoël, M.; Ylieff, M.; Poncin, P. Video multitracking of fish behaviour: A synthesis and future perspectives. Fish Fish. 2013, 14, 186–204. [Google Scholar] [CrossRef]
- Han, M.; Lyu, Z.; Qiu, T.; Xu, M. A Review on Intelligence Dehazing and Color Restoration for Underwater Images. IEEE Trans. Syst. Man Cybern. Syst. 2020, 50, 1820–1832. [Google Scholar] [CrossRef]
- Goodwin, M.; Halvorsen, K.T.; Jiao, L.; Knausgård, K.M.; Martin, A.H.; Moyano, M.; Oomen, R.A.; Rasmussen, J.H.; Sørdalen, T.K.; Thorbjørnsen, S.H. Unlocking the potential of deep learning for marine ecology: Overview, applications, and outlook. ICES J. Mar. Sci. 2022, 79, 319–336. [Google Scholar] [CrossRef]
- Li, D.; Wang, Z.; Wu, S.; Miao, Z.; Du, L.; Duan, Y. Automatic recognition methods of fish feeding behavior in aquaculture: A review. Aquaculture 2020, 528, 735508. [Google Scholar] [CrossRef]
- Li, D.; Hao, Y.; Duan, Y. Nonintrusive methods for biomass estimation in aquaculture with emphasis on fish: A review. Rev. Aquac. 2020, 12, 1390–1411. [Google Scholar] [CrossRef]
- Li, D.; Du, L. Recent advances of deep learning algorithms for aquacultural machine vision systems with emphasis on fish. Artif. Intell. Rev. 2022, 55, 4077–4116. [Google Scholar] [CrossRef]
- Li, D.; Wang, Q.; Li, X.; Niu, M.; Wang, H.; Liu, C. Recent advances of machine vision technology in fish classification. ICES J. Mar. Sci. 2022, 79, 263–284. [Google Scholar] [CrossRef]
- Li, D.; Wang, G.; Du, L.; Zheng, Y.; Wang, Z. Recent advances in intelligent recognition methods for fish stress behavior. Aquac. Eng. 2022, 96, 102222. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, H.; Wu, J.; Cui, Z.; Zhang, F. Survey of Fish Behavior Analysis by Computer Vision. J. Aquac. Res. Dev. 2018, 9, 534. [Google Scholar] [CrossRef]
- Saleh, A.; Sheaves, M.; Jerry, D.; Azghadi, M.R. Applications of Deep Learning in Fish Habitat Monitoring: A Tutorial and Survey. arXiv 2022, arXiv:2206.05394. [Google Scholar] [CrossRef]
- Sheaves, M.; Bradley, M.; Herrera, C.; Mattone, C.; Lennard, C.; Sheaves, J.; Konovalov, D.A. Optimizing video sampling for juvenile fish surveys: Using deep learning and evaluation of assumptions to produce critical fisheries parameters. Fish Fish. 2020, 21, 1259–1276. [Google Scholar] [CrossRef]
- Shortis, M.R.; Ravanbakhsh, M.; Shafait, F.; Mian, A. Progress in the Automated Identification, Measurement, and Counting of Fish in Underwater Image Sequences. Mar. Technol. Soc. J. 2016, 50, 4–16. [Google Scholar] [CrossRef]
- Ubina, N.A.; Cheng, S.C. A Review of Unmanned System Technologies with Its Application to Aquaculture Farm Monitoring and Management. Drones 2022, 6, 12. [Google Scholar] [CrossRef]
- Wang, C.; Li, Z.; Wang, T.; Xu, X.; Zhang, X.; Li, D. Intelligent fish farm—The future of aquaculture. Aquac. Int. 2021, 29, 2681–2711. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Fu, L.; Liu, Z.; Liu, H.; Chen, L.; Liu, Y. Aquatic Toxic Analysis by Monitoring Fish Behavior Using Computer Vision: A Recent Progress. J. Toxicol. 2018, 2018, 2591924. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, S.; Liu, J.; Gao, Q.; Dong, S.; Zhou, C. Deep learning for smart fish farming: Applications, opportunities and challenges. Rev. Aquac. 2021, 13, 66–90. [Google Scholar] [CrossRef]
- Yang, L.; Liu, Y.; Yu, H.; Fang, X.; Song, L.; Li, D.; Chen, Y. Computer Vision Models in Intelligent Aquaculture with Emphasis on Fish Detection and Behavior Analysis: A Review. Arch. Comput. Methods Eng. 2021, 28, 2785–2816. [Google Scholar] [CrossRef]
- Application of machine learning in intelligent fish aquaculture: A review. Aquaculture 2021, 540, 736724. [CrossRef]
- Zion, B. The use of computer vision technologies in aquaculture—A review. Computers and Electronics in Agriculture 2012, 88, 125–132. [Google Scholar] [CrossRef]
- Bock, C.H.; Pethybridge, S.J.; Barbedo, J.G.A.; Esker, P.D.; Mahlein, A.K.; Ponte, E.M.D. A phytopathometry glossary for the twenty-first century: Towards consistency and precision in intra- and inter-disciplinary dialogues. Trop. Plant Pathol. 2022, 47, 14–24. [Google Scholar] [CrossRef]
