Properties of Protein Hydrolysates and Bioinformatics Prediction of Peptides Derived from Thermal and Enzymatic Process of Skipjack Tuna (Katsuwonus pelamis) Roe
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Tuna Roe
2.2. Preparation of Hydrolysate Powder from Tuna Roe
2.2.1. Preparation of Hydrolysate from Tuna Roe by Enzymes
2.2.2. Preparation of Hydrolysate from Tuna Roe Using Autoclave
2.3. Preparation of Hydrolysate Powder
2.4. Determination of Properties of Tuna Roe Hydrolysate-Powder
2.4.1. Amino Acid Profile
2.4.2. Antioxidant Activity
DPPH Radical Scavenging Activity
ABTS Radical Scavenging Activity
Metal Chelating Activity
2.4.3. Protein Solubility
2.4.4. Foaming Capacity and Foam Stability
2.5. Proteomic Analysis of the Hydrolysate Peptides
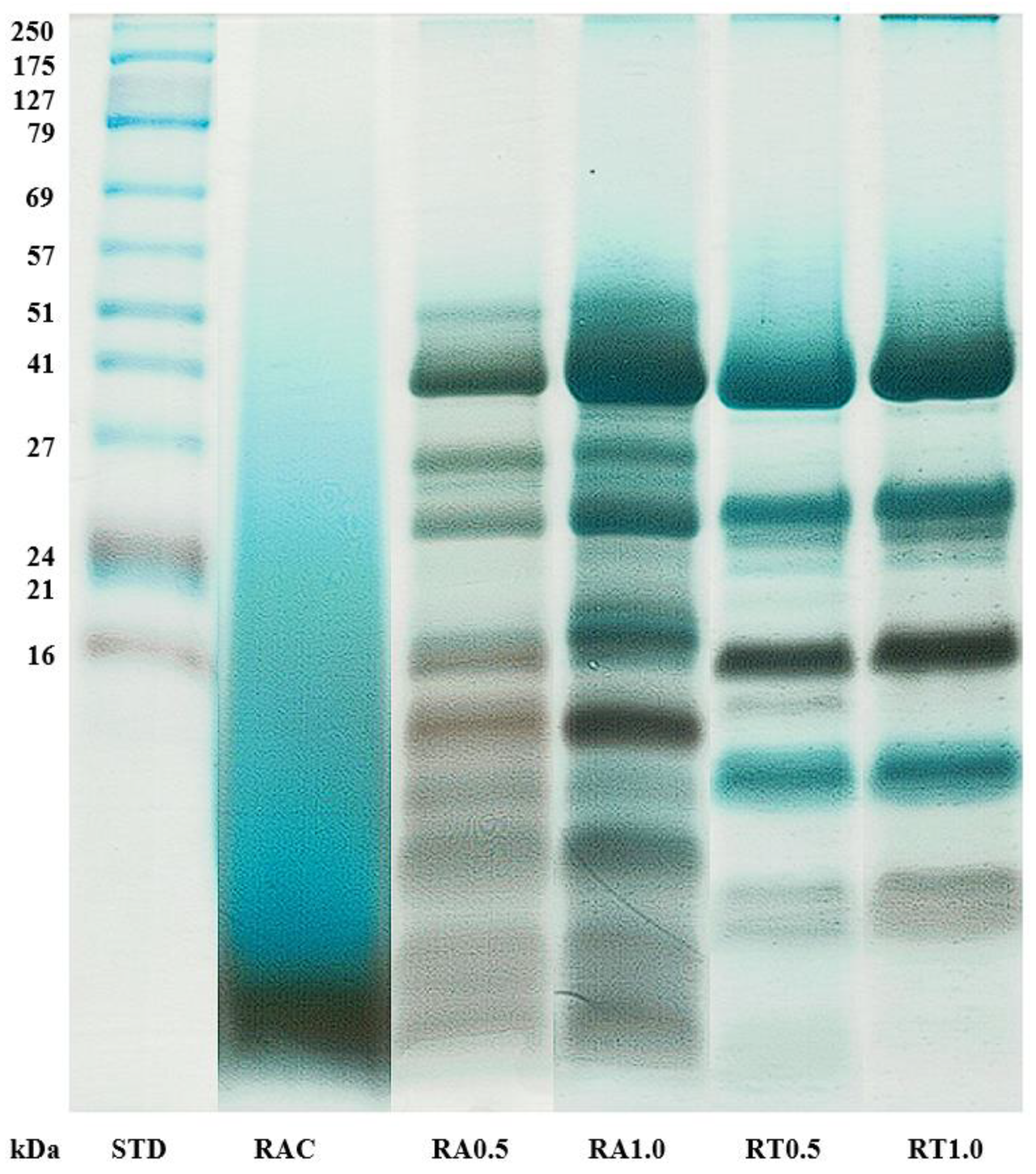
2.5.1. Protein and Peptide Separation Using 1D SDS-PAGE
2.5.2. LC-ESI-MS/MS Analysis
2.5.3. Bioinformatics Prediction of the Identified Peptides
2.6. Statistical Analysis
3. Results and Discussion
3.1. Amino Acid Composition of Hydrolysates from Tuna Roes
3.2. Protein and Peptide Profiles of Hydrolysates from Tuna Roes
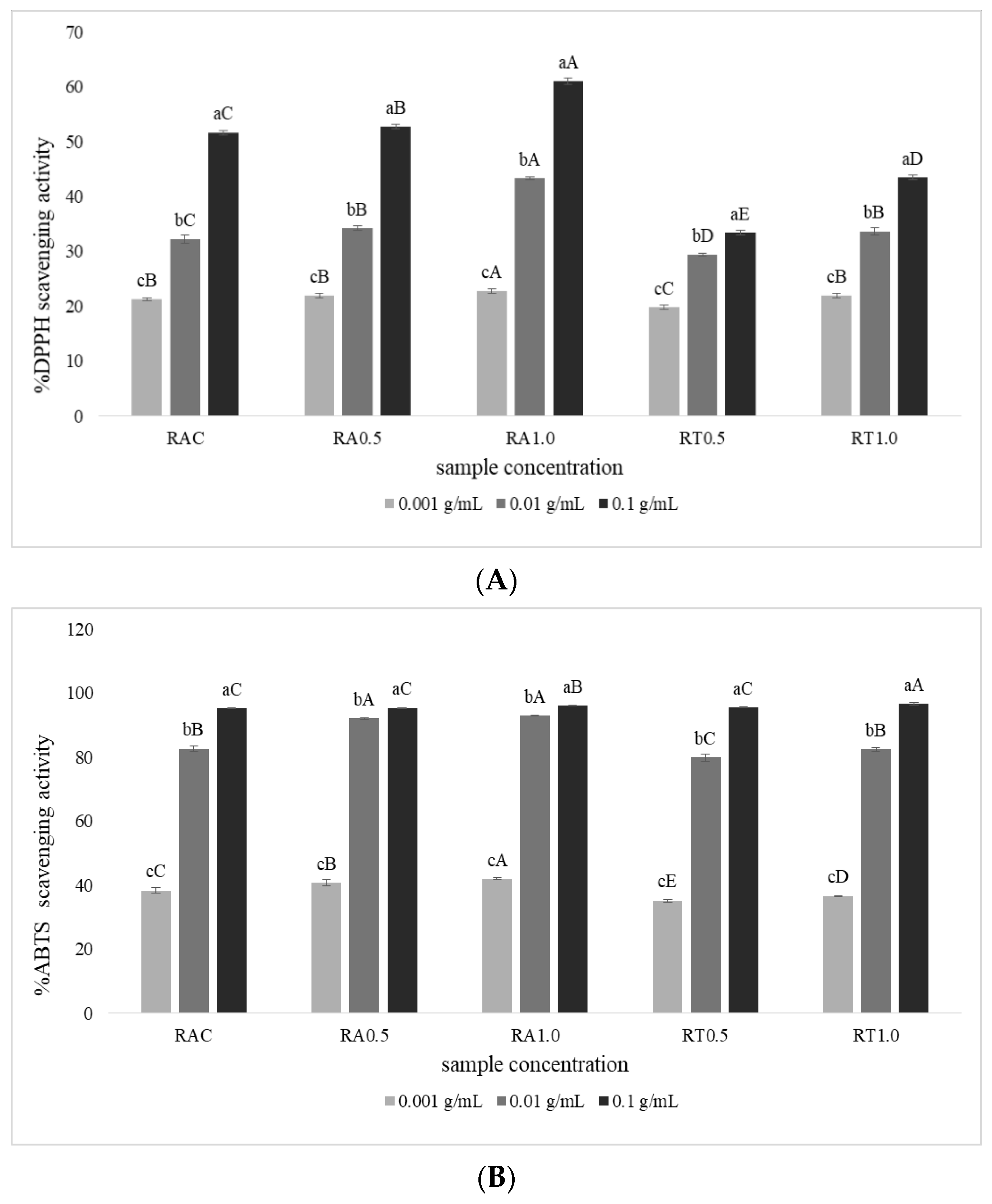
3.3. Antioxidant Activity of Hydrolysates from Tuna Roes
3.3.1. DPPH Radical Scavenging Activity
3.3.2. ABTS Radical Scavenging Activity
3.3.3. Metal Chelating Activity
3.4. Functional Properties of Hydrolysates from Tuna Roes
3.4.1. Solubility
3.4.2. Foaming Properties
3.5. Putative Bioactive Peptides of Hydrolysates from Tuna Roes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsu, K.-C. Purification of antioxidative peptides prepared from enzymatic hydrolysates of tuna dark muscle by-product. Food Chem. 2010, 122, 42–48. [Google Scholar] [CrossRef]
- Stevens, J.R.; Newton, R.W.; Tlusty, M.; Little, D.C. The rise of aquaculture by-products: Increasing food production, value, and sustainability through strategic utilisation. Mar. Policy 2018, 90, 115–124. [Google Scholar] [CrossRef]
- Klomklao, S.; Benjakul, S. Utilization of Tuna Processing Byproducts: Protein Hydrolysate from Skipjack Tuna (Katsuwonus pelamis) Viscera. J. Food Process. Preserv. 2017, 41, e12970. [Google Scholar] [CrossRef]
- Chalamaiah, M.; Jyothirmayi, T.; Diwan, P.V.; Dinesh Kumar, B. Antioxidant activity and functional properties of enzymatic protein hydrolysates from common carp (Cyprinus carpio) roe (egg). J. Food Sci. Technol. 2015, 52, 5817–5825. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A.S. A review of fish-derived antioxidant and antimicrobial peptides: Their production, assessment, and applications. Peptides 2012, 33, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Intarasirisawat, R.; Benjakul, S.; Visessanguan, W.; Wu, J. Antioxidative and functional properties of protein hydrolysate from defatted skipjack (Katsuwonous pelamis) roe. Food Chem. 2012, 135, 3039–3048. [Google Scholar] [CrossRef] [PubMed]
- Rajabzadeh, M.; Pourashouri, P.; Shabanpour, B.; Alishahi, A. Amino acid composition, antioxidant and functional properties of protein hydrolysates from the roe of rainbow trout (Oncorhynchus mykiss). Int. J. Food Sci. Technol. 2018, 53, 313–319. [Google Scholar] [CrossRef]
- Chen, W.; Wei, J.; Zhang, L.; Chen, J.; Li, Y.; Pei, D.; Wang, N.; Liu, Y.; Di, D. Fish Roe Polypeptide Exerts Hypoglycemia Activity via Regulating Insulin Secretion Mediated by Nrf2/ERK Signaling. Int. J. Pept. Res. Ther. 2020, 27, 543–553. [Google Scholar] [CrossRef]
- Kim, S.Y.; Je, J.Y.; Kim, S.K. Purification and characterization of antioxidant peptide from hoki (Johnius belengerii) frame protein by gastrointestinal digestion. J. Nutr. Biochem. 2007, 18, 31–38. [Google Scholar] [CrossRef]
