Effects of Glutamine Starvation on SHVV Replication by Quantitative Proteomics Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Cells
2.2. CCO Cell Culture and SHVV Infection
2.3. Protein Extraction and Quantitation
2.4. Protein Alkylation and Trypsin Digestion
2.5. Label-Free Lc-Ms/MS Analysis
2.6. Database Search and DEPs Screen
2.7. Bioinformatics Analysis
2.8. Statistical Analysis
3. Results
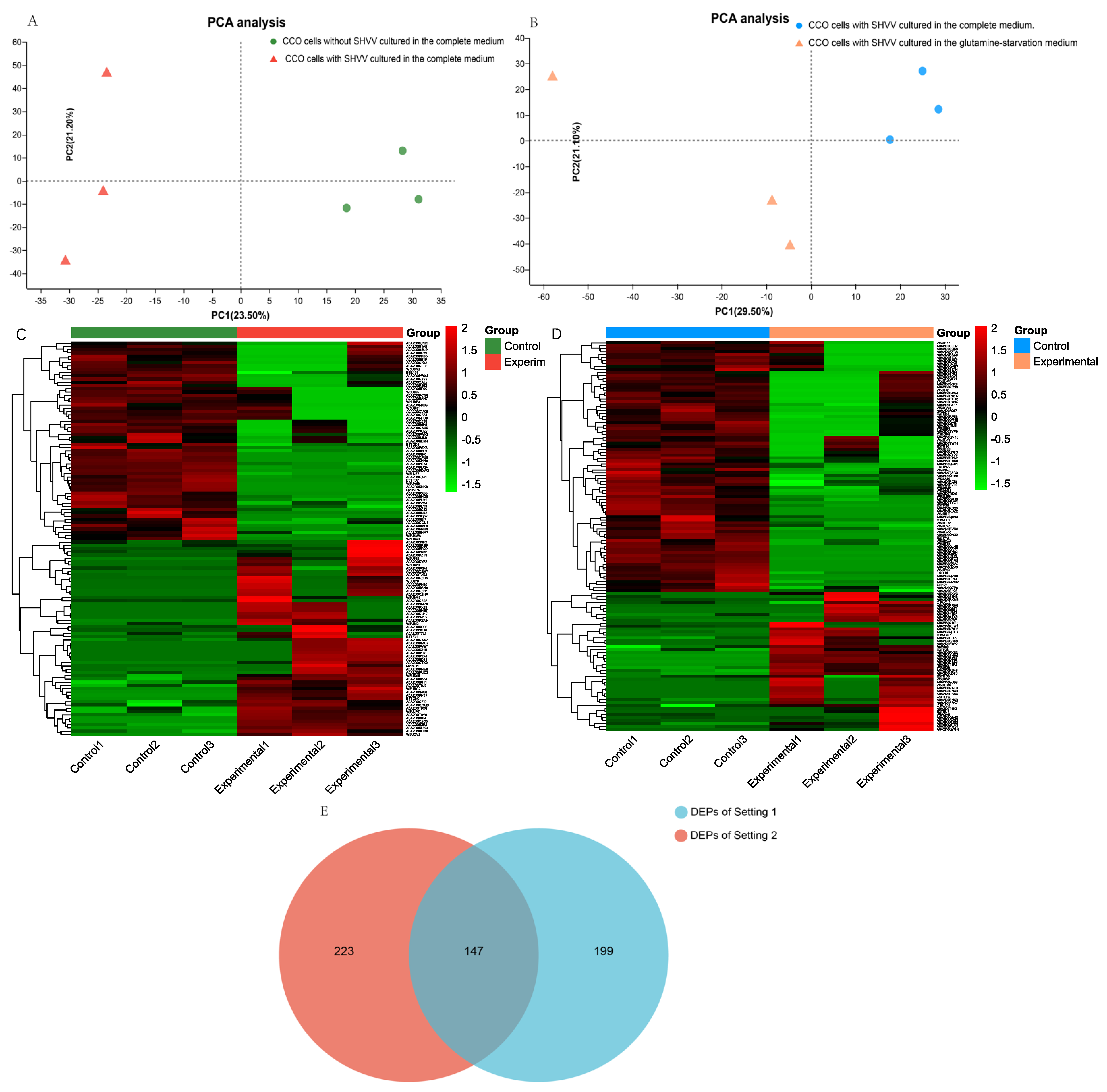
3.1. Experimental Design and the Identification of Differentially Expressed Proteins (DEPs)
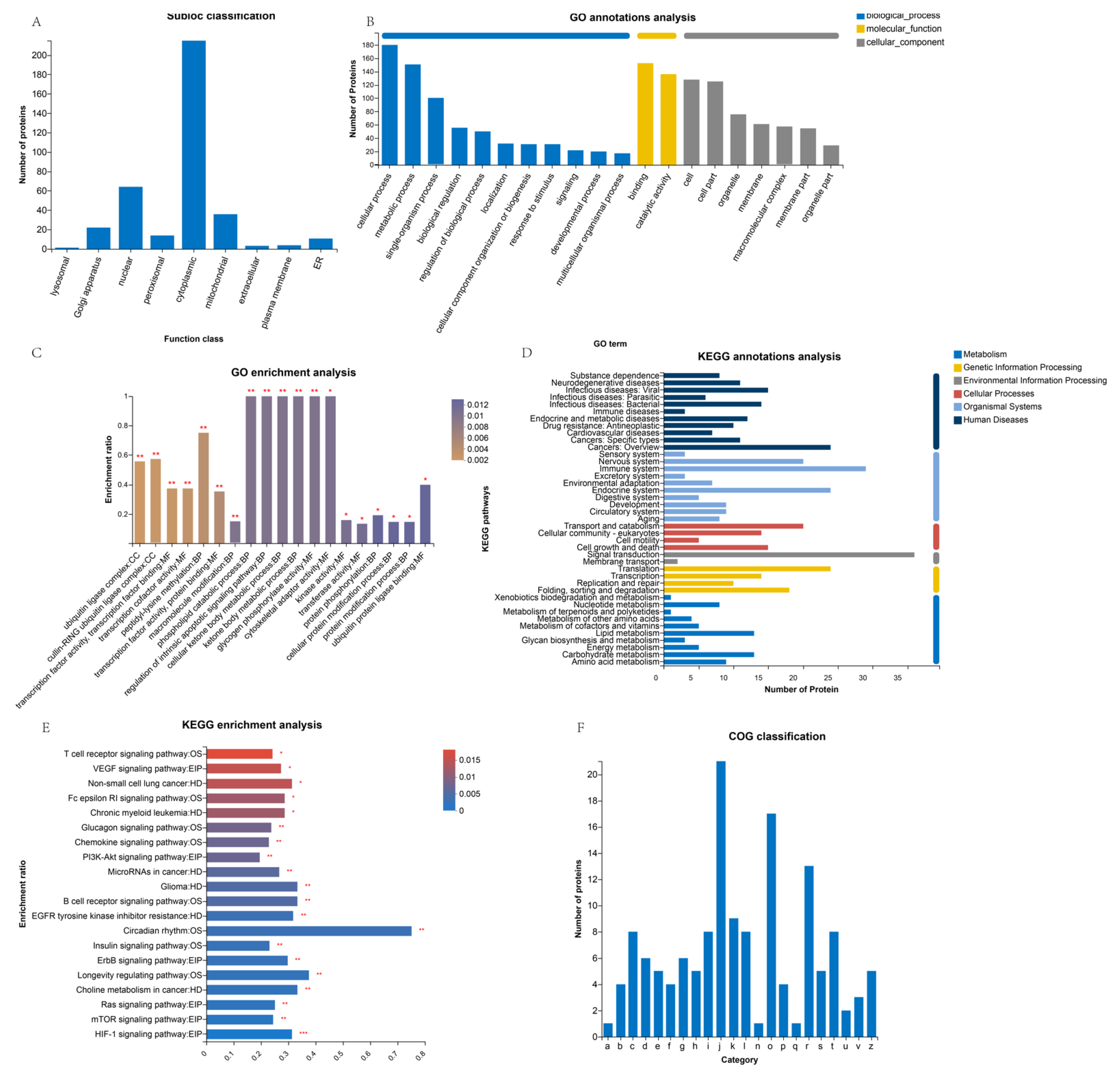
3.2. Subcellular Localization of Differentially Expressed Proteins (DEPs)
3.3. GO Analysis of Differentially Expressed Proteins (DEPs)
3.4. KEGG Pathway Analysis of Differentially Expressed Proteins (DEPs)
3.5. COG Analysis of Differentially Expressed Proteins (DEPs)
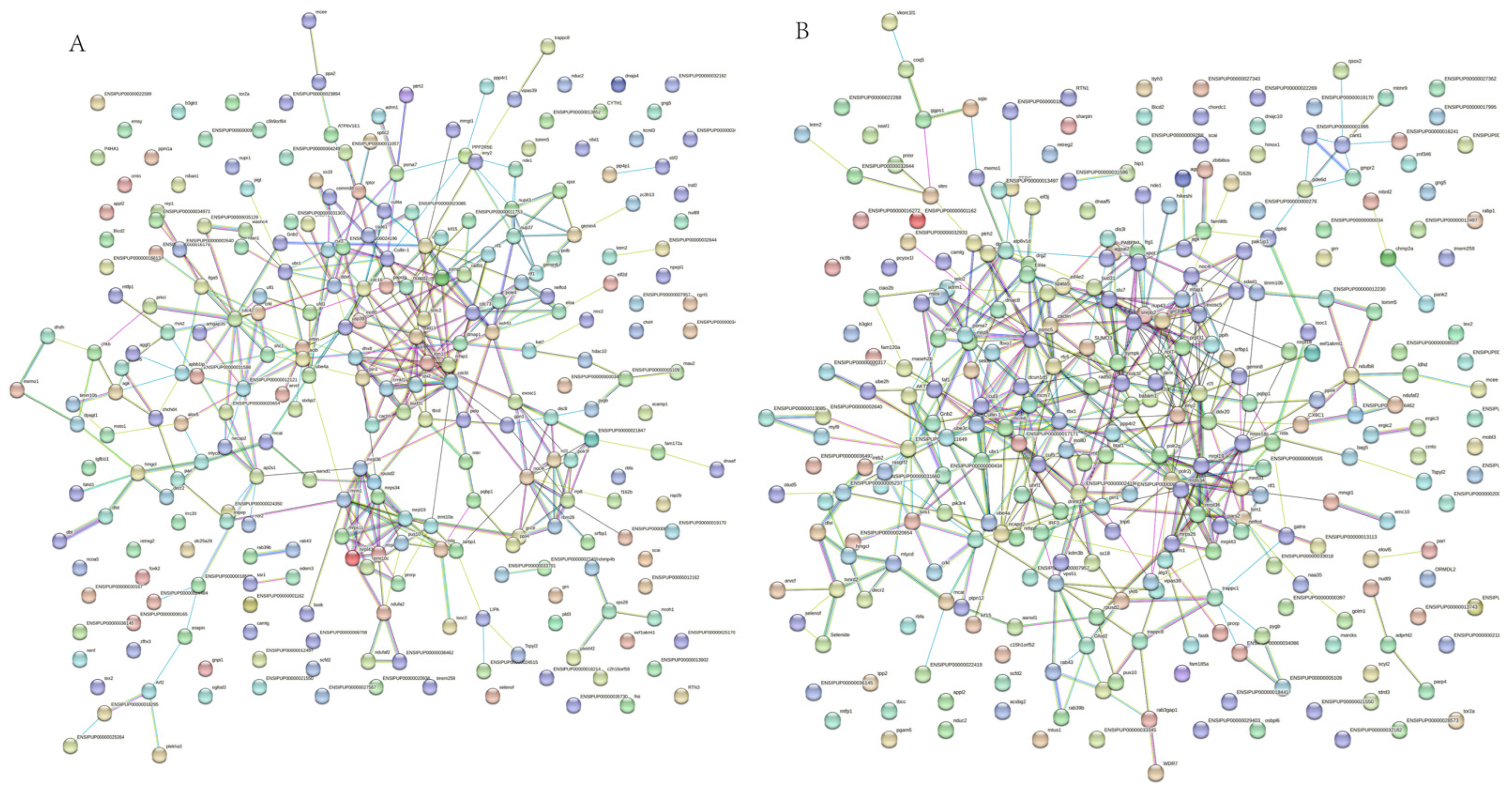
3.6. Protein–Protein Interaction Network of Differentially Expressed Proteins (DEPs)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RIPA Lysis Buffer | Radio Immunoprecipitation Assay Lysis buffer |
| Tris-HCl | TRIS hydrochloride |
| EDTA | Ethylene Diamine Tetraacetic Acid |
| NP-40 | Nonidet P 40 |
| SDS | Sodium dodecyl sulfate |
| PMSF | Phenylmethylsulfonyl fluoride |
| DTT | Dithiothreitol |
References
- Wu, C.Y.; Jeng, K.S.; Lai, M.M. The SUMOylation of matrix protein M1 modulates the assembly and morphogenesis of influenza A virus. J. Virol. 2011, 85, 6618–6628. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Lim, Y.S.; Park, E.M.; Baek, S.H.; Hwang, S.B. SUMOylation of nonstructural 5A protein regulates hepatitis C virus replication. J. Viral Hepat. 2014, 21, e108–e117. [Google Scholar] [CrossRef] [PubMed]
- Konig, R.; Stertz, S.; Zhou, Y.; Inoue, A.; Hoffmann, H.H.; Bhattacharyya, S.; Alamares, J.G.; Tscherne, D.M.; Ortigoza, M.B.; Liang, Y.; et al. Human host factors required for influenza virus replication. Nature 2010, 463, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Shapira, S.D.; Gat-Viks, I.; Shum, B.O.; Dricot, A.; de Grace, M.M.; Wu, L.; Gupta, P.B.; Hao, T.; Silver, S.J.; Root, D.E.; et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 2009, 139, 1255–1267. [Google Scholar] [CrossRef]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- Chandra, V.; Bhagyaraj, E.; Parkesh, R.; Gupta, P. Transcription factors and cognate signalling cascades in the regulation of autophagy. Biol. Rev. Camb. Philos. Soc. 2016, 91, 429–451. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, X.; Liu, D.; Fan, Z.; Hu, X.; Yan, J.; Wang, M.; Gao, G.F. Autophagy is involved in influenza A virus replication. Autophagy 2009, 5, 321–328. [Google Scholar] [CrossRef]
- Sun, Y.; Li, C.; Shu, Y.; Ju, X.; Zou, Z.; Wang, H.; Rao, S.; Guo, F.; Liu, H.; Nan, W.; et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci. Signal. 2012, 5, ra16. [Google Scholar] [CrossRef]
- Garcia-Valtanen, P.; Ortega-Villaizan Mdel, M.; Martinez-Lopez, A.; Medina-Gali, R.; Perez, L.; Mackenzie, S.; Figueras, A.; Coll, J.M.; Estepa, A. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy 2014, 10, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Cottet, L.; Rivas-Aravena, A.; Cortez-San Martin, M.; Sandino, A.M.; Spencer, E. Infectious salmon anemia virus--genetics and pathogenesis. Virus Res. 2011, 155, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhu, H.; Xu, X.; Li, L.; Tan, H.; Cai, X. Inactivated Sendai virus induces apoptosis and autophagy via the PI3K/Akt/mTOR/p70S6K pathway in human non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 2015, 465, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Manuse, M.J.; Briggs, C.M.; Parks, G.D. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 Requires TLR7 and autophagy pathways. Virology 2010, 405, 383–389. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, C.; Sun, L.; Lin, H.; Qin, Z.; Tu, J.; Li, J.; Chen, K.; Babu, V.S.; Lin, L. Glutamine starvation inhibits snakehead vesiculovirus replication via inducing autophagy associated with the disturbance of endogenous glutathione pool. Fish Shellfish Immunol. 2019, 86, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Curi, R.; Lagranha, C.J.; Doi, S.Q.; Sellitti, D.F.; Procopio, J.; Pithon-Curi, T.C.; Corless, M.; Newsholme, P. Molecular mechanisms of glutamine action. J. Cell. Physiol. 2005, 204, 392–401. [Google Scholar] [CrossRef]