- Follana-Berná, G.; Palmer, M.; Lekanda-Guarrotxena, A.; Grau, A.; Arechavala-Lopez, P. Fish density estimation using unbaited cameras: Accounting for environmental-dependent detectability. J. Exp. Mar. Biol. Ecol. 2020, 527, 151376. [Google Scholar] [CrossRef]
- Jeong, S.J.; Yang, Y.S.; Lee, K.; Kang, J.G.; Lee, D.G. Vision-based Automatic System for Non-contact Measurement of Morphometric Characteristics of Flatfish. J. Electr. Eng. Technol. 2013, 8, 1194–1201. [Google Scholar] [CrossRef]
- Abe, S.; Takagi, T.; Torisawa, S.; Abe, K.; Habe, H.; Iguchi, N.; Takehara, K.; Masuma, S.; Yagi, H.; Yamaguchi, T.; et al. Development of fish spatio-temporal identifying technology using SegNet in aquaculture net cages. Aquac. Eng. 2021, 93, 102146. [Google Scholar] [CrossRef]
- Labao, A.B.; Naval, P.C. Cascaded deep network systems with linked ensemble components for underwater fish detection in the wild. Ecol. Inform. 2019, 52, 103–121. [Google Scholar] [CrossRef]
- French, G.; Fisher, M.; Mackiewicz, M.; Needle, C. Convolutional Neural Networks for Counting Fish in Fisheries Surveillance Video. In Machine Vision of Animals and their Behaviour (MVAB); Amaral, T., Matthews, S., Fisher, R., Eds.; BMVA Press: Dundee, UK, 2015; pp. 7.1–7.10. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, S.; Lu, J.; Wang, H.; Feng, Y.; Shi, C.; Li, D.; Zhao, R. A lightweight dead fish detection method based on deformable convolution and YOLOV4. Comput. Electron. Agric. 2022, 198, 107098. [Google Scholar] [CrossRef]
- Huang, P.; Boom, B.J.; Fisher, R.B. Hierarchical classification with reject option for live fish recognition. Mach. Vis. Appl. 2015, 26, 89–102. [Google Scholar] [CrossRef]
- Aliyu, I.; Gana, K.J.; Musa, A.A.; Adegboye, M.A.; Lim, C.G. Incorporating Recognition in Catfish Counting Algorithm Using Artificial Neural Network and Geometry. KSII Trans. Internet Inf. Syst. 2020, 14, 4866–4888. [Google Scholar] [CrossRef]
- Boudhane, M.; Nsiri, B. Underwater image processing method for fish localization and detection in submarine environment. J. Vis. Commun. Image Represent. 2016, 39, 226–238. [Google Scholar] [CrossRef]
- Coro, G.; Walsh, M.B. An intelligent and cost-effective remote underwater video device for fish size monitoring. Ecol. Inform. 2021, 63, 101311. [Google Scholar] [CrossRef]
- Coronel, L.; Badoy, W.; Namoco, C.S., Jr. Identification of an efficient filtering-segmentation technique for automated counting of fish fingerlings. Int. Arab. J. Inf. Technol. 2018, 15, 708–714. [Google Scholar]
- Costa, C.S.; Zanoni, V.A.G.; Curvo, L.R.V.; de Araújo Carvalho, M.; Boscolo, W.R.; Signor, A.; dos Santos de Arruda, M.; Nucci, H.H.P.; Junior, J.M.; Gonçalves, W.N.; et al. Deep learning applied in fish reproduction for counting larvae in images captured by smartphone. Aquac. Eng. 2022, 97, 102225. [Google Scholar] [CrossRef]
- Ditria, E.M.; Connolly, R.M.; Jinks, E.L.; Lopez-Marcano, S. Annotated Video Footage for Automated Identification and Counting of Fish in Unconstrained Seagrass Habitats. Front. Mar. Sci. 2021, 8, 629485. [Google Scholar] [CrossRef]
- Jiang, L.; Quan, H.; Xie, T.; Qian, J. Fish recognition in complex underwater scenes based on targeted sample transfer learning. Multimed. Tools Appl. 2022, 81, 25303–25317. [Google Scholar] [CrossRef]
- Laradji, I.H.; Saleh, A.; Rodriguez, P.; Nowrouzezahrai, D.; Azghadi, M.R.; Vazquez, D. Weakly supervised underwater fish segmentation using affinity LCFCN. Sci. Rep. 2021, 11, 17379. [Google Scholar] [CrossRef]
- Lee, J.V.; Loo, J.L.; Chuah, Y.D.; Tang, P.Y.; Tan, Y.C.; Goh, W.J. The Use of Vision in a Sustainable Aquaculture Feeding System. Res. J. Appl. Sci. Eng. Technol. 2013, 6, 3658–3669. [Google Scholar] [CrossRef]
- Li, J.; Zhu, K.; Wang, F.; Jiang, F. Deep neural network-based real time fish detection method in the scene of marine fishing supervision. J. Intell. Fuzzy Syst. 2021, 41, 4527–4532. [Google Scholar] [CrossRef]
- Li, J.; Liu, C.; Lu, X.; Wu, B. CME-YOLOv5: An Efficient Object Detection Network for Densely Spaced Fish and Small Targets. Water 2022, 14, 2412. [Google Scholar] [CrossRef]
- Lin, B.; Jiang, K.; Xu, Z.; Li, F.; Li, J.; Mou, C.; Gong, X.; Duan, X. Feasibility Research on Fish Pose Estimation Based on Rotating Box Object Detection. Fishes 2021, 6, 65. [Google Scholar] [CrossRef]