- Ahn, C.B.; Kim, J.G.; Je, J.Y. Purification and antioxidant properties of octapeptide from salmon byproduct protein hydrolysate by gastrointestinal digestion. Food Chem. 2014, 147, 78–83. [Google Scholar] [CrossRef]
- Cai, L.; Wu, X.; Zhang, Y.; Li, X.; Ma, S.; Li, J. Purification and characterization of three antioxidant peptides from protein hydrolysate of grass carp (Ctenopharyngodon idella) skin. J. Funct. Foods 2015, 16, 234–242. [Google Scholar] [CrossRef]
- Chi, C.-F.; Wang, B.; Hu, F.-Y.; Wang, Y.-M.; Zhang, B.; Deng, S.-G.; Wu, C.-W. Purification and identification of three novel antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) skin. Food Res. Int. 2015, 73, 124–129. [Google Scholar] [CrossRef]
- Chi, C.-F.; Wang, B.; Wang, Y.-M.; Zhang, B.; Deng, S.-G. Isolation and characterization of three antioxidant peptides from protein hydrolysate of bluefin leatherjacket (Navodon septentrionalis) heads. J. Funct. Foods 2015, 12, 1–10. [Google Scholar] [CrossRef]
- Gajanan, P.G.; Elavarasan, K.; Shamasundar, B.A. Bioactive and functional properties of protein hydrolysates from fish frame processing waste using plant proteases. Environ. Sci. Pollut Res. Int. 2016, 23, 24901–24911. [Google Scholar] [CrossRef]
- Picot, L.; Bordenave, S.; Didelot, S.; Fruitier-Arnaudin, I.; Sannier, F.; Thorkelsson, G.; Bergé, J.P.; Guérard, F.; Chabeaud, A.; Piot, J.M. Antiproliferative activity of fish protein hydrolysates on human breast cancer cell lines. Process Biochem. 2006, 41, 1217–1222. [Google Scholar] [CrossRef]
- Song, R.; Wei, R.; Zhang, B.; Yang, Z.; Wang, D. Antioxidant and antiproliferative activities of heated sterilized pepsin hydrolysate derived from half-fin anchovy (Setipinna taty). Mar. Drugs 2011, 9, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Cudennec, B.; Ravallec-Plé, R.; Courois, E.; Fouchereau-Peron, M. Peptides from fish and crustacean by-products hydrolysates stimulate cholecystokinin release in STC-1 cells. Food Chem. 2008, 111, 970–975. [Google Scholar] [CrossRef]
- Ennaas, N.; Hammami, R.; Beaulieu, L.; Fliss, I. Purification and characterization of four antibacterial peptides from protamex hydrolysate of Atlantic mackerel (Scomber scombrus) by-products. Biochem. Biophys. Res. Commun. 2015, 462, 195–200. [Google Scholar] [CrossRef]
- Danquah, M.K.; Agyei, D. Pharmaceutical applications of bioactive peptides. OA Biotechnol. 2012, 1, 1–7. [Google Scholar] [CrossRef]
- Olga, M.; Larisa, B.; Natalya, M.; Svetlana, A.; Vladimir, V.; Vasilij, V.; Thomas, G.; Axel, H. High-temperature Hydrolysis As A Method For Complex Processing Of Sprat (sprattus Sprattus Balticus) By-products. JNTM 2022, 12, 8–14. [Google Scholar]
- Yang, J.-I.; Liang, W.-S.; Chow, C.-J.; Siebert, K.J. Process for the production of tilapia retorted skin gelatin hydrolysates with optimized antioxidative properties. Process Biochem. 2009, 44, 1152–1157. [Google Scholar] [CrossRef]
- Wang, W.; Li, Z.; Liu, J.; Wang, Y.; Liu, S.; Sun, M. Comparison between Thermal Hydrolysis and Enzymatic Proteolysis Processes for the Preparation of Tilapia Skin Collagen Hydrolysates. Czech J. Food Sci. 2013, 31, 1–4. [Google Scholar] [CrossRef]
- Ji, C.; Han, J.; Zhang, J.; Hu, J.; Fu, Y.; Qi, H.; Sun, Y.; Yu, C. Omics-prediction of bioactive peptides from the edible cyanobacterium Arthrospira platensis proteome. J. Sci. Food Agric. 2018, 98, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Li-Chan, E.C.Y. Bioactive peptides and protein hydrolysates: Research trends and challenges for application as nutraceuticals and functional food ingredients. Curr. Opin. Food Sci. 2015, 1, 28–37. [Google Scholar] [CrossRef]