- Porcheray, F.; Leone, C.; Samah, B.; Rimaniol, A.C.; Dereuddre-Bosquet, N.; Gras, G. Glutamate metabolism in HIV-infected macrophages: Implications for the CNS. Am. J. Physiol. Cell Physiol. 2006, 291, C618–C626. [Google Scholar] [CrossRef]
- Fu, X.; Hu, X.; Li, N.; Zheng, F.; Dong, X.; Duan, J.; Lin, Q.; Tu, J.; Zhao, L.; Huang, Z.; et al. Glutamine and glutaminolysis are required for efficient replication of infectious spleen and kidney necrosis virus in Chinese perch brain cells. Oncotarget 2017, 8, 2400–2412. [Google Scholar] [CrossRef]
- Li, C.Y.; Wang, Y.J.; Huang, S.W.; Cheng, C.S.; Wang, H.C. Replication of the Shrimp Virus WSSV Depends on Glutamate-Driven Anaplerosis. PLoS ONE 2016, 11, e0146902. [Google Scholar] [CrossRef]
- Asim, M.; Jiang, S.; Yi, L.; Chen, W.; Sun, L.; Zhao, L.; Khan Khattak, M.N.; Tu, J.; Lin, L. Glutamine is required for red-spotted grouper nervous necrosis virus replication via replenishing the tricarboxylic acid cycle. Virus Res. 2017, 227, 245–248. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Takemoto, Y.; Sumi, S.; Inoue, D.; Kishimoto, N.; Takamune, N.; Shoji, S.; Suzu, S.; Misumi, S. ATP generation in a host cell in early-phase infection is increased by upregulation of cytochrome c oxidase activity via the p2 peptide from human immunodeficiency virus type 1 Gag. Retrovirology 2015, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Chi, P.I.; Huang, W.R.; Chiu, H.C.; Li, J.Y.; Nielsen, B.L.; Liu, H.J. Avian reovirus sigmaA-modulated suppression of lactate dehydrogenase and upregulation of glutaminolysis and the mTOC1/eIF4E/HIF-1alpha pathway to enhance glycolysis and the TCA cycle for virus replication. Cell Microbiol. 2018, 20, e12946. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.E.; Calantone, N.; Lichon, D.; Hudson, H.; Clerc, I.; Campbell, E.M.; D’Aquila, R.T. mTOR Overcomes Multiple Metabolic Restrictions to Enable HIV-1 Reverse Transcription and Intracellular Transport. Cell Rep. 2020, 31, 107810. [Google Scholar] [CrossRef]
- Ye, C.; Liu, S.; Li, N.; Zuo, S.; Niu, Y.; Lin, Q.; Liang, H.; Luo, X.; Fu, X. Mandarin Fish (Siniperca chuatsi) p53 Regulates Glutaminolysis Induced by Virus via the p53/miR145-5p/c-Myc Pathway in Chinese Perch Brain Cells. Microbiol. Spectr. 2022, 10, e0272721. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wen, Y.; Hu, X.; Wang, W.; Liang, X.; Li, J.; Vakharia, V.; Lin, L. Breaking the host range: Mandarin fish is susceptible to a vesiculovirus derived from snakehead fish. J. Gen. Virol. 2015, 96, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Su, J.; Lin, L.; Tu, J. Development of a reverse genetics system for snakehead vesiculovirus (SHVV). Virology 2019, 526, 32–37. [Google Scholar] [CrossRef]
- Zhang, C.; Feng, S.; Zhang, W.; Chen, N.; Hegazy, A.M.; Chen, W.; Liu, X.; Zhao, L.; Li, J.; Lin, L.; et al. MicroRNA miR-214 Inhibits Snakehead Vesiculovirus Replication by Promoting IFN-alpha Expression via Targeting Host Adenosine 5′-Monophosphate-Activated Protein Kinase. Front. Immunol. 2017, 8, 1775. [Google Scholar] [CrossRef]
- Slivac, I.; Gaurina Srcek, V.; Radosevic, K.; Porobic, I.; Bilic, K.; Fumic, K.; Kniewald, Z. Growth characteristics of channel catfish ovary cells-influence of glucose and glutamine. Cytotechnology 2008, 57, 273–278. [Google Scholar] [CrossRef]
- Fontaine, K.A.; Camarda, R.; Lagunoff, M. Vaccinia virus requires glutamine but not glucose for efficient replication. J. Virol. 2014, 88, 4366–4374. [Google Scholar] [CrossRef]