- Liu, H.; Liu, T.; Gu, Y.; Li, P.; Zhai, F.; Huang, H.; He, S. A high-density fish school segmentation framework for biomass statistics in a deep-sea cage. Ecol. Inform. 2021, 64, 101367. [Google Scholar] [CrossRef]
- Marini, S.; Corgnati, L.; Mantovani, C.; Bastianini, M.; Ottaviani, E.; Fanelli, E.; Aguzzi, J.; Griffa, A.; Poulain, P.M. Automated estimate of fish abundance through the autonomous imaging device GUARD1. Measurement 2018, 126, 72–75. [Google Scholar] [CrossRef]
- Marini, S.; Fanelli, E.; Sbragaglia, V.; Azzurro, E.; Fernandez, J.D.R.; Aguzzi, J. Tracking Fish Abundance by Underwater Image Recognition. Sci. Rep. 2018, 8, 13748. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kang, C. A Study on Enhancement of Fish Recognition Using Cumulative Mean of YOLO Network in Underwater Video Images. J. Mar. Sci. Eng. 2020, 8, 952. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Wang, Y.; Zhao, Z.; Liu, J.; Liu, Y.; Sun, C.; Zhou, C. Automatic Fish Population Counting by Machine Vision and a Hybrid Deep Neural Network Model. Animals 2020, 10, 364. [Google Scholar] [CrossRef]
- Li, C.Y.; Guo, J.C.; Cong, R.M.; Pang, Y.W.; Wang, B. Underwater Image Enhancement by Dehazing With Minimum Information Loss and Histogram Distribution Prior. IEEE Trans. Image Process. 2016, 25, 5664–5677. [Google Scholar] [CrossRef]
- Ditria, E.M.; Jinks, E.L.; Connolly, R.M. Automating the analysis of fish grazing behaviour from videos using image classification and optical flow. Anim. Behav. 2021, 177, 31–37. [Google Scholar] [CrossRef]
- Qian, Z.M.; Cheng, X.E.; Chen, Y.Q. Automatically Detect and Track Multiple Fish Swimming in Shallow Water with Frequent Occlusion. PLoS ONE 2014, 9, e106506. [Google Scholar] [CrossRef]
- Atienza-Vanacloig, V.; Andreu-García, G.; López-García, F.; Valiente-González, J.M.; Puig-Pons, V. Vision-based discrimination of tuna individuals in grow-out cages through a fish bending model. Comput. Electron. Agric. 2016, 130, 142–150. [Google Scholar] [CrossRef]
- Muñoz-Benavent, P.; Andreu-García, G.; Valiente-González, J.M.; Atienza-Vanacloig, V.; Puig-Pons, V.; Espinosa, V. Enhanced fish bending model for automatic tuna sizing using computer vision. Comput. Electron. Agric. 2018, 150, 52–61. [Google Scholar] [CrossRef]
- Palmer, M.; Álvarez Ellacuría, A.; Moltó, V.; Catalán, I.A. Automatic, operational, high-resolution monitoring of fish length and catch numbers from landings using deep learning. Fish. Res. 2022, 246, 106166. [Google Scholar] [CrossRef]
- Bravata, N.; Kelly, D.; Eickholt, J.; Bryan, J.; Miehls, S.; Zielinski, D. Applications of deep convolutional neural networks to predict length, circumference, and weight from mostly dewatered images of fish. Ecol. Evol. 2020, 10, 9313–9325. [Google Scholar] [CrossRef] [PubMed]
- Al-Jubouri, Q.; Al-Nuaimy, W.; Al-Taee, M.; Young, I. An automated vision system for measurement of zebrafish length using low-cost orthogonal web cameras. Aquac. Eng. 2017, 78, 155–162. [Google Scholar] [CrossRef]
- Alshdaifat, N.F.F.; Talib, A.Z.; Osman, M.A. Improved deep learning framework for fish segmentation in underwater videos. Ecol. Inform. 2020, 59, 101121. [Google Scholar] [CrossRef]
- Baloch, A.; Ali, M.; Gul, F.; Basir, S.; Afzal, I. Fish Image Segmentation Algorithm (FISA) for Improving the Performance of Image Retrieval System. Int. J. Adv. Comput. Sci. Appl. 2017, 8. [Google Scholar] [CrossRef]
- Fernandes, A.F.; Turra, E.M.; Érika, R. de Alvarenga.; Passafaro, T.L.; Lopes, F.B.; Alves, G.F.; Singh, V.; Rosa, G.J. Deep Learning image segmentation for extraction of fish body measurements and prediction of body weight and carcass traits in Nile tilapia. Comput. Electron. Agric. 2020, 170, 105274. [Google Scholar] [CrossRef]
- Garcia, R.; Prados, R.; Quintana, J.; Tempelaar, A.; Gracias, N.; Rosen, S.; Vågstøl, H.; Løvall, K. Automatic segmentation of fish using deep learning with application to fish size measurement. ICES J. Mar. Sci. 2020, 77, 1354–1366. [Google Scholar] [CrossRef]
- Konovalov, D.; Saleh, A.; Domingos, J.; White, R.; Jerry, D. Estimating Mass of Harvested Asian Seabass Lates calcarifer from Images. World J. Eng. Technol. 2018, 6, 15–23. [Google Scholar] [CrossRef]