- Cottage, Y.J.K.E.A. Bioinformatics methods to predict protein structure and function. A practical approach. Mol. Biotechnol. 2003, 23, 139–166. [Google Scholar] [CrossRef]
- Tachapuripunya, V.; Roytrakul, S.; Chumnanpuen, P. Unveiling Putative Functions of Mucus Proteins and Their Tryptic Peptides in Seven Gastropod Species Using Comparative Proteomics and Machine Learning-Based Bioinformatics Predictions. Molecules 2021, 26, 3475. [Google Scholar] [CrossRef] [PubMed]
- Latimer, G.W., Jr. Official Methods of Analysis of AOAC International, 20th ed.; AOAC: Rockville, ML, USA, 2016. [Google Scholar]
- Phetchthumrongchai, T.; Chuchird, N.; Roytrakul, S.; Chintong, S.; Klaypradit, W. Physical, chemical composition and umami compound of dried immature and mature roes of skipjack tuna (Katsuwonus pelamis). Fish. Aquat. Sci. 2022, 25, 390–402. [Google Scholar] [CrossRef]
- Kedare, S.B.; Singh, R.P. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 2011, 48, 412–422. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-evans, C. Antioxidant activity applying an improved abts radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, Q.; Zhu, X.; Lin, T.; Hao, D.; Xu, J. Antioxidant activities of Clerodendrum cyrtophyllum Turcz leaf extracts and their major components. PLoS ONE 2020, 15, e0234435. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, H.J.; Yoon, I.S.; Lee, G.W.; Kim, J.S.; Heu, M.S. Protein functionality of concentrates prepared from yellowfin tuna (Thunnus albacares) roe by cook-dried process. Food Sci. Biotechnol. 2016, 25, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Wubshet, S.G.; Lindberg, D.; Veiseth-Kent, E.; Kristoffersen, K.A.; Böcker, U.; Washburn, K.E.; Afseth, N.K. Bioanalytical Aspects in Enzymatic Protein Hydrolysis of By-Products. In Proteins: Sustainable Source, Processing and Applications; Academic Press: Cambridge, MA, USA, 2019; pp. 225–258. [Google Scholar]
- Kiettiolarn, M.; Kitsanayanyong, L.; Maneerote, J.; Unajak, S.; Tepwong, P. Optimization and production of protein hydrolysate containing antioxidant activity from tuna cooking juice concentrate by response surface methodology. Fish. Aquat. Sci. 2022, 25, 335–349. [Google Scholar] [CrossRef]
- Lee, H.J.; Roy, V.C.; Ho, T.C.; Park, J.S.; Jeong, Y.R.; Lee, S.C.; Kim, S.Y.; Chun, B.S. Amino Acid Profiles and Biopotentiality of Hydrolysates Obtained from Comb Penshell (Atrina pectinata) Viscera Using Subcritical Water Hydrolysis. Mar. Drugs 2021, 19, 137. [Google Scholar] [CrossRef] [PubMed]
- Kohlmeier, M. Amino Acids and Nitrogen Compounds. In Nutrient Metabolism; Academic Press: Cambridge, MA, USA, 2015; pp. 265–477. [Google Scholar]
- Papet, I.; Rémond, D.; Dardevet, D.; Mosoni, L.; Polakof, S.; Peyron, M.-A.; Savary-Auzeloux, I. Sulfur Amino Acids and Skeletal Muscle. In Nutrition and Skeletal Muscle; Academic Press: Cambridge, MA, USA, 2019; pp. 335–363. [Google Scholar]
- He, X.Q.; Cao, W.H.; Pan, G.K.; Yang, L.; Zhang, C.H. Enzymatic hydrolysis optimization of Paphia undulata and lymphocyte proliferation activity of the isolated peptide fractions. J. Sci. Food Agric. 2015, 95, 1544–1553. [Google Scholar] [CrossRef]
- Chi, C.F.; Hu, F.Y.; Wang, B.; Li, Z.R.; Luo, H.Y. Influence of Amino Acid Compositions and Peptide Profiles on Antioxidant Capacities of Two Protein Hydrolysates from Skipjack Tuna (Katsuwonus pelamis) Dark Muscle. Mar. Drugs 2015, 13, 2580–2601. [Google Scholar] [CrossRef]