- Walker, J.M. The bicinchoninic acid (BCA) assay for protein quantitation. Methods Mol. Biol. 1994, 32, 5–8. [Google Scholar] [PubMed]
- Westermeier, R.; Marouga, R. Protein detection methods in proteomics research. Biosci. Rep. 2005, 25, 19–32. [Google Scholar] [CrossRef]
- Biarc, J.; Gonzalo, P.; Mikaelian, I.; Fattet, L.; Deygas, M.; Gillet, G.; Lemoine, J.; Rimokh, R. Combination of a discovery LC-MS/MS analysis and a label-free quantification for the characterization of an epithelial-mesenchymal transition signature. J. Proteom. 2014, 110, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Huan, T. Comparison of Full-Scan, Data-Dependent, and Data-Independent Acquisition Modes in Liquid Chromatography-Mass Spectrometry Based Untargeted Metabolomics. Anal. Chem. 2020, 92, 8072–8080. [Google Scholar] [CrossRef] [PubMed]
- Orsburn, B.C. Proteome Discoverer-A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 2021, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38, D142–D148. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramirez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Blum, T.; Briesemeister, S.; Kohlbacher, O. MultiLoc2: Integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinform. 2009, 10, 274. [Google Scholar] [CrossRef] [PubMed]
- Pinto, G.; Alhaiek, A.A.; Godovac-Zimmermann, J. Proteomics reveals the importance of the dynamic redistribution of the subcellular location of proteins in breast cancer cells. Expert Rev. Proteom. 2015, 12, 61–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hongzhan, H.; Shukla, H.D.; Cathy, W.; Satya, S. Challenges and solutions in proteomics. Curr. Genom. 2007, 8, 21–28. [Google Scholar]
- Chou, K.C.; Shen, H.B. Recent progress in protein subcellular location prediction. Anal. Biochem. 2007, 370, 1–16. [Google Scholar] [CrossRef]
- Albertini, A.A.; Ruigrok, R.W.; Blondel, D. Rabies virus transcription and replication. Adv. Virus Res. 2011, 79, 1–22. [Google Scholar]
- Guu, T.S.; Zheng, W.; Tao, Y.J. Bunyavirus: Structure and replication. Adv. Exp. Med. Biol. 2012, 726, 245–266. [Google Scholar] [CrossRef]
- Bitzer, M.; Armeanu, S.; Lauer, U.M.; Neubert, W.J. Sendai virus vectors as an emerging negative-strand RNA viral vector system. J. Gene Med. 2003, 5, 543–553. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, K.; Zhang, Q.; Meng, S.; Ding, C. Autophagy in Negative-Strand RNA Virus Infection. Front. Microbiol. 2018, 9, 206. [Google Scholar] [CrossRef]
- Yang, Y.; Xue, J.; Teng, Q.; Li, X.; Bu, Y.; Zhang, G. Mechanisms and consequences of Newcastle disease virus W protein subcellular localization in the nucleus or mitochondria. J. Virol. 2021, 95, e02087-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.E.; Park, A.; Lake, M.; Pentecost, M.; Torres, B.; Yun, T.E.; Wolf, M.C.; Holbrook, M.R.; Freiberg, A.N.; Lee, B. Ubiquitin-regulated nuclear-cytoplasmic trafficking of the Nipah virus matrix protein is important for viral budding. PLoS Pathog. 2010, 6, e1001186. [Google Scholar] [CrossRef]
- Whittaker, G.R. Virus nuclear import. Adv. Drug Deliv. Rev. 2003, 55, 733–747. [Google Scholar] [CrossRef]
- Te Velthuis, A.J.; Fodor, E. Influenza virus RNA polymerase: Insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Zhang, K.; Min, J.; Chen, C.; Cao, Y.; Ding, C.; Liu, W.; Li, J. Metabolomic Analysis of Influenza A Virus A/WSN/1933 (H1N1) Infected A549 Cells during First Cycle of Viral Replication. Viruses 2019, 11, 1007. [Google Scholar] [CrossRef] [PubMed]
- Appelberg, S.; Gupta, S.; Svensson Akusjarvi, S.; Ambikan, A.T.; Mikaeloff, F.; Saccon, E.; Vegvari, A.; Benfeitas, R.; Sperk, M.; Stahlberg, M.; et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes Infect. 2020, 9, 1748–1760. [Google Scholar] [CrossRef] [PubMed]
