Transcriptome Analysis of Immune Response against Streptococcus agalactiae Infection in the Nile Tilapia GIFT Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sampling
2.2. Paraffin Sections of the Gill
2.3. RNA Isolate and Sequencing
2.4. RNA Reads Processing and Mapping
2.5. Annotation
2.6. Differential Expression Analysis and Enrichment Analysis
2.7. PPI (Protein–Protein Interaction)
2.8. Real-Time RT-PCR Confirmation of Illumina Sequencing Data
3. Results
3.1. RNA Sequencing Results
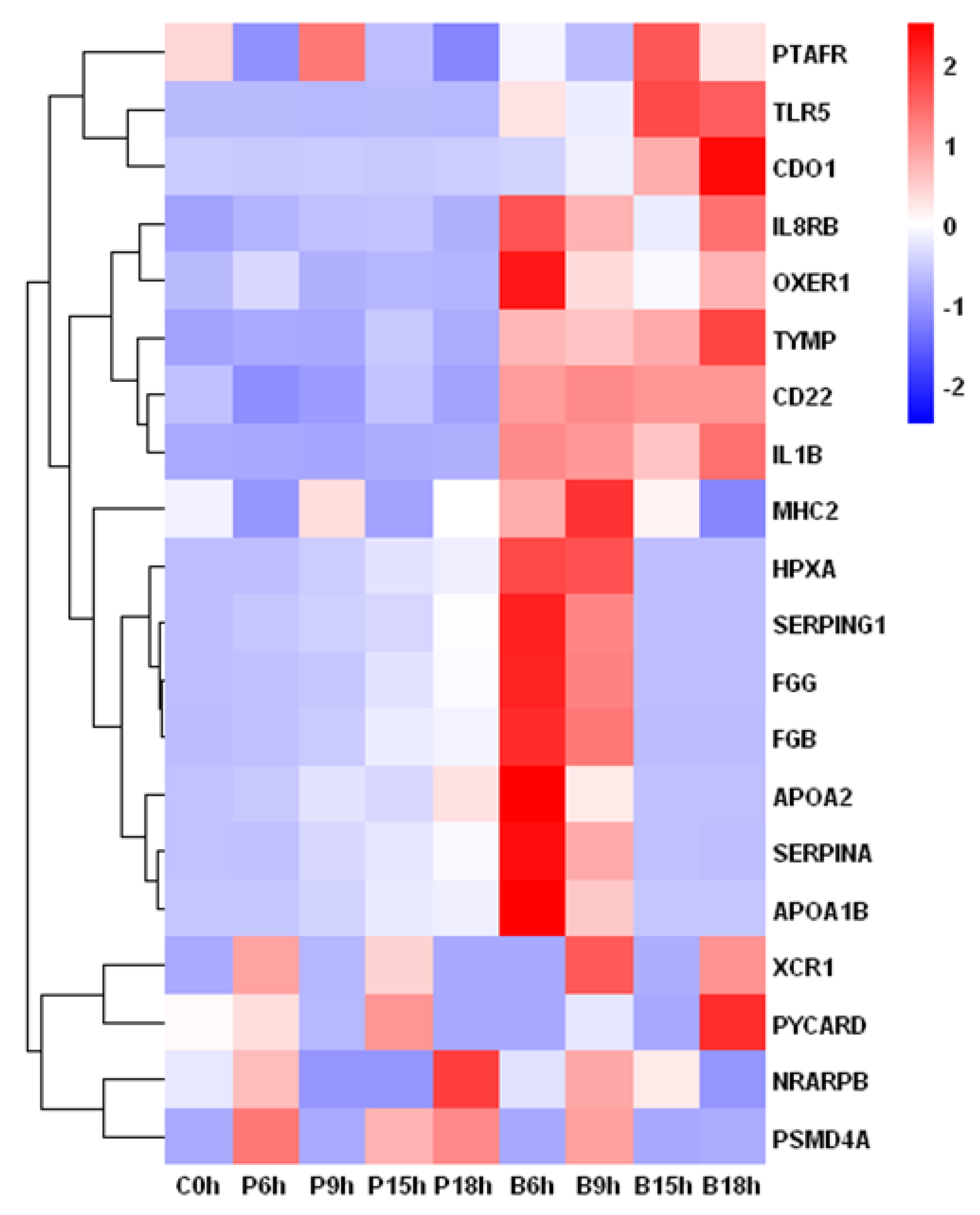
3.2. Differential Gene Expression Analysis
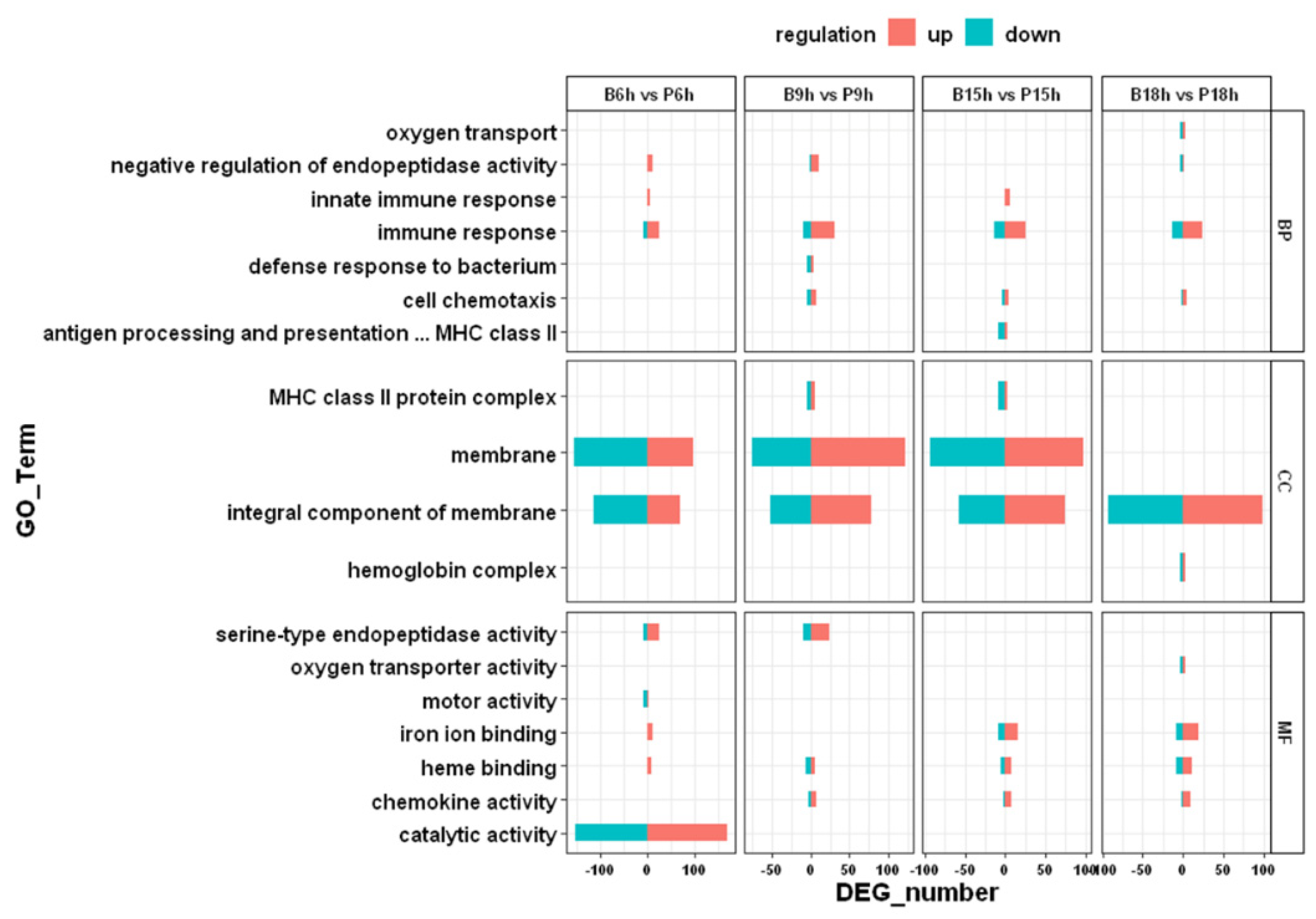
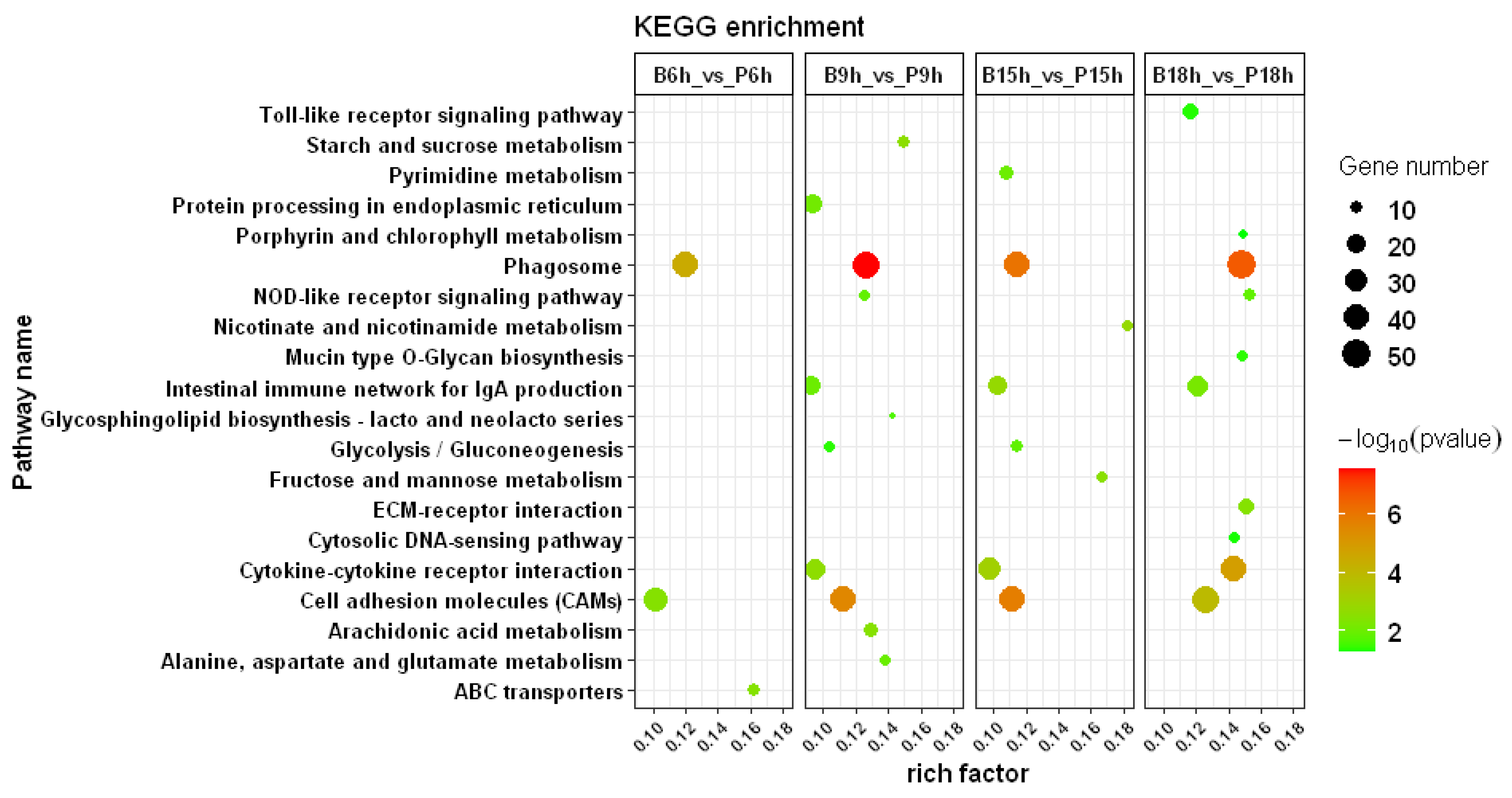
3.3. GO and KEGG Enrichment Analysis
3.4. Immune-Related Pathways after Infection
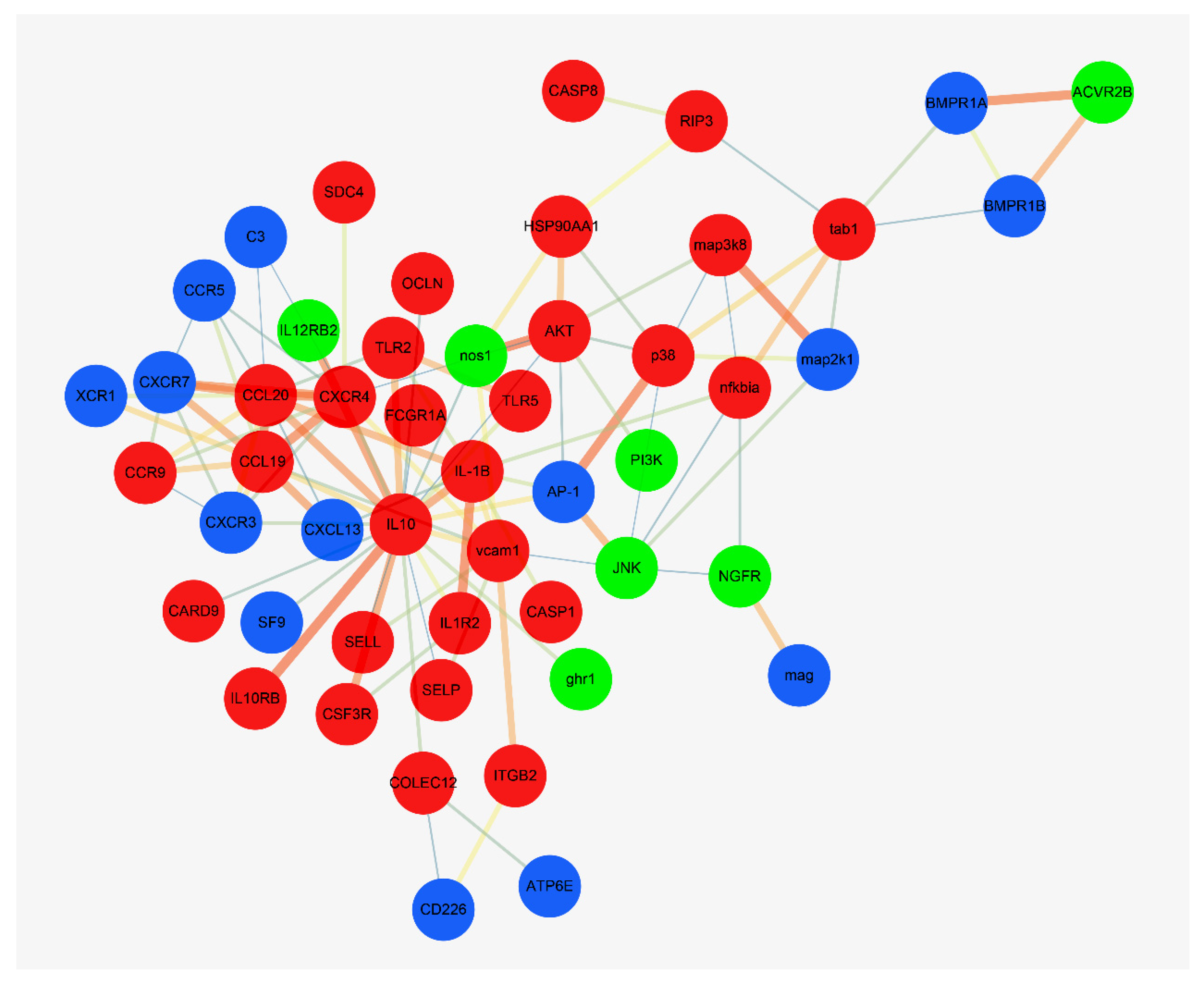
3.5. Interaction Network Analysis of DEGs
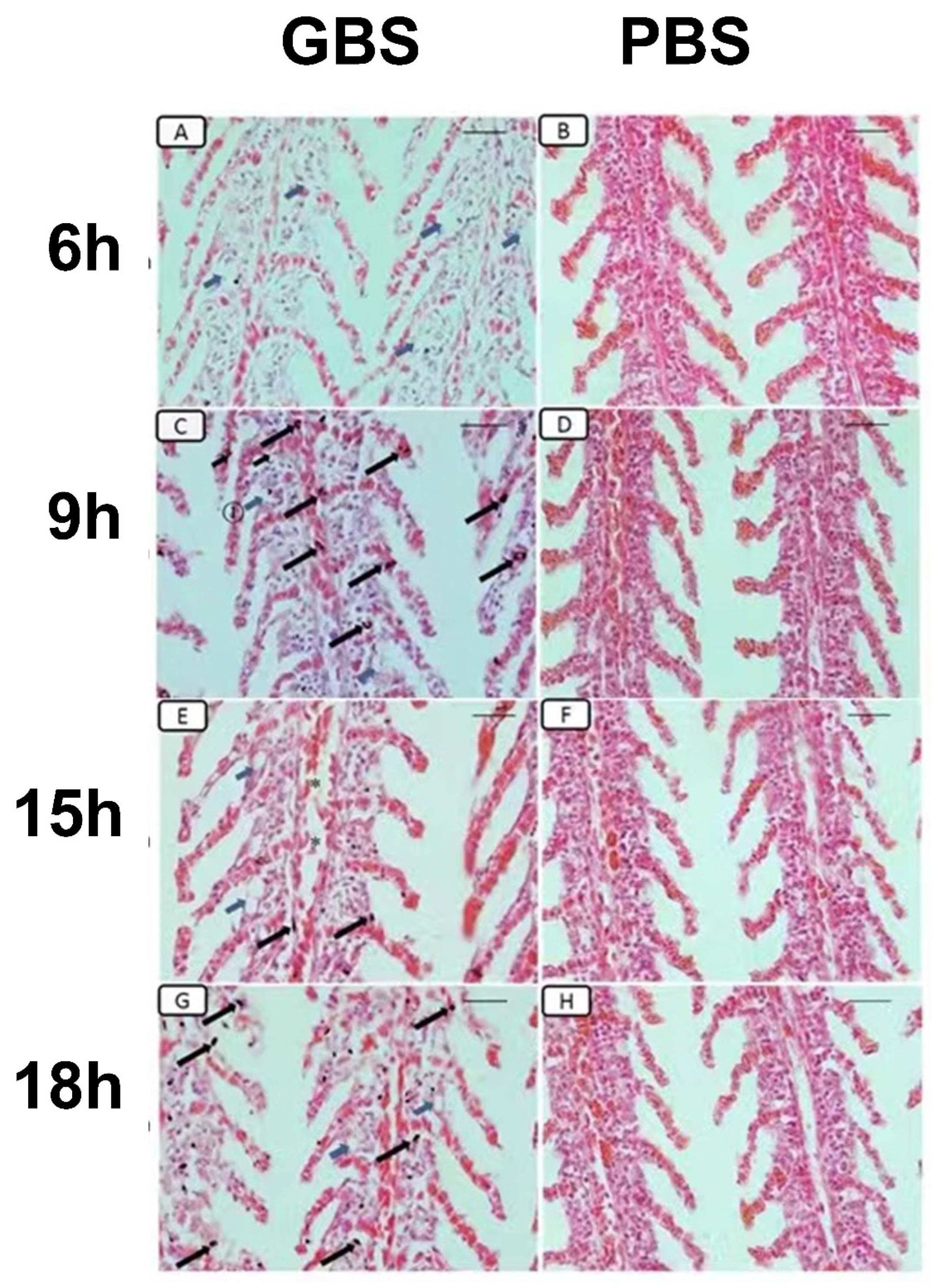
3.6. Histopathological Changes Induced by S. agalactiae Infection
3.7. qRT-PCR of Immune-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdullah, S.; Omar, N.; Yusoff, S.M.; Obukwho, B.; Nwunuji, T.P.; Hanan, L.; Samad, J. Clinicopathological Features and Immunohistochemical Detection of Antigens in Acute Experimental Streptococcus Agalactiae Infection in Red Tilapia (Oreochromis spp.). SpingerPlus 2013, 2, 286. [Google Scholar] [CrossRef]