- Klompong, V.; Benjakul, S.; Kantachote, D.; Shahidi, F. Antioxidative activity and functional properties of protein hydrolysate of yellow stripe trevally (Selaroides leptolepis) as influenced by the degree of hydrolysis and enzyme type. Food Chem. 2007, 102, 1317–1327. [Google Scholar] [CrossRef]
- Nalinanon, S.; Benjakul, S.; Kishimura, H.; Shahidi, F. Functionalities and antioxidant properties of protein hydrolysates from the muscle of ornate threadfin bream treated with pepsin from skipjack tuna. Food Chem. 2011, 124, 1354–1362. [Google Scholar] [CrossRef]
- Marcet, I.; Alvarez, C.; Paredes, B.; Diaz, M. The use of sub-critical water hydrolysis for the recovery of peptides and free amino acids from food processing wastes. Review of sources and main parameters. Waste Manag. 2016, 49, 364–371. [Google Scholar] [CrossRef]
- Lee, H.J.; Park, S.H.; Yoon, I.S.; Lee, G.-W.; Kim, Y.J.; Kim, J.-S.; Heu, M.S. Chemical composition of protein concentrate prepared from Yellowfin tuna Thunnus albacares roe by cook-dried process. Fish. Aquat. Sci. 2016, 19, 12. [Google Scholar] [CrossRef]
- Al-holy, M.A.; Rasco, B.A. Characterization of salmon (Oncorhynchus keta) and sturgeon (Acipenser transmontanus) caviar proteins. J. Food Biochem. 2006, 30, 422–428. [Google Scholar] [CrossRef]
- Chalamaiah, M.; Jyothirmayi, T.; Bhaskarachary, K.; Vajreswari, A.; Hemalatha, R.; Dinesh Kumar, B. Chemical composition, molecular mass distribution and antioxidant capacity of rohu (Labeo rohita) roe (egg) protein hydrolysates prepared by gastrointestinal proteases. Food Res. Int. 2013, 52, 221–229. [Google Scholar] [CrossRef]
- Thiansilakul, Y.; Benjakul, S.; Shahidi, F. Compositions, functional properties and antioxidative activity of protein hydrolysates prepared from round scad (Decapterus maruadsi). Food Chem. 2007, 103, 1385–1394. [Google Scholar] [CrossRef]
- Benjakul, S.; Yarnpakdee, S.; Senphan, T.; Halldorsdottir, S.M.; Kristinsson, H.G. Fish protein hydrolysates: Production, bioactivities, and applications. In Antioxidants and Functional Components in Aquatic Foods; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014. [Google Scholar]
- Yang, P.; Ke, H.; Hong, P.; Zeng, S.; Cao, W. Antioxidant activity of bigeye tuna (Thunnus obesus) head protein hydrolysate prepared with Alcalase. Int. J. Food Sci. Technol. 2011, 46, 2460–2466. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.M.; Li, L.Y.; Chi, C.F.; Wang, B. Twelve Antioxidant Peptides From Protein Hydrolysate of Skipjack Tuna (Katsuwonus pelamis) Roe Prepared by Flavourzyme: Purification, Sequence Identification, and Activity Evaluation. Front Nutr. 2021, 8, 813780. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, S.; Tachibana, Y. Active oxygen scavenging activity of egg-yolk protein hydrolysates and their effects on lipid oxidation in beef and tuna homogenates. Food Chem. 2006, 95, 243–249. [Google Scholar] [CrossRef]
- Binsan, W.; Benjakul, S.; Visessanguan, W.; Roytrakul, S.; Tanaka, M.; Kishimura, H. Antioxidative activity of Mungoong, an extract paste, from the cephalothorax of white shrimp (Litopenaeus vannamei). Food Chem. 2008, 106, 185–193. [Google Scholar] [CrossRef]
- Senphan, T.; Benjakul, S. Antioxidative activities of hydrolysates from seabass skin prepared using protease from hepatopancreas of Pacific white shrimp. J. Funct. Foods 2014, 6, 147–156. [Google Scholar] [CrossRef]
- Idowu, A.T.; Benjakul, S.; Sinthusamran, S.; Sookchoo, P.; Kishimura, H. Protein hydrolysate from salmon frames: Production, characteristics and antioxidative activity. J. Food Biochem. 2019, 43, e12734. [Google Scholar] [CrossRef] [PubMed]