- Holder, K.A.; Russell, R.S.; Grant, M.D. Natural killer cell function and dysfunction in hepatitis C virus infection. Biomed. Res. Int. 2014, 2014, 903764. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Koyama, S.; Ishii, K.J.; Coban, C.; Akira, S. Innate immune response to viral infection. Cytokine 2008, 43, 336–341. [Google Scholar] [CrossRef]
- Diebold, S.S. Recognition of viral single-stranded RNA by Toll-like receptors. Adv. Drug Deliv. Rev. 2008, 60, 813–823. [Google Scholar] [CrossRef]
- Nimmerjahn, F.; Dudziak, D.; Dirmeier, U.; Hobom, G.; Riedel, A.; Schlee, M.; Staudt, L.M.; Rosenwald, A.; Behrends, U.; Bornkamm, G.W.; et al. Active NF-kappaB signalling is a prerequisite for influenza virus infection. J. Gen. Virol. 2004, 85, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J.; Nguyen, T.L.; Arguello, M.; Nakhaei, P.; Paz, S. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene 2006, 25, 6844–6867. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Marjuki, H.; Wolff, T.; Nurnberg, B.; Planz, O.; Pleschka, S.; Ludwig, S. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 2006, 8, 1336–1348. [Google Scholar] [CrossRef] [PubMed]
- Dunn, E.F.; Connor, J.H. HijAkt: The PI3K/Akt pathway in virus replication and pathogenesis. Prog. Mol. Biol. Transl. Sci. 2012, 106, 223–250. [Google Scholar]
- Barber, S.A.; Bruett, L.; Douglass, B.R.; Herbst, D.S.; Zink, M.C.; Clements, J.E. Visna virus-induced activation of MAPK is required for virus replication and correlates with virus-induced neuropathology. J. Virol. 2002, 76, 817–828. [Google Scholar] [CrossRef]
- Zhan, Y.; Yu, S.; Yang, S.; Qiu, X.; Meng, C.; Tan, L.; Song, C.; Liao, Y.; Liu, W.; Sun, Y.; et al. Newcastle Disease virus infection activates PI3K/Akt/mTOR and p38 MAPK/Mnk1 pathways to benefit viral mRNA translation via interaction of the viral NP protein and host eIF4E. PLoS Pathog. 2020, 16, e1008610. [Google Scholar] [CrossRef]
- Pan, W.; Bodempudi, V.; Esfandyari, T.; Farassati, F. Utilizing ras signaling pathway to direct selective replication of herpes simplex virus-1. PLoS ONE 2009, 4, e6514. [Google Scholar] [CrossRef]
- Gretton, S.; Hughes, M.; Harris, M. Hepatitis C virus RNA replication is regulated by Ras-Erk signalling. J. Gen. Virol. 2010, 91, 671–680. [Google Scholar] [CrossRef]
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [Google Scholar] [CrossRef]
- Shin, Y.K.; Liu, Q.; Tikoo, S.K.; Babiuk, L.A.; Zhou, Y. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J. Gen. Virol. 2007, 88, 942–950. [Google Scholar] [CrossRef]
- Ayllon, J.; Hale, B.G.; Garcia-Sastre, A. Strain-specific contribution of NS1-activated phosphoinositide 3-kinase signaling to influenza A virus replication and virulence. J. Virol. 2012, 86, 5366–5370. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Yuan, R.; Zhao, X.; Xiang, B.; Gao, S.; Gao, P.; Dai, X.; Feng, M.; Li, Y.; Xie, P.; et al. Transient activation of the PI3K/Akt pathway promotes Newcastle disease virus replication and enhances anti-apoptotic signaling responses. Oncotarget 2017, 8, 23551–23563. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.L.; McDonald, N.J.; Sheng, J.; Shaw, M.W.; Hodge, T.W.; Rubin, D.H.; O’Brien, W.A.; Smee, D.F. Inhibition of influenza A virus replication by antagonism of a PI3K-AKT-mTOR pathway member identified by gene-trap insertional mutagenesis. Antivir. Chem. Chemother. 2012, 22, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.S.; Yen, H.R.; Chang, C.W.; Peng, T.Y.; Hsieh, C.F.; Chen, C.J.; Lin, T.Y.; Horng, J.T. Mechanism of action of the suppression of influenza virus replication by Ko-Ken Tang through inhibition of the phosphatidylinositol 3-kinase/Akt signaling pathway and viral RNP nuclear export. J. Ethnopharmacol. 2011, 134, 614–623. [Google Scholar] [CrossRef]
- Zhirnov, O.P.; Klenk, H.D. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 2007, 12, 1419–1432. [Google Scholar] [CrossRef]