- Monkman, G.G.; Hyder, K.; Kaiser, M.J.; Vidal, F.P. Using machine vision to estimate fish length from images using regional convolutional neural networks. Methods Ecol. Evol. 2019, 10, 2045–2056. [Google Scholar] [CrossRef]
- Rasmussen, J.H.; Moyano, M.; Fuiman, L.A.; Oomen, R.A. FishSizer: Software solution for efficiently measuring larval fish size. Ecol. Evol. 2022, 12, e8672. [Google Scholar] [CrossRef]
- Ravanbakhsh, M.; Shortis, M.R.; Shafait, F.; Mian, A.; Harvey, E.S.; Seager, J.W. Automated Fish Detection in Underwater Images Using Shape-Based Level Sets. Photogramm. Rec. 2015, 30, 46–62. [Google Scholar] [CrossRef]
- Rico-Díaz, J.; Rabuñal, J.R.; Gestal, M.; Mures, O.A.; Puertas, J. An Application of Fish Detection Based on Eye Search with Artificial Vision and Artificial Neural Networks. Water 2020, 12, 3013. [Google Scholar] [CrossRef]
- Shafait, F.; Harvey, E.S.; Shortis, M.R.; Mian, A.; Ravanbakhsh, M.; Seager, J.W.; Culverhouse, P.F.; Cline, D.E.; Edgington, D.R. Towards automating underwater measurement of fish length: A comparison of semi-automatic and manual stereo–video measurements. ICES J. Mar. Sci. 2017, 74, 1690–1701. [Google Scholar] [CrossRef]
- Tseng, C.H.; Hsieh, C.L.; Kuo, Y.F. Automatic measurement of the body length of harvested fish using convolutional neural networks. Biosyst. Eng. 2020, 189, 36–47. [Google Scholar] [CrossRef]
- White, D.; Svellingen, C.; Strachan, N. Automated measurement of species and length of fish by computer vision. Fish. Res. 2006, 80, 203–210. [Google Scholar] [CrossRef]
- Yao, H.; Duan, Q.; Li, D.; Wang, J. An improved K-means clustering algorithm for fish image segmentation. Math. Comput. Model. 2013, 58, 790–798. [Google Scholar] [CrossRef]
- Yu, C.; Fan, X.; Hu, Z.; Xia, X.; Zhao, Y.; Li, R.; Bai, Y. Segmentation and measurement scheme for fish morphological features based on Mask R-CNN. Inf. Process. Agric. 2020, 7, 523–534. [Google Scholar] [CrossRef]
- Yu, C.; Hu, Z.; Han, B.; Wang, P.; Zhao, Y.; Wu, H. Intelligent Measurement of Morphological Characteristics of Fish Using Improved U-Net. Electronics 2021, 10, 1426. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, J.; Duan, Q. Estimation for fish mass using image analysis and neural network. Comput. Electron. Agric. 2020, 173, 105439. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, C.; Bao, Z. DPANet: Dual Pooling-aggregated Attention Network for fish segmentation. IET Comput. Vis. 2022, 16, 67–82. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, S.; Ren, Y.; Zhang, Y.; Fu, J.; Fan, D.; Lin, J.; Wang, Q. Atrous Pyramid GAN Segmentation Network for Fish Images with High Performance. Electronics 2022, 11, 911. [Google Scholar] [CrossRef]
- Qian, Z.M.; Wang, S.H.; Cheng, X.E.; Chen, Y.Q. An effective and robust method for tracking multiple fish in video image based on fish head detection. BMC Bioinform. 2016, 17, 251. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Muhammad, A.; Liu, C.; Du, L.; Li, D. Automatic Recognition of Fish Behavior with a Fusion of RGB and Optical Flow Data Based on Deep Learning. Animals 2021, 11, 2774. [Google Scholar] [CrossRef] [PubMed]
- Anas, O.; Wageeh, Y.; Mohamed, H.E.D.; Fadl, A.; ElMasry, N.; Nabil, A.; Atia, A. Detecting Abnormal Fish Behavior Using Motion Trajectories in Ubiquitous Environments. Procedia Comput. Sci. 2020, 175, 141–148. [Google Scholar] [CrossRef]
- Han, F.; Zhu, J.; Liu, B.; Zhang, B.; Xie, F. Fish Shoals Behavior Detection Based on Convolutional Neural Network and Spatiotemporal Information. IEEE Access 2020, 8, 126907–126926. [Google Scholar] [CrossRef]
- Xu, W.; Zhu, Z.; Ge, F.; Han, Z.; Li, J. Analysis of Behavior Trajectory Based on Deep Learning in Ammonia Environment for Fish. Sensors 2020, 20, 4425. [Google Scholar] [CrossRef]
- Zhao, X.; Yan, S.; Gao, Q. An Algorithm for Tracking Multiple Fish Based on Biological Water Quality Monitoring. IEEE Access 2019, 7, 15018–15026. [Google Scholar] [CrossRef]
- Iqbal, U.; Li, D.; Akhter, M. Intelligent Diagnosis of Fish Behavior Using Deep Learning Method. Fishes 2022, 7, 201. [Google Scholar] [CrossRef]