- Shi, Y.H.; Chen, K.; Ma, W.J.; Chen, J. Ayu C-Reactive Protein/Serum Amyloid P Agglutinates Bacteria and Inhibits Complement-Mediated Opsonophagocytosis by Monocytes/Macrophages. Fish Shellfish Immunol. 2018, 76, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, A.; Nikookhah, F.; Nikpour, S.; Majiian, F.; Gholami, M. Isolation of Streptococci from Milk Samples of Normal, Acute and Subclinical Mastitis Cows and Determination of Their Antibiotic Susceptibility Patterns. Pak. J. Biol. Sci. 2008, 11, 148–150. [Google Scholar] [CrossRef]
- Lämmler, C.; Abdulmawjood, A.; Weiss, R. Properties of Serological Group B Streptococci of Dog, Cat and Monkey Origin. Zent. Vet. B 1998, 45, 561–566. [Google Scholar] [CrossRef]
- Santi, M.; Egan, S.A.; Leigh, J.A.; Verner-Jeffreys, D.W.; Blanchard, A.M. Complete Genome Sequence of Streptococcus Agalactiae Strain 01173, Isolated from Kuwaiti Wild Fish. Microbiol. Resour. Announc. 2021, 9, e00674-20. [Google Scholar] [CrossRef]
- Rosa-Fraile, M.; Sampedro, A.; Varela, J.; Garcia-Peña, M.; Gimenez-Gallego, G. Identification of a Peptide from Mammal Albumins Responsible for Enhanced Pigment Production by Group B Streptococci. Clin. Diagn. Lab. Immunol. 1999, 6, 425–426. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Ulett, G.C. Evaluation of Hematogenous Spread and Ascending Infection in the Pathogenesis of Acute Pyelonephritis Due to Group B Streptococcus in Mice. Microb. Pathog. 2020, 138, 103796. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, A.O.; Lämmler, C.; Weiss, R. Identification and Characterization of Streptococcus Agalactiae Isolated from Horses. Vet. Microbiol. 2002, 85, 31–35. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Luo, Z.; Wang, R.; Huang, T.; Liang, W.; Gu, Q.; Yu, F.; Chen, M. Arginine Deiminase and Biotin Metabolism Signaling Pathways Play an Important Role in Human-Derived Serotype V, ST1 Streptococcus Agalactiae Virulent Strain upon Infected Tilapia. Animals 2020, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.J.; Klesius, P.H.; Gilbert, P.M.; Shoemaker, C.A.; al Sarawi, M.A.; Landsberg, J.; Duremdez, R.; al Marzouk, A.; al Zenki, S. Characterization of β-Haemolytic Group B Streptococcus Agalactiae in Cultured Seabream, Sparus auratus L., and Wild Mullet, Liza Klunzingeri (Day), in Kuwait. J. Fish Dis. 2002, 25, 505–513. [Google Scholar] [CrossRef]
- Hernandez, E.; Figueroa, J.; Iregui, C. Streptococcosis on a Red Tilapia, Oreochromis sp., Farm: A Case Study. J. Fish Dis. 2009, 32, 247–252. [Google Scholar] [CrossRef]
- Pulido Bravo, E.; Iregui, C.; Figueroa, J.; Klesius, P. Streptococosis in cultured Tilapias (Oreochromis spp.) in Colombia. Revista AquaTIC 2004, 20, 97–106. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2014; FAO: Rome, Italy, 2011; Volume 4, pp. 40–41. [Google Scholar]
- Zhang, W.B.; Murray, F.J.; Liu, L.P.; Little, D.C. A Comparative Analysis of Four Internationally Traded Farmed Seafood Commodities in China: Domestic and International Markets as Key Drivers. Rev. Aquac. 2017, 9, 157–178. [Google Scholar] [CrossRef]
- Basri, L.; Nor, R.M.; Salleh, A.; Md Yasin, I.S.; Saad, M.Z.; Abd Rahaman, N.Y.; Barkham, T.; Amal, M.N.A. Co-Infections of Tilapia Lake Virus, Aeromonas Hydrophila and Streptococcus Agalactiae in Farmed Red Hybrid Tilapia. Animals 2020, 10, 2141. [Google Scholar] [CrossRef] [PubMed]