- Bonizzi, G.; Karin, M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25, 280–288. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Wang, J.; Mena, I.; White, K.; Metreveli, G.; Sakthivel, R.; Mata, M.A.; Munoz-Moreno, R.; Chen, X.; Krammer, F.; et al. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog. 2017, 13, e1006635. [Google Scholar] [CrossRef]
- Bonjardim, C.A. Viral exploitation of the MEK/ERK pathway—A tale of vaccinia virus and other viruses. Virology 2017, 507, 267–275. [Google Scholar] [CrossRef]
- Gack, M.U.; Shin, Y.C.; Joo, C.H.; Urano, T.; Liang, C.; Sun, L.; Takeuchi, O.; Akira, S.; Chen, Z.; Inoue, S.; et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 2007, 446, 916–920. [Google Scholar] [CrossRef]
- Franz, K.M.; Neidermyer, W.J.; Tan, Y.J.; Whelan, S.P.J.; Kagan, J.C. STING-dependent translation inhibition restricts RNA virus replication. Proc. Natl. Acad. Sci. USA 2018, 115, E2058–E2067. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, Y.; Xu, X.; Tang, T.; Sun, L.; Wang, H.; Zhou, W.; Fang, L.; Li, Q.; Xie, P. miR-146a promotes Borna disease virus 1 replication through IRAK1/TRAF6/NF-kappaB signaling pathway. Virus Res. 2019, 271, 197671. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, T.; Yamayoshi, S.; Noda, T.; Kawaoka, Y. G Protein Pathway Suppressor 1 Promotes Influenza Virus Polymerase Activity by Activating the NF-kappaB Signaling Pathway. mBio 2019, 10, e02867-19. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, X.; Zhou, Y. Ribosomal protein L18 is an essential factor that promote rice stripe virus accumulation in small brown planthopper. Virus Res. 2018, 247, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Huang, C.; Lokugamage, K.; Kamitani, W.; Ikegami, T.; Tseng, C.T.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008, 82, 4471–4479. [Google Scholar] [CrossRef]
- Li, Y.; Fang, L.; Zhou, Y.; Tao, R.; Wang, D.; Xiao, S. Porcine Reproductive and Respiratory Syndrome Virus Infection Induces both eIF2alpha Phosphorylation-Dependent and -Independent Host Translation Shutoff. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Helenius, A. Virus entry at a glance. J. Cell Sci. 2013, 126, 1289–1295. [Google Scholar] [CrossRef]
- Marsh, M.; Helenius, A. Virus entry: Open sesame. Cell 2006, 124, 729–740. [Google Scholar] [CrossRef]
- Perrin-Cocon, L.; Diaz, O.; Jacquemin, C.; Barthel, V.; Ogire, E.; Ramiere, C.; Andre, P.; Lotteau, V.; Vidalain, P.O. The current landscape of coronavirus-host protein-protein interactions. J. Transl. Med. 2020, 18, 319. [Google Scholar] [CrossRef]
- Sajidah, E.S.; Lim, K.; Wong, R.W. How SARS-CoV-2 and Other Viruses Build an Invasion Route to Hijack the Host Nucleocytoplasmic Trafficking System. Cells 2021, 10, 1424. [Google Scholar] [CrossRef] [PubMed]
- Fassati, A. HIV infection of non-dividing cells: A divisive problem. Retrovirology 2006, 3, 74. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, O.G.; Fodor, E. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 2006, 16, 329–345. [Google Scholar] [CrossRef]
- Kluge, M.A.; Fetterman, J.L.; Vita, J.A. Mitochondria and endothelial function. Circ. Res. 2013, 112, 1171–1188. [Google Scholar] [CrossRef]
- West, A.P.; Shadel, G.S.; Ghosh, S. Mitochondria in innate immune responses. Nat. Rev. Immunol. 2011, 11, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Chaubey, G.; Chen, J.Y.; Suravajhala, P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell Physiol. 2020, 319, C258–C267. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Gao, G.; Wong, J.; Wang, Y.; Zhang, J.; Luo, H. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS ONE 2008, 3, e2585. [Google Scholar] [CrossRef]
- Zinngrebe, J.; Montinaro, A.; Peltzer, N.; Walczak, H. Ubiquitin in the immune system. EMBO Rep. 2014, 15, 28–45. [Google Scholar] [CrossRef]