- Duarte, S.; Reig, L.; Oca, J. Measurement of sole activity by digital image analysis. Aquac. Eng. 2009, 41, 22–27. [Google Scholar] [CrossRef]
- Pinkiewicz, T.; Purser, G.; Williams, R. A computer vision system to analyse the swimming behaviour of farmed fish in commercial aquaculture facilities: A case study using cage-held Atlantic salmon. Aquac. Eng. 2011, 45, 20–27. [Google Scholar] [CrossRef]
- Papadakis, V.M.; Papadakis, I.E.; Lamprianidou, F.; Glaropoulos, A.; Kentouri, M. A computer-vision system and methodology for the analysis of fish behavior. Aquac. Eng. 2012, 46, 53–59. [Google Scholar] [CrossRef]
- Papadakis, V.M.; Glaropoulos, A.; Kentouri, M. Sub-second analysis of fish behavior using a novel computer-vision system. Aquac. Eng. 2014, 62, 36–41. [Google Scholar] [CrossRef]
- Boom, B.J.; He, J.; Palazzo, S.; Huang, P.X.; Beyan, C.; Chou, H.M.; Lin, F.P.; Spampinato, C.; Fisher, R.B. A research tool for long-term and continuous analysis of fish assemblage in coral-reefs using underwater camera footage. Ecol. Inform. 2014, 23, 83–97. [Google Scholar] [CrossRef]
- Cheng, X.E.; Du, S.S.; Li, H.Y.; Hu, J.F.; Chen, M.L. Obtaining three-dimensional trajectory of multiple fish in water tank via video tracking. Multimed. Tools Appl. 2018, 77, 24499–24519. [Google Scholar] [CrossRef]
- Delcourt, J.; Becco, C.; Vandewalle, N.; Poncin, P. A video multitracking system for quantification of individual behavior in a large fish shoal: Advantages and limits. Behav. Res. Methods 2009, 41, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.W.; Hwang, J.N.; Romain, S.; Wallace, F. Fish Tracking and Segmentation From Stereo Videos on the Wild Sea Surface for Electronic Monitoring of Rail Fishing. IEEE Trans. Circuits Syst. Video Technol. 2019, 29, 3146–3158. [Google Scholar] [CrossRef]
- Li, W.; Li, F.; Li, Z. CMFTNet: Multiple fish tracking based on counterpoised JointNet. Comput. Electron. Agric. 2022, 198, 107018. [Google Scholar] [CrossRef]
- Liu, X.; Yue, Y.; Shi, M.; Qian, Z.M. 3-D Video Tracking of Multiple Fish in a Water Tank. IEEE Access 2019, 7, 145049–145059. [Google Scholar] [CrossRef]
- Pérez-Escudero, A.; Vicente-Page, J.; Hinz, R.C.; Arganda, S.; de Polavieja, G.G. idTracker: Tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 2014, 11, 743–748. [Google Scholar] [CrossRef]
- Saberioon, M.; Cisar, P. Automated multiple fish tracking in three-Dimension using a Structured Light Sensor. Comput. Electron. Agric. 2016, 121, 215–221. [Google Scholar] [CrossRef]
- Sadoul, B.; Evouna Mengues, P.; Friggens, N.; Prunet, P.; Colson, V. A new method for measuring group behaviours of fish shoals from recorded videos taken in near aquaculture conditions. Aquaculture 2014, 430, 179–187. [Google Scholar] [CrossRef]
- Sun, L.; Luo, B.; Liu, T.; Liu, Y.; Wei, Y. Algorithm of Adaptive Fast Clustering for Fish Swarm Color Image Segmentation. IEEE Access 2019, 7, 178753–178762. [Google Scholar] [CrossRef]
- Teles, L.O.; Fernandes, M.; Amorim, J.; Vasconcelos, V. Video-tracking of zebrafish (Danio rerio) as a biological early warning system using two distinct artificial neural networks: Probabilistic neural network (PNN) and self-organizing map (SOM). Aquat. Toxicol. 2015, 165, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Wageeh, Y.; Mohamed, H.E.D.; Fadl, A.; Anas, O.; ElMasry, N.; Nabil, A.; Atia, A. YOLO fish detection with Euclidean tracking in fish farms. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 5–12. [Google Scholar] [CrossRef]
- Wang, S.H.; Zhao, J.W.; Chen, Y.Q. Robust tracking of fish schools using CNN for head identification. Multimed. Tools Appl. 2017, 76, 23679–23697. [Google Scholar] [CrossRef]
- Xia, C.; Chon, T.S.; Liu, Y.; Chi, J.; Lee, J.M. Posture tracking of multiple individual fish for behavioral monitoring with visual sensors. Ecol. Inform. 2016, 36, 190–198. [Google Scholar] [CrossRef]
- Villon, S.; Mouillot, D.; Chaumont, M.; Darling, E.S.; Subsol, G.; Claverie, T.; Villéger, S. A Deep learning method for accurate and fast identification of coral reef fishes in underwater images. Ecol. Inform. 2018, 48, 238–244. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, W.; Zhu, Y.; Han, W.; Huang, Y.; Li, J. Research on fish identification in tropical waters under unconstrained environment based on transfer learning. Earth Sci. Inform. 2022, 15, 1155–1166. [Google Scholar] [CrossRef]