- Maina, J.N. Structure, Function and Evolution of the Gas Exchangers: Comparative Perspectives. J. Anat. 2002, 201, 281–304. [Google Scholar] [CrossRef] [PubMed]
- Koppang, E.O.; Kvellestad, A.; Fischer, U. Fish Mucosal Immunity: Gill. In Mucosal Health Aquac; Elsevier Inc: Amsterdam, The Netherlands, 2015; pp. 93–133. [Google Scholar] [CrossRef]
- Wilson, J.M.; Laurent, P. Fish Gill Morphology: Inside Out. J. Exp. Zool. 2002, 293, 192–213. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The Mucosal Immune System of Fish: The Evolution of Tolerating Commensals While Fighting Pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Baums, C.G.; Hermeyer, K.; Leimbach, S.; Adamek, M.; Czerny, C.P.; Hörstgen-Schwark, G.; Valentin-Weigand, P.; Baumgärtner, W.; Steinhagen, D. Establishment of a Model of Streptococcus Iniae Meningoencephalitis in Nile Tilapia (Oreochromis Niloticus). J. Comp. Pathol. 2013, 149, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Huang, P.Y.; Chen, H.M.; Wang, Y.H.; Tsai, I.A.; Lu, C.C.; Chen, C.C. Genetic and Pathogenic Difference between Streptococcus Agalactiae Serotype Ia Fish and Human Isolates. BMC Microbiol. 2016, 16, 175. [Google Scholar] [CrossRef] [PubMed]
- Junior, J.A.F.; Leal, C.A.G.; de Oliveira, T.F.; Nascimento, K.A.; de Macêdo, J.T.S.A.; Pedroso, P.M.O. Anatomopathological Characterization and Etiology of Lesions on Nile Tilapia Fillets (Oreochromis niloticus) Caused by Bacterial Pathogens. Aquaculture 2020, 526, 735387. [Google Scholar] [CrossRef]