- Park, E.S.; Dezhbord, M.; Lee, A.R.; Kim, K.H. The Roles of Ubiquitination in Pathogenesis of Influenza Virus Infection. Int. J. Mol. Sci. 2022, 23, 4593. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, L.; Sun, J.; Chen, W.; Li, S.; Wang, Q.; Yu, H.; Xia, Z.; Jin, X.; Wang, C. Endoplasmic Reticulum Protein SCAP Inhibits Dengue Virus NS2B3 Protease by Suppressing Its K27-Linked Polyubiquitylation. J. Virol. 2017, 91, e02234-16. [Google Scholar] [CrossRef]
- Liao, T.L.; Wu, C.Y.; Su, W.C.; Jeng, K.S.; Lai, M.M. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J. 2010, 29, 3879–3890. [Google Scholar] [CrossRef] [PubMed]
- Kitab, B.; Satoh, M.; Ohmori, Y.; Munakata, T.; Sudoh, M.; Kohara, M.; Tsukiyama-Kohara, K. Ribonucleotide reductase M2 promotes RNA replication of hepatitis C virus by protecting NS5B protein from hPLIC1-dependent proteasomal degradation. J. Biol. Chem. 2019, 294, 5759–5773. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Vargas-Cuartas, O.; Gallego-Gomez, J.C.; Shi, P.Y.; Padilla-Sanabria, L.; Castano-Osorio, J.C.; Rajsbaum, R. K48-linked polyubiquitination of dengue virus NS1 protein inhibits its interaction with the viral partner NS4B. Virus Res. 2018, 246, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.C.; Jeng, K.S.; Lai, M.M.C. CNOT4-Mediated Ubiquitination of Influenza A Virus Nucleoprotein Promotes Viral RNA Replication. mBio 2017, 8, e00597-17. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef]
- Yau, R.; Rape, M. The increasing complexity of the ubiquitin code. Nat. Cell Biol. 2016, 18, 579–586. [Google Scholar] [CrossRef]
- Nakayama, K.I.; Nakayama, K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin. Cell Dev. Biol. 2005, 16, 323–333. [Google Scholar] [CrossRef]
- Bornstein, G.; Ganoth, D.; Hershko, A. Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. USA 2006, 103, 11515–11520. [Google Scholar] [CrossRef]
- Thompson, L.L.; Rutherford, K.A.; Lepage, C.C.; McManus, K.J. The SCF Complex Is Essential to Maintain Genome and Chromosome Stability. Int. J. Mol. Sci. 2021, 22, 8544. [Google Scholar] [CrossRef]
- Nakayama, K.I.; Nakayama, K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 2006, 6, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Ly, H.J.; Ikegami, T. Rift Valley fever virus NSs protein functions and the similarity to other bunyavirus NSs proteins. Virol. J. 2016, 13, 118. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.A.; Tu, J. Hsp90 Is Required for Snakehead Vesiculovirus Replication via Stabilization of the Viral L Protein. J. Virol. 2021, 95, e0059421. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, M.S.; Gallo, E.S.; Borenstein, R. Multifaceted Role of AMPK in Viral Infections. Cells 2021, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Mankouri, J.; Harris, M. Viruses and the fuel sensor: The emerging link between AMPK and virus replication. Rev. Med. Virol. 2011, 21, 205–212. [Google Scholar] [CrossRef]
- Song, G.; Ouyang, G.; Bao, S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med. 2005, 9, 59–71. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef]
- Wu, Z.; Li, X.; Guo, D.; Li, P.; Zhang, Y.; Zou, D.; Wang, X.; Xu, J.; Wu, X.; Shen, Y.; et al. Lipid raft-associated PI3K/Akt/SREBP1 signaling regulates coxsackievirus A16 (CA16) replication. Vet. Microbiol. 2021, 252, 108921. [Google Scholar] [CrossRef]
- Chen, H.H.; Chen, C.C.; Lin, Y.S.; Chang, P.C.; Lu, Z.Y.; Lin, C.F.; Chen, C.L.; Chang, C.P. AR-12 suppresses dengue virus replication by down-regulation of PI3K/AKT and GRP78. Antivir. Res. 2017, 142, 158–168. [Google Scholar] [CrossRef]