- Klomklao, S.; Benjakul, S. Protein Hydrolysates Prepared from the Viscera of Skipjack Tuna (Katsuwonus pelmamis): Antioxidative Activity and Functional Properties. Turk. J. Fish. Aquat. Sci. 2018, 18, 69–79. [Google Scholar] [CrossRef]
- Gómez-Ruiz, J.Á.; López-Expósito, I.; Pihlanto, A.; Ramos, M.; Recio, I. Antioxidant activity of ovine casein hydrolysates: Identification of active peptides by HPLC–MS/MS. Eur. Food Res. Technol. 2008, 227, 1061–1067. [Google Scholar] [CrossRef]
- Avelar, Z.; Vicente, A.A.; Saraiva, J.A.; Rodrigues, R.M. The role of emergent processing technologies in tailoring plant protein functionality: New insights. Trends Food Sci. Technol. 2021, 113, 219–231. [Google Scholar] [CrossRef]
- Klomklao, S.; Benjakul, S.; Kishimura, H. Functional properties and antioxidative activity of protein hydrolysates from toothed ponyfish muscle treated with viscera extract from hybrid catfish. Int. J. Food Sci. Technol. 2013, 48, 1483–1489. [Google Scholar] [CrossRef]
- McClements, D.J.; Grossmann, L. The science of plant-based foods: Constructing next-generation meat, fish, milk, and egg analogs. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4049–4100. [Google Scholar] [CrossRef] [PubMed]
- Wouters, A.G.B.; Rombouts, I.; Fierens, E.; Brijs, K.; Delcour, J.A. Relevance of the Functional Properties of Enzymatic Plant Protein Hydrolysates in Food Systems. Compr. Rev. Food Sci. Food Saf. 2016, 15, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Vogelsang-O’Dwyer, M.; Sahin, A.W.; Arendt, E.K.; Zannini, E. Enzymatic Hydrolysis of Pulse Proteins as a Tool to Improve Techno-Functional Properties. Foods 2022, 11, 1307. [Google Scholar] [CrossRef]
- Mokni Ghribi, A.; Maklouf Gafsi, I.; Sila, A.; Blecker, C.; Danthine, S.; Attia, H.; Bougatef, A.; Besbes, S. Effects of enzymatic hydrolysis on conformational and functional properties of chickpea protein isolate. Food Chem. 2015, 187, 322–330. [Google Scholar] [CrossRef] [PubMed]
- García Arteaga, V.; Apéstegui Guardia, M.; Muranyi, I.; Eisner, P.; Schweiggert-Weisz, U. Effect of enzymatic hydrolysis on molecular weight distribution, techno-functional properties and sensory perception of pea protein isolates. Innov. Food Sci. Emerg. Technol. 2020, 65, 102449. [Google Scholar] [CrossRef]
- Foegeding, E.A.; Davis, J.P. Food protein functionality: A comprehensive approach. Food Hydrocoll. 2011, 25, 1853–1864. [Google Scholar] [CrossRef]
- Liu, Q.; Kong, B.; Xiong, Y.L.; Xia, X. Antioxidant activity and functional properties of porcine plasma protein hydrolysate as influenced by the degree of hydrolysis. Food Chem. 2010, 118, 403–410. [Google Scholar] [CrossRef]
- Zayas, J.F. Foaming Properties of Proteins. In Functionality of Proteins in Food; Springer: Berlin/Heidelberg, Germany, 1997. [Google Scholar]
- Mauer, L. PROTEIN|Heat Treatment for Food Proteins. In Encyclopedia of Food Science and Nutrition, 2nd ed.; Caballero, B., Ed.; Academic Press: Oxford, UK, 2003; pp. 4868–4872. [Google Scholar]
- Nongonierma, A.; FitzGerald, R.; Nongonierma, A.B.; FitzGerald, R.J. Enzymes exogenous to milk in dairy technology|Proteinases. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 289–296. [Google Scholar]
- Daliri, E.B.; Oh, D.H.; Lee, B.H. Bioactive Peptides. Foods 2017, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Ucak, I.; Afreen, M.; Montesano, D.; Carrillo, C.; Tomasevic, I.; Simal-Gandara, J.; Barba, F.J. Functional and Bioactive Properties of Peptides Derived from Marine Side Streams. Mar. Drugs 2021, 19, 71. [Google Scholar] [CrossRef]