- Liang, F.-R.; Wang, Q.-Q.; Jiang, Y.-L.; Yue, B.-Y.; Zhou, Q.-Z.; Wang, J.-H. Characterization of Matrix Metalloprotease-9 Gene from Nile Tilapia (Oreochromis niloticus) and Its High-Level Expression Induced by the Streptococcus Agalactiae Challenge. Biomolecules 2020, 10, 76. [Google Scholar] [CrossRef]
- Phan-Aram, P.; Mahasri, G.; Kayansamruaj, P.; Amparyup, P.; Srisapoome, P. Immune Regulation, but Not Antibacterial Activity, Is a Crucial Function of Hepcidins in Resistance against Pathogenic Bacteria in Nile Tilapia (Oreochromis niloticus Linn.). Biomolecules 2020, 10, 1132. [Google Scholar] [CrossRef]
- Mann, E.R.; Li, X. Intestinal Antigen-Presenting Cells in Mucosal Immune Homeostasis: Crosstalk between Dendritic Cells, Macrophages and B-Cells. World J. Gastroenterol. 2014, 20, 9653–9664. [Google Scholar] [CrossRef]
- Young, M.D.; Smyth, G.K.; Oshlack, A.; Wakefield, M.J. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated Genome Annotation and Pathway Identification Using the KEGG Orthology (KO) as a Controlled Vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Dezfuli, B.S.; Giari, L.; Lui, A.; Lorenzoni, M.; Noga, E.J. Mast Cell Responses to Ergasilus (Copepoda), a Gill Ectoparasite of Sea Bream. Fish Shellfish Immunol. 2011, 30, 1087–1094. [Google Scholar] [CrossRef]