- Luo, H.; Yanagawa, B.; Zhang, J.; Luo, Z.; Zhang, M.; Esfandiarei, M.; Carthy, C.; Wilson, J.E.; Yang, D.; McManus, B.M. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. J. Virol. 2002, 76, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Li, X.; Cai, Q.; Li, C.; Tian, L.; Chen, J.; Xing, X.; Gan, Y.; Ouyang, W.; Yang, Z. The PI3K/Akt/mTOR pathway is involved in CVB3-induced autophagy of HeLa cells. Int. J. Mol. Med. 2017, 40, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.S.; Lee, S.A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Geng, P.; Liu, Y.; Wu, J.; Qiao, H.; Xie, Y.; Yin, N.; Chen, L.; Lin, X.; Liu, Y.; et al. Rotavirus-encoded virus-like small RNA triggers autophagy by targeting IGF1R via the PI3K/Akt/mTOR pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, N.; Hegazy, A.M.; Liu, X.; Wu, Z.; Liu, X.; Zhao, L.; Qin, Q.; Lan, J.; Lin, L. Autophagy induced by snakehead fish vesiculovirus inhibited its replication in SSN-1 cell line. Fish Shellfish Immunol. 2016, 55, 415–422. [Google Scholar] [CrossRef]
- Blanco, J.; Cameirao, C.; Lopez, M.C.; Munoz-Barroso, I. Phosphatidylinositol-3-kinase-Akt pathway in negative-stranded RNA virus infection: A minireview. Arch. Virol. 2020, 165, 2165–2176. [Google Scholar] [CrossRef]
- Sun, M.; Fuentes, S.M.; Timani, K.; Sun, D.; Murphy, C.; Lin, Y.; August, A.; Teng, M.N.; He, B. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J. Virol. 2008, 82, 105–114. [Google Scholar] [CrossRef]

| KEGG Description | p-Value |
|---|---|
| MicroRNAs in cancer | 0.0011 |
| ErbB signaling pathway | 0.0022 |
| Glioma | 0.0042 |
| Natural killer cell mediated cytotoxicity | 0.0056 |
| Non-small cell lung cancer | 0.0120 |
| Alcoholism | 0.0123 |
| Chemokine signaling pathway | 0.0168 |
| Focal adhesion | 0.0176 |
| Insulin signaling pathway | 0.0183 |
| Proteoglycans in cancer | 0.0367 |
| Chronic myeloid leukemia | 0.0383 |
| Endocrine resistance | 0.0383 |
| Ras signaling pathway | 0.0448 |
| D-Glutamine and D-glutamate metabolism | 0.0449 |
| Phospholipase D signaling pathway | 0.0452 |
| VEGF signaling pathway | 0.0459 |
| EGFR tyrosine kinase inhibitor resistance | 0.0459 |
| Proteins | Gene | Protein Fold Change | Subpathway | KEGG Subpathway p-Value | Superpathway |
|---|---|---|---|---|---|
| A0A2D0PVW4 | ACTB_G1 ↑ | >32 | RIG-I-like receptor signaling pathway | 0.4424 | immune system |
| A0A2D0QBZ9 | TRAF2 ↓ | <0.00001 | |||
| A0A2D0RKM8 | AKT ↑ | >32 | Toll-like receptor signaling pathway | 0.9202 | |
| A0A2D0R0K4 | SHC1 ↑ | 3.652 | MAPK signaling pathway | 0.1960 | Signal transduction |
| A0A2D0QNJ3 | PPM1A, PP2CA ↑ | 2.457 | |||
| A0A2D0QFC3 | CASP3 ↑ | >32 | |||
| A0A2D0RBZ2 | PPP3C, CAN ↑ | >32 | |||
| A0A2D0RKM8 | AKT ↑ | >32 | |||
| A0A2D0PU15 | GRB2 ↓ | <0.00001 | |||
| A0A2D0QBZ9 | TRAF2 ↓ | <0.00001 | |||
| A0A2D0SZV8 | CRK, CRKII ↓ | <0.00001 | |||
| A0A2D0T445 | ERC1, CAST2, ELKS ↓ | <0.00001 | NF-kappa B signaling pathway | 0.1781 | |
| A0A2D0SJ10 | PLCG1 ↑ | >32 | |||
| A0A2D0QBZ9 | TRAF2 ↓ | <0.00001 | |||
| A0A2D0SN73 | ERBB2IP, ERBIN ↓ | <0.00001 | |||
| W5UJ57 | ITGA5, CD49e ↓ | 0.4468 | PI3K-Akt signaling pathway | 0.3448 | |
| A0A2D0RKM8 | AKT ↑ | >32 | |||
| A0A2D0PSX8 | PTK2, FAK ↓ | <0.00001 | |||
| A0A2D0QAU5 | GNB2 ↓ | <0.00001 | |||
| A0A2D0S1J2 | RAPTOR ↓ | <0.00001 | |||
| E3TE89 | GNG5 ↓ | <0.00001 | |||
| W5UHZ2 | PPP2R5 ↓ | <0.00001 |
| KEGG Description | p-Value |
|---|---|
| HIF-1 signaling pathway | 0.0005 |
| mTOR signaling pathway | 0.0017 |
| Ras signaling pathway | 0.0022 |
| Choline metabolism in cancer | 0.0025 |
| Longevity regulating pathway | 0.0026 |
| ErbB signaling pathway | 0.0028 |
| Insulin signaling pathway | 0.0030 |
| Circadian rhythm | 0.0033 |
| EGFR tyrosine kinase inhibitor resistance | 0.0033 |
| B cell receptor signaling pathway | 0.0051 |
| Glioma | 0.0051 |
| MicroRNAs in cancer | 0.0058 |
| PI3K-Akt signaling pathway | 0.0072 |
| Chemokine signaling pathway | 0.0073 |
| Glucagon signaling pathway | 0.0081 |
| Chronic myeloid leukemia | 0.0116 |
| Fc epsilon RI signaling pathway | 0.0116 |
| Non-small cell lung cancer | 0.0141 |
| VEGF signaling pathway | 0.0147 |
| T cell receptor signaling pathway | 0.0171 |
| Proteins | Gene | Protein Fold Change | Subpathway | KEGG Subpathway p-Value | Superpathway |