- Zhang, D.; Lee, D.J.; Zhang, M.; Tippetts, B.J.; Lillywhite, K.D. Object recognition algorithm for the automatic identification and removal of invasive fish. Biosyst. Eng. 2016, 145, 65–75. [Google Scholar] [CrossRef]
- Ovalle, J.C.; Vilas, C.; Antelo, L.T. On the use of deep learning for fish species recognition and quantification on board fishing vessels. Mar. Policy 2022, 139, 105015. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Aurpa, T.T.; Azad, M.A.K. Fish Disease Detection Using Image Based Machine Learning Technique in Aquaculture. J. King Saud Univ.—Comput. Inf. Sci. 2022, 34, 5170–5182. [Google Scholar] [CrossRef]
- Allken, V.; Handegard, N.O.; Rosen, S.; Schreyeck, T.; Mahiout, T.; Malde, K. Fish species identification using a convolutional neural network trained on synthetic data. ICES J. Mar. Sci. 2019, 76, 342–349. [Google Scholar] [CrossRef]
- Alsmadi, M.K.; Tayfour, M.; Alkhasawneh, R.A.; Badawi, U.; Almarashdeh, I.; Haddad, F. Robust feature extraction methods for general fish classification. Int. J. Electr. Comput. Eng. 2019, 9, 5192–5204. [Google Scholar] [CrossRef]
- Alsmadi, M.K. Hybrid Genetic Algorithm with Tabu Search with Back-Propagation Algorithm for Fish Classification: Determining the Appropriate Feature Set. Int. J. Appl. Eng. Res. 2019, 14, 4387–4396. [Google Scholar]
- Banan, A.; Nasiri, A.; Taheri-Garavand, A. Deep learning-based appearance features extraction for automated carp species identification. Aquac. Eng. 2020, 89, 102053. [Google Scholar] [CrossRef]
- Banerjee, A.; Das, A.; Behra, S.; Bhattacharjee, D.; Srinivasan, N.T.; Nasipuri, M.; Das, N. Carp-DCAE: Deep convolutional autoencoder for carp fish classification. Comput. Electron. Agric. 2022, 196, 106810. [Google Scholar] [CrossRef]
- Chuang, M.C.; Hwang, J.N.; Williams, K. A Feature Learning and Object Recognition Framework for Underwater Fish Images. IEEE Trans. Image Process. 2016, 25, 1862–1872. [Google Scholar] [CrossRef]
- Hernández-Serna, A.; Jiménez-Segura, L.F. Automatic identification of species with neural networks. PeerJ 2014, 2, e563. [Google Scholar] [CrossRef]
- Hsiao, Y.H.; Chen, C.C.; Lin, S.I.; Lin, F.P. Real-world underwater fish recognition and identification, using sparse representation. Ecol. Inform. 2014, 23, 13–21. [Google Scholar] [CrossRef]
- Hu, J.; Li, D.; Duan, Q.; Han, Y.; Chen, G.; Si, X. Fish species classification by color, texture and multi-class support vector machine using computer vision. Comput. Electron. Agric. 2012, 88, 133–140. [Google Scholar] [CrossRef]
- Iqbal, M.A.; Wang, Z.; Ali, Z.A.; Riaz, S. Automatic Fish Species Classification Using Deep Convolutional Neural Networks. Wirel. Pers. Commun. 2021, 116, 1043–1053. [Google Scholar] [CrossRef]
- Ismail, H.; Ayob, A.F.M.; Muslim, A.M.S.M.; Zulkifi, M.F.R. Convolutional neural network architectures performance evaluation for fish species classification. J. Sustain. Sci. Manag. 2021, 16, 124–139. [Google Scholar] [CrossRef]
- Jalal, A.; Salman, A.; Mian, A.; Shortis, M.; Shafait, F. Fish detection and species classification in underwater environments using deep learning with temporal information. Ecol. Inform. 2020, 57, 101088. [Google Scholar] [CrossRef]
- Joo, D.; Kwan, Y.s.; Song, J.; Pinho, C.; Hey, J.; Won, Y.J. Identification of Cichlid Fishes from Lake Malawi Using Computer Vision. PLOS ONE 2013, 8, e77686. [Google Scholar] [CrossRef]
- Ju, Z.; Xue, Y. Fish species recognition using an improved AlexNet model. Optik 2020, 223, 165499. [Google Scholar] [CrossRef]
- Knausgård, K.M.; Wiklund, A.; Sørdalen, T.K.; Halvorsen, K.T.; Kleiven, A.R.; Jiao, L.; Goodwin, M. Temperate fish detection and classification: A deep learning based approach. Appl. Intell. 2022, 52, 6988–7001. [Google Scholar] [CrossRef]
- Kutlu, Y.; Iscimen, B.; Turan, C. Multi-Stage Fish Classification System Using Morphometry. Fresenius Environ. Bull. 2017, 26, 1911–1917. [Google Scholar]