- Yamamoto-Furusho, J.K.; Barnich, N.; Hisamatsu, T.; Podolsky, D.K. MDP-NOD2 Stimulation Induces HNP-1 Secretion, Which Contributes to NOD2 Antibacterial Function. Inflamm. Bowel Dis. 2010, 16, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Park, S.J.; Kwon, M.J.; Jeong, T.S.; Bok, S.H.; Choi, W.Y.; Jeong, W.I.; Ryu, S.Y.; Do, S.H.; Lee, C.S.; et al. Quercetin Suppresses Proinflammatory Cytokines Production through MAP Kinases AndNF-KappaB Pathway in Lipopolysaccharide-Stimulated Macrophage. Mol. Cell. Biochem. 2003, 243, 153–160. [Google Scholar] [CrossRef]
- Abrahams, V.M. The Role of the Nod-like Receptor Family in Trophoblast Innate Immune Responses. J. Reprod. Immunol. 2011, 88, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Feng, L.; Jiang, W.D.; Wu, P.; Kuang, S.Y.; Jiang, J.; Tang, L.; Tang, W.N.; Zhang, Y.A.; Liu, Y.; et al. Copper-Induced Tight Junction MRNA Expression Changes, Apoptosis and Antioxidant Responses via NF-KappaB, TOR and Nrf2 Signaling Molecules in the Gills of Fish: Preventive Role of Arginine. Aquat. Toxicol. 2015, 158, 125–137. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, Y.; Luo, Y.; Zhong, H.; Huang, T.; Liang, W.; Xiao, J.; Wu, W.; Li, L.; Chen, M. Large-Scale Profiling of the Proteome and Dual Transcriptome in Nile Tilapia (Oreochromis niloticus) Challenged with Low- and High-Virulence Strains of Streptococcus Agalactiae. Fish Shellfish Immunol. 2020, 100, 386–396. [Google Scholar] [CrossRef]
- Zubair, S.; de Villiers, E.P.; Younan, M.; Andersson, G.; Tettelin, H.; Riley, D.R.; Jores, J.; Bongcam-Rudloff, E.; Bishop, R.P. Genome Sequences of Two Pathogenic Streptococcus Agalactiae Isolates from the One-Humped Camel Camelus Dromedarius. Genome Announc. 2013, 1, e00515-13. [Google Scholar] [CrossRef]