- Panjaitan, F.C.A.; Gomez, H.L.R.; Chang, Y.W. In Silico Analysis of Bioactive Peptides Released from Giant Grouper (Epinephelus lanceolatus) Roe Proteins Identified by Proteomics Approach. Molecules 2018, 23, 2910. [Google Scholar] [CrossRef]
- Song, R.; Wei, R.; Zhang, B.; Wang, D. Optimization of the Antibacterial Activity of Half-Fin Anchovy (Setipinna taty) Hydrolysates. Food Bioproc. Technol. 2011, 5, 1979–1989. [Google Scholar] [CrossRef]
- Bergsson, G.; Agerberth, B.; Jornvall, H.; Gudmundsson, G.H. Isolation and identification of antimicrobial components from the epidermal mucus of Atlantic cod (Gadus morhua). FEBS J. 2005, 272, 4960–4969. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Go, H.J.; Kim, Y.J.; Park, N.G. Antimicrobial function of the GAPDH-related antimicrobial peptide in the skin of skipjack tuna, Katsuwonus pelamis. Fish Shellfish Immunol. 2014, 36, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Natrah, F.M.; Defoirdt, T.; Sorgeloos, P.; Bossier, P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar. Biotechnol. 2011, 13, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef]
- Kondo, E.; Iioka, H.; Saito, K. Tumor-homing peptide and its utility for advanced cancer medicine. Cancer Sci. 2021, 112, 2118–2125. [Google Scholar] [CrossRef] [PubMed]
- Parn, K.; Eriste, E.; Langel, U. The Antimicrobial and Antiviral Applications of Cell-Penetrating Peptides. Methods Mol. Biol. 2015, 1324, 223–245. [Google Scholar] [CrossRef]
- Athira, P.P.; Anooja, V.V.; Anju, M.V.; Neelima, S.; Archana, K.; Muhammed Musthafa, S.; Antony, S.P.; Bright Singh, I.S.; Philip, R. A hepatic antimicrobial peptide, hepcidin from Indian major carp, Catla catla: Molecular identification and functional characterization. J. Genet. Eng. Biotechnol. 2022, 20, 49. [Google Scholar] [CrossRef]
- Cheung, R.C.; Ng, T.B.; Wong, J.H. Marine Peptides: Bioactivities and Applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef]
- Roomi, M.W.; Shanker, N.; Niedzwiecki, A.; Rath, M. Induction of Apoptosis in the Human Prostate Cancer Cell Line DU-145 by a Novel Micronutrient Formulation. Open J. Apoptosis 2015, 4, 11–21. [Google Scholar] [CrossRef][Green Version]
- Hsu, K.-C.; Li-Chan, E.C.Y.; Jao, C.-L. Antiproliferative activity of peptides prepared from enzymatic hydrolysates of tuna dark muscle on human breast cancer cell line MCF-7. Food Chem. 2011, 126, 617–622. [Google Scholar] [CrossRef]
- Ishak, N.H.; Sarbon, N.M. A Review of Protein Hydrolysates and Bioactive Peptides Deriving from Wastes Generated by Fish Processing. Food Bioproc. Technol. 2017, 11, 2–16. [Google Scholar] [CrossRef]
- Nurdiani, R.; Vasiljevic, T.; Singh, T.K.; Donkor, O.N. Bioactive peptides from fish by-products with anticarcinogenic potential. Int. Food Res. J. 2016, 24, 1840–1849. [Google Scholar]
- Kemp, D.C.; Kwon, J.Y. Fish and Shellfish-Derived Anti-Inflammatory Protein Products: Properties and Mechanisms. Molecules 2021, 26, 3225. [Google Scholar] [CrossRef]
- Calder, P.C.; Ahluwalia, N.; Albers, R.; Bosco, N.; Bourdet-Sicard, R.; Haller, D.; Holgate, S.T.; Jonsson, L.S.; Latulippe, M.E.; Marcos, A.; et al. A consideration of biomarkers to be used for evaluation of inflammation in human nutritional studies. Br. J. Nutr. 2013, 109 (Suppl. S1), S1–S34. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Shu, W.; Shen, Y.; Sun, Q.; Bai, F.; Wang, J.; Li, D.; Li, Y.; Jin, W.; Yuan, L. Sturgeon protein-derived peptides exert anti-inflammatory effects in LPS-stimulated RAW264.7 macrophages via the MAPK pathway. J. Funct. Foods 2020, 72, 104044. [Google Scholar] [CrossRef]