- Li, D.; Su, H.; Jiang, K.; Liu, D.; Duan, X. Fish Face Identification Based on Rotated Object Detection: Dataset and Exploration. Fishes 2022, 7, 219. [Google Scholar] [CrossRef]
- Liu, Z.; Li, X.; Fan, L.; Lu, H.; Liu, L.; Liu, Y. Measuring feeding activity of fish in RAS using computer vision. Aquac. Eng. 2014, 60, 20–27. [Google Scholar] [CrossRef]
- Lu, Y.C.; Tung, C.; Kuo, Y.F. Identifying the species of harvested tuna and billfish using deep convolutional neural networks. ICES J. Mar. Sci. 2020, 77, 1318–1329. [Google Scholar] [CrossRef]
- Måløy, H.; Aamodt, A.; Misimi, E. A spatio-temporal recurrent network for salmon feeding action recognition from underwater videos in aquaculture. Comput. Electron. Agric. 2019, 167, 105087. [Google Scholar] [CrossRef]
- Mana, S.C.; Sasipraba, T. An Intelligent Deep Learning Enabled Marine Fish Species Detection and Classification Model. Int. J. Artif. Intell. Tools 2022, 31, 2250017. [Google Scholar] [CrossRef]
- Mathur, M.; Vasudev, D.; Sahoo, S.; Jain, D.; Goel, N. Crosspooled FishNet: Transfer learning based fish species classification model. Multimed. Tools Appl. 2020, 79, 31625–31643. [Google Scholar] [CrossRef]
- Meng, L.; Hirayama, T.; Oyanagi, S. Underwater-Drone With Panoramic Camera for Automatic Fish Recognition Based on Deep Learning. IEEE Access 2018, 6, 17880–17886. [Google Scholar] [CrossRef]
- Pramunendar, R.A.; Prabowo, D.P.; Pergiwati, D.; Sari, Y.; Andono, P.N.; Soeleman, M.A. New Workflow for Marine Fish Classification Based on Combination Features and CLAHE Enhancement Technique. Int. J. Intell. Eng. Syst. 2020, 13, 293–304. [Google Scholar] [CrossRef]
- Qin, H.; Li, X.; Liang, J.; Peng, Y.; Zhang, C. DeepFish: Accurate underwater live fish recognition with a deep architecture. Neurocomputing 2016, 187, 49–58. [Google Scholar] [CrossRef]
- Qiu, C.; Zhang, S.; Wang, C.; Yu, Z.; Zheng, H.; Zheng, B. Improving Transfer Learning and Squeeze- and-Excitation Networks for Small-Scale Fine-Grained Fish Image Classification. IEEE Access 2018, 6, 78503–78512. [Google Scholar] [CrossRef]
- Rauf, H.T.; Lali, M.I.U.; Zahoor, S.; Shah, S.Z.H.; Rehman, A.U.; Bukhari, S.A.C. Visual features based automated identification of fish species using deep convolutional neural networks. Comput. Electron. Agric. 2019, 167, 105075. [Google Scholar] [CrossRef]
- Rohani, A.; Taki, M.; Bahrami, G. Application of artificial intelligence for separation of live and dead rainbow trout fish eggs. Artif. Intell. Agric. 2019, 1, 27–34. [Google Scholar] [CrossRef]
- Saberioon, M.; Císař, P.; Labbé, L.; Souček, P.; Pelissier, P.; Kerneis, T. Comparative Performance Analysis of Support Vector Machine, Random Forest, Logistic Regression and k-Nearest Neighbours in Rainbow Trout (Oncorhynchus Mykiss) Classification Using Image-Based Features. Sensors 2018, 18, 1027. [Google Scholar] [CrossRef]
- Saitoh, T.; Shibata, T.; Miyazono, T. Feature Points based Fish Image Recognition. Int. J. Comput. Inf. Syst. Ind. Manag. Appl. 2016, 8, 12–22. [Google Scholar]
- Salman, A.; Jalal, A.; Shafait, F.; Mian, A.; Shortis, M.; Seager, J.; Harvey, E. Fish species classification in unconstrained underwater environments based on deep learning. Limnol. Oceanogr. Methods 2016, 14, 570–585. [Google Scholar] [CrossRef]
- dos Santos, A.A.; Gonçalves, W.N. Improving Pantanal fish species recognition through taxonomic ranks in convolutional neural networks. Ecol. Inform. 2019, 53, 100977. [Google Scholar] [CrossRef]
- Sharmin, I.; Islam, N.F.; Jahan, I.; Joye, T.A.; Rahman, M.R.; Habib, M.T. Machine vision based local fish recognition. SN Appl. Sci. 2019, 1, 1529. [Google Scholar] [CrossRef]
- Siddiqui, S.A.; Salman, A.; Malik, M.I.; Shafait, F.; Mian, A.; Shortis, M.R.; Harvey, E.S. Automatic fish species classification in underwater videos: Exploiting pre-trained deep neural network models to compensate for limited labelled data. ICES J. Mar. Sci. 2018, 75, 374–389. [Google Scholar] [CrossRef]
- Smadi, A.A.; Mehmood, A.; Abugabah, A.; Almekhlafi, E.; Al-Smadi, A.M. Deep convolutional neural network-based system for fish classification. Int. J. Electr. Comput. Eng. 2022, 12, 2026–2039. [Google Scholar] [CrossRef]