- Chen, L.; Feng, L.; Jiang, W.D.; Jiang, J.; Wu, P.; Zhao, J.; Kuang, S.Y.; Tang, L.; Tang, W.N.; Zhang, Y.A.; et al. Dietary Riboflavin Deficiency Decreases Immunity and Antioxidant Capacity, and Changes Tight Junction Proteins and Related Signaling Molecules MRNA Expression in the Gills of Young Grass Carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2015, 45, 307–320. [Google Scholar] [CrossRef]
- Evans, J.J.; Shoemaker, C.A.; Klesius, P.H. Density and Dose: Factors Affecting Mortality of Streptococcus Iniaeinfected Tilapia (Oreochromis niloticus). Aquaculture 2000, 188, 229–235. [Google Scholar]
- Wu, W.; Li, L.; Liu, Y.; Huang, T.; Liang, W.; Chen, M. Multiomics Analyses Reveal That NOD-like Signaling Pathway Plays an Important Role against Streptococcus agalactiae in the Spleen of Tilapia. Fish Shellfish Immunol. 2019, 95, 336–348. [Google Scholar] [CrossRef]
- Bai, H.; Zhou, T.; Zhao, J.; Chen, B.; Xu, P. Transcriptome Analysis Reveals the Temporal Gene Expression Patterns in Skin of Large Yellow Croaker (Larimichthys crocea) in Response to Cryptocaryon Irritans Infection. Fish Shellfish Immunol. 2020, 99, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Lacey, C.A.; Mitchell, W.J.; Dadelahi, A.S.; Skyberg, J.A. Caspase-1 and Caspase-11 Mediate Pyroptosis, Inflammation, and Control of Brucella Joint Infection. Infect. Immun. 2018, 86, e00361-18. [Google Scholar] [CrossRef]