- Phadke, G.G.; Rathod, N.B.; Ozogul, F.; Elavarasan, K.; Karthikeyan, M.; Shin, K.H.; Kim, S.K. Exploiting of Secondary Raw Materials from Fish Processing Industry as a Source of Bioactive Peptide-Rich Protein Hydrolysates. Mar. Drugs 2021, 19, 480. [Google Scholar] [CrossRef]
- Ahn, C.B.; Cho, Y.S.; Je, J.Y. Purification and anti-inflammatory action of tripeptide from salmon pectoral fin byproduct protein hydrolysate. Food Chem. 2015, 168, 151–156. [Google Scholar] [CrossRef]
- Suo, S.K.; Zheng, S.L.; Chi, C.F.; Luo, H.Y.; Wang, B. Novel angiotensin-converting enzyme inhibitory peptides from tuna byproducts-milts: Preparation, characterization, molecular docking study, and antioxidant function on H2O2-damaged human umbilical vein endothelial cells. Front. Nutr. 2022, 9, 957778. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Wan, P.; Chen, H.; Huang, J.; Ye, Z.; Chen, D.; Pan, J. Purification and Identification of Novel Myeloperoxidase Inhibitory Antioxidant Peptides from Tuna (Thunnas albacares) Protein Hydrolysates. Molecules 2022, 27, 2681. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.L.; Luo, Q.B.; Suo, S.K.; Zhao, Y.Q.; Chi, C.F.; Wang, B. Preparation, Identification, Molecular Docking Study and Protective Function on HUVECs of Novel ACE Inhibitory Peptides from Protein Hydrolysate of Skipjack Tuna Muscle. Mar. Drugs 2022, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Behera, A.; Das, R.; Patnaik, P.; Mohanty, J.; Mohanty, G. A review on fish peptides isolated from fish waste with their potent bioactivities. J. Appl. Biol. Biotechnol. 2022, 10, 195–209. [Google Scholar] [CrossRef]


| Amino Acid | Amino Acid Content (g/100 g Protein) | Group | |||||
|---|---|---|---|---|---|---|---|
| RAC | RA0.5 | RA1.0 | RT0.5 | RT1.0 | |||
| Essential amino acids (EAA) | |||||||
| Histidine | HIS | 2.98 ± 0.03 | 3.47 ± 0.10 | 3.52 ± 0.07 | 3.22 ± 0.13 | 3.26 ± 0.09 | HPL |
| Isoleucine | ILE | 1.69 ± 0.01 | 3.11 ± 0.01 | 3.05 ± 0.03 | 2.56 ± 0.03 | 3.18 ± 0.15 | HPB |
| Leucine | LEU | 5.30 ± 0.03 | 7.81 ± 0.01 | 8.03 ± 0.02 | 7.17 ± 0.05 | 7.71 ± 0.29 | HPB |
| Lysine | LYS | 5.46 ± 0.03 | 6.23 ± 0.09 | 6.01 ± 0.04 | 6.27 ± 0.05 | 6.02 ± 0.03 | HPL |
| Methionine | MET | 2.19 ± 0.01 | 2.76 ± 0.01 | 2.84 ± 0.08 | 2.62 ± 0.04 | 2.65 ± 0.06 | HPB |
| Phenylalanine | PHE | 2.96 ± 0.00 | 4.04 ± 0.01 | 4.28 ± 0.03 | 4.24 ± 0.06 | 4.26 ± 0.05 | HPB |
| Threonine | THR | 2.07 ± 0.02 | 2.44 ± 0.01 | 2.29 ± 0.19 | 2.09 ± 0.03 | 2.09 ± 0.08 | HPL |
| Valine | VAL | 5.44 ± 0.02 | 3.79 ± 0.06 | 3.64 ± 0.06 | 3.84 ± 0.03 | 3.64 ± 0.01 | HPB |
| Tryptophan | TRP | 2.15 ± 0.01 | 1.94 ± 0.09 | 2.22 ± 0.09 | 1.95 ± 0.13 | 2.31 ± 0.19 | HPB |
| Non-essential amino acids (NEAA) | |||||||
| Alanine | ALA | 5.19 ± 0.01 | 5.90 ± 0.01 | 5.98 ± 0.01 | 5.58 ± 0.05 | 5.85 ± 0.17 | HPB |
| Arginine | ARG | 10.87 ± 0.05 | 11.20 ± 0.07 | 11.18 ± 0.07 | 12.08 ± 0.09 | 11.82 ± 0.06 | HPL |
| Aspartic | ASP | 2.70 ± 0.05 | 2.80 ± 0.00 | 2.89 ± 0.02 | 2.81 ± 0.04 | 3.10 ± 0.02 | HPL |
| Cysteine | CYS | 20.65 ± 0.14 | 18.14 ± 0.20 | 17.53 ± 0.22 | 18.13 ± 0.23 | 16.26 ± 0.24 | HPB |
| Glutamic | GLU | 9.04 ± 0.03 | 9.03 ± 0.01 | 9.03 ± 0.03 | 8.97 ± 0.10 | 9.83 ± 0.01 | HPL |
| Glycine | GLY | 13.61 ± 0.04 | 10.88 ± 0.03 | 10.67 ± 0.13 | 10.97 ± 0.05 | 10.80 ± 0.04 | HPL |
| Serine | SER | 5.25 ± 0.01 | 4.61 ± 0.00 | 4.74 ± 0.02 | 4.98 ± 0.05 | 4.77 ± 0.08 | HPL |