- Spampinato, C.; Palazzo, S.; Joalland, P.H.; Paris, S.; Glotin, H.; Blanc, K.; Lingrand, D.; Precioso, F. Fine-grained object recognition in underwater visual data. Multimed. Tools Appl. 2016, 75, 131–142. [Google Scholar] [CrossRef]
- Štifanić, D.; Musulin, J.; Car, Z.; Čep, R. Use of Convolutional Neural Network for Fish Species Classification. Pomor. Zb. 2020, 59, 131–142. [Google Scholar] [CrossRef]
- Storbeck, F.; Daan, B. Fish species recognition using computer vision and a neural network. Fish. Res. 2001, 51, 11–15. [Google Scholar] [CrossRef]
- Tharwat, A.; Hemedan, A.A.; Hassanien, A.E.; Gabel, T. A biometric-based model for fish species classification. Fish. Res. 2018, 204, 324–336. [Google Scholar] [CrossRef]
- Ubina, N.; Cheng, S.C.; Chang, C.C.; Chen, H.Y. Evaluating fish feeding intensity in aquaculture with convolutional neural networks. Aquac. Eng. 2021, 94, 102178. [Google Scholar] [CrossRef]
- Wishkerman, A.; Boglino, A.; Darias, M.J.; Andree, K.B.; Estévez, A.; Gisbert, E. Image analysis-based classification of pigmentation patterns in fish: A case study of pseudo-albinism in Senegalese sole. Aquaculture 2016, 464, 303–308. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Duan, Q. Transfer learning and SE-ResNet152 networks-based for small-scale unbalanced fish species identification. Comput. Electron. Agric. 2021, 180, 105878. [Google Scholar] [CrossRef]
- Zhou, C.; Xu, D.; Chen, L.; Zhang, S.; Sun, C.; Yang, X.; Wang, Y. Evaluation of fish feeding intensity in aquaculture using a convolutional neural network and machine vision. Aquaculture 2019, 507, 457–465. [Google Scholar] [CrossRef]
- Zion, B.; Shklyar, A.; Karplus, I. In-vivo fish sorting by computer vision. Aquac. Eng. 2000, 22, 165–179. [Google Scholar] [CrossRef]
- Zion, B.; Shklyar, A.; Karplus, I. Sorting fish by computer vision. Comput. Electron. Agric. 1999, 23, 175–187. [Google Scholar] [CrossRef]
- Zhou, C.; Yang, X.; Zhang, B.; Lin, K.; Xu, D.; Guo, Q.; Sun, C. An adaptive image enhancement method for a recirculating aquaculture system. Sci. Rep. 2017, 7, 6243. [Google Scholar] [CrossRef]
- Villon, S.; Mouillot, D.; Chaumont, M.; Subsol, G.; Claverie, T.; Villéger, S. A new method to control error rates in automated species identification with deep learning algorithms. Sci. Rep. 2020, 10, 10972. [Google Scholar] [CrossRef]
- Islam, M.J.; Xia, Y.; Sattar, J. Fast Underwater Image Enhancement for Improved Visual Perception. IEEE Robot. Autom. Lett. 2020, 5, 3227–3234. [Google Scholar] [CrossRef]
- Ma, J.; Fan, X.; Yang, S.X.; Zhang, X.; Zhu, X. Contrast Limited Adaptive Histogram Equalization-Based Fusion in YIQ and HSI Color Spaces for Underwater Image Enhancement. Int. J. Pattern Recognit. Artif. Intell. 2018, 32, 1854018. [Google Scholar] [CrossRef]
- Abdul Ghani, A.S.; Mat Isa, N.A. Underwater image quality enhancement through integrated color model with Rayleigh distribution. Appl. Soft Comput. 2015, 27, 219–230. [Google Scholar] [CrossRef]
- Ancuti, C.O.; Ancuti, C.; De Vleeschouwer, C.; Bekaert, P. Color Balance and Fusion for Underwater Image Enhancement. IEEE Trans. Image Process. 2018, 27, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Tarling, P.; Cantor, M.; Clapés, A.; Escalera, S. Deep learning with self-supervision and uncertainty regularization to count fish in underwater images. PLoS ONE 2022, 17, e0267759. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, K.; Xu, D.; Chen, L.; Guo, Q.; Sun, C.; Yang, X. Near infrared computer vision and neuro-fuzzy model-based feeding decision system for fish in aquaculture. Comput. Electron. Agric. 2018, 146, 114–124. [Google Scholar] [CrossRef]
- Irwin, A. Citizen Science: A Study of People, Expertise and Sustainable Development, 1st ed.; Routledge: London, UK, 2002. [Google Scholar]
- Barbedo, J.G.A. Deep learning applied to plant pathology: The problem of data representativeness. Trop. Plant Pathol. 2022, 47, 85–94. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Deng, Y. Deep learning on mobile devices: A review. In Mobile Multimedia/Image Processing, Security, and Applications 2019; Agaian, S.S., Asari, V.K., DelMarco, S.P., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2019; Volume 10993, p. 109930A. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).