|---|---|---|---|---|---|
| A0A2D0PSX8 | PTK2, FAK ↑ | >32 | PI3K-Akt signaling pathway | 0.0072 | Signal transduction |
| A0A2D0QAU5 | GNB2 ↑ | >32 | |||
| E3TE89 | GNG5 ↑ | >32 | |||
| A0A2D0Q294 | PPP2R5 ↓ | <0.00001 | |||
| A0A2D0QC90 | PRKAA, AMPK ↓ | <0.00001 | |||
| A0A2D0QGD6 | EIF4E ↓ | <0.00001 | |||
| A0A2D0QH87 | RELA ↓ | <0.00001 | |||
| A0A2D0QR45 | EIF4E ↓ | <0.00001 | |||
| A0A2D0QVE7 | PDPK1 ↓ | <0.00001 | |||
| A0A2D0RKM8 | AKT ↓ | <0.00001 | |||
| A0A2D0RSR0 | GNB4 ↓ | <0.00001 | |||
| W5U9B8 | MLST8, GBL ↓ | <0.00001 | |||
| A0A2D0SZV8 | CRK, CRKII ↑ | >32 | MAPK signaling pathway | 0.2262 | |
| A0A2D0PJF0 | RASGRF2 ↓ | <0.00001 | |||
| A0A2D0QH87 | RELA ↓ | <0.00001 | |||
| A0A2D0R0K4 | SHC1 ↓ | <0.00001 | |||
| A0A2D0RBZ2 | PPP3C, CAN ↓ | <0.00001 | |||
| A0A2D0RKM8 | AKT ↓ | <0.00001 | |||
| E3TEE7 | RBX1, ROC1 ↓ | <0.00001 | |||
| E3TE71 | ATPeV1D, ATP6M ↑ | 2.076 | mTOR signaling pathway | 0.0017 | |
| A0A2D0PUT4 | MIOS, MIO ↓ | <0.00001 | |||
| A0A2D0QC90 | PRKAA, AMPK ↓ | <0.00001 | |||
| A0A2D0QGD6 | EIF4E ↓ | <0.00001 | |||
| A0A2D0QL93 | TELO2, TEL2 ↓ | <0.00001 | |||
| A0A2D0QR45 | EIF4E ↓ | <0.00001 | |||
| A0A2D0QVE7 | PDPK1 ↓ | <0.00001 | |||
| A0A2D0RKM8 | AKT ↓ | <0.00001 | |||
| E3TF29 | RRAGC_D ↓ | <0.00001 | |||
| W5U9B8 | MLST8, GBL ↓ | <0.00001 | |||
| A0A2D0QH87 | RELA ↓ | <0.00001 | NF-kappa B signaling pathway | 0.4632 | Immune system |
| A0A2D0SJ10 | PLCG1 ↓ | <0.00001 | |||
| A0A2D0Q3G1 | PIN1 ↓ | <0.00001 | RIG-I-like receptor signaling pathway | 0.0599 | |
| A0A2D0QH87 | RELA ↓ | <0.00001 | |||
| A0A2D0R0L0 | OTUD5, DUBA ↓ | <0.00001 | |||
| W5U5F5 | IRF3 ↓ | <0.00001 |
| UniProtKB | Protein Name | Gene | Regulation in Group 1 | Protein | Regulation in Group 2 | Protein | ||
|---|---|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | |||||
| A0A2D0RR20 | cell division cycle 5-like protein isoform X1 | CDC5L | Up | >32 | <0.00001 | Down | <0.00001 | <0.00001 |
| A0A2D0PN16 | guanine nucleotide-binding protein-like 3-like protein | GNL3L | Up | >32 | <0.00001 | Down | <0.00001 | <0.00001 |
| A0A2D0RJY2 | Hydroxymethylglutaryl-CoA lyase | HMGCL | Up | >32 | <0.00001 | Down | <0.00001 | <0.00001 |
| A0A2D0PW54 | nucleoporin Nup43 | NUP43 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| W5U919 | Proteasome subunit alpha type | PSMA7 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| A0A2D0RMF5 | intersectin-2-like isoform X1 | ITSN | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| A0A2D0PSX8 | Non-specific protein-tyrosine kinase | PTK2 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| W5U6X2 | 28S ribosomal protein S34, mitochondrial | MRPS34 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| A0A2D0SA67 | 39S ribosomal protein L43, mitochondrial | MRPL43 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
| A0A2D0T9T5 | cullin-1 | CUL1 | Down | <0.00001 | <0.00001 | Up | >32 | <0.00001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhang, Y.; Liu, X.; Zhang, H.; Liu, Y.; Chen, K.; Tang, M.; Sun, L. Effects of Glutamine Starvation on SHVV Replication by Quantitative Proteomics Analysis. Fishes 2022, 7, 315. https://doi.org/10.3390/fishes7060315
Liu J, Zhang Y, Liu X, Zhang H, Liu Y, Chen K, Tang M, Sun L. Effects of Glutamine Starvation on SHVV Replication by Quantitative Proteomics Analysis. Fishes. 2022; 7(6):315. https://doi.org/10.3390/fishes7060315
Chicago/Turabian StyleLiu, Junlin, Yulei Zhang, Xiaoyan Liu, Hantao Zhang, Yi Liu, Keping Chen, Min Tang, and Lindan Sun. 2022. "Effects of Glutamine Starvation on SHVV Replication by Quantitative Proteomics Analysis" Fishes 7, no. 6: 315. https://doi.org/10.3390/fishes7060315
APA StyleLiu, J., Zhang, Y., Liu, X., Zhang, H., Liu, Y., Chen, K., Tang, M., & Sun, L. (2022). Effects of Glutamine Starvation on SHVV Replication by Quantitative Proteomics Analysis. Fishes, 7(6), 315. https://doi.org/10.3390/fishes7060315