| Tyrosine | TYR | 2.45 ± 0.00 | 1.85 ± 0.01 | 2.10 ± 0.02 | 2.52 ± 0.03 | 2.45 ± 0.08 | HPB |
| Total (%) | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | ||
| EAA (%) | 30.24 | 35.59 | 35.88 | 33.96 | 35.12 | ||
| NEAA (%) | 69.76 | 64.41 | 64.12 | 66.04 | 64.88 | ||
| HPB (%) | 48.02 | 49.34 | 49.67 | 48.61 | 48.31 | ||
| HPL (%) | 51.98 | 50.66 | 50.33 | 51.39 | 51.69 | ||
| Sample | Solubility (%) | Foaming Capacity (FC) (%) | Foam Stability (FS) (%) | ||
|---|---|---|---|---|---|
| FS 15 (min) | FS 30 (min) | FS 45 (min) | |||
| RAC | 95.15 ± 0.83 a | 148.44 ± 1.02 a | 123.80 ± 0.65 aA | 99.03 ± 1.43 aB | 57.05 ± 1.57 bC |
| RA0.5 | 91.57 ± 0.77 b | 141.78 ± 1.02 b | 77.84 ± 0.78 bA | 77.16 ± 1.03 bA | 69.52 ± 1.64 aB |
| RA1.0 | 92.72 ± 0.65 b | 147.33 ± 0.67 a | 79.36 ± 0.66 bA | 78.59 ± 0.67 bA | 70.18 ± 1.43 aB |
| RT0.5 | 89.65 ± 1.26 c | 120.89 ± 1.68 c | 41.80 ± 1.92 cA | 41.24 ± 1.92 cA | 38.45 ± 1.95 cA |
| RT1.0 | 91.73 ± 0.94 b | 119.33 ± 1.33 c | 41.57 ± 2.74 cA | 40.97 ± 2.08 cA | 38.58 ± 0.00 cA |
| Sample | Property | Number of Peptide | |
|---|---|---|---|
| RAC | Anti-hypertensive | 111 | (20.98%) |
| RA0.5 | 95 | (17.96%) | |
| RA1.0 | 92 | (17.39%) | |
| RT0.5 | 184 | (34.78%) | |
| RT1.0 | 192 | (36.29%) | |
| RAC | Anti-virus | 46 | (8.70%) |
| RA0.5 | 43 | (8.13%) | |
| RA1.0 | 40 | (7.56%) | |
| RT0.5 | 75 | (14.18%) | |
| RT1.0 | 80 | (15.12%) | |
| RAC | Anti-parasite | 18 | (3.40%) |
| RA0.5 | 26 | (4.91%) | |
| RA1.0 | 18 | (3.40%) | |
| RT0.5 | 49 | (9.26%) | |
| RT1.0 | 46 | (8.70%) | |
| Type of Peptide | Amino Acid Sequence | Property |
|---|---|---|
| Octa-peptides | MLRASAMR | Anti-hypertensive Anti-parasite Drug delivering |
| Deca-peptides | CDSTSTLCLR | Anti-hypertensive Cell communicating Tumor homing |
| SENQDAQMEK | Anti-hypertensive Anti-parasite Cell communicating | |
| EHKSLTGTAR | Anti-hypertensive Drug delivering Anti-quorum sensing | |
| Tetradeca-peptides | ADVIFKNESLYSHR | Anti-cancer Anti-hypertensive Anti-parasite |
| Pentadeca-peptides | MNLGDATTRPPVGRR | Anti-hypertensive Cell communicating Anti-quorum sensing |
| Docosa-peptides | LHIQWLEAQEQHQQQEAQLSSR | Anti-hypertensive Drug delivering Tumor homing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phetchthumrongchai, T.; Tachapuripunya, V.; Chintong, S.; Roytrakul, S.; E-kobon, T.; Klaypradit, W. Properties of Protein Hydrolysates and Bioinformatics Prediction of Peptides Derived from Thermal and Enzymatic Process of Skipjack Tuna (Katsuwonus pelamis) Roe. Fishes 2022, 7, 255. https://doi.org/10.3390/fishes7050255
Phetchthumrongchai T, Tachapuripunya V, Chintong S, Roytrakul S, E-kobon T, Klaypradit W. Properties of Protein Hydrolysates and Bioinformatics Prediction of Peptides Derived from Thermal and Enzymatic Process of Skipjack Tuna (Katsuwonus pelamis) Roe. Fishes. 2022; 7(5):255. https://doi.org/10.3390/fishes7050255
Chicago/Turabian StylePhetchthumrongchai, Thithi, Viroj Tachapuripunya, Sutasinee Chintong, Sittiruk Roytrakul, Teerasak E-kobon, and Wanwimol Klaypradit. 2022. "Properties of Protein Hydrolysates and Bioinformatics Prediction of Peptides Derived from Thermal and Enzymatic Process of Skipjack Tuna (Katsuwonus pelamis) Roe" Fishes 7, no. 5: 255. https://doi.org/10.3390/fishes7050255
APA StylePhetchthumrongchai, T., Tachapuripunya, V., Chintong, S., Roytrakul, S., E-kobon, T., & Klaypradit, W. (2022). Properties of Protein Hydrolysates and Bioinformatics Prediction of Peptides Derived from Thermal and Enzymatic Process of Skipjack Tuna (Katsuwonus pelamis) Roe. Fishes, 7(5), 255. https://doi.org/10.3390/fishes7050255