- He, Y.; Wang, K.Y.; Xiao, D.; Chen, D.F.; Huang, L.; Liu, T.; Wang, J.; Geng, Y.; Wang, E.L.; Yang, Q. A Recombinant Truncated Surface Immunogenic Protein (TSip) plus Adjuvant FIA Confers Active Protection against Group B Streptococcus Infection in Tilapia. Vaccine 2014, 32, 7025–7032. [Google Scholar] [CrossRef]
- Bergsbaken, T.; Fink, S.L.; Cookson, B.T. Pyroptosis: Host Cell Death and Inflammation. Nat. Rev. Microbiol. 2009, 7, 99–109. [Google Scholar] [CrossRef]
- Whidbey, C.; Vornhagen, J.; Gendrin, C.; Boldenow, E.; Samson, J.M.; Doering, K.; Ngo, L.; Ezekwe, E.A.D.; Gundlach, J.H.; Elovitz, M.A.; et al. A Streptococcal Lipid Toxin Induces Membrane Permeabilization and Pyroptosis Leading to Fetal Injury. EMBO Mol. Med. 2015, 7, 488–505. [Google Scholar] [CrossRef]
- Vogel, S.N. How Discovery of Toll-Mediated Innate Immunity in Drosophila Impacted Our Understanding of TLR Signaling (and Vice Versa). J. Immunol. 2012, 188, 5207–5209. [Google Scholar] [CrossRef]
- Carranza, C.; Chavez-Galan, L. Several Routes to the Same Destination: Inhibition of Phagosome-Lysosome Fusion by Mycobacterium tuberculosis. Am. J. Med. Sci. 2019, 357, 184–194. [Google Scholar] [CrossRef]
- Raeesi, V.; Ehsani, A.; Torshizi, R.V. Genomic Loci Associated with Antibody-Mediated Immune Responses in an F2 Chicken Population. Animal 2019, 13, 1341–1349. [Google Scholar] [CrossRef]
- Feinberg, H.; Taylor, M.E.; Weis, W.I. Scavenger Receptor C-Type Lectin Binds to the Leukocyte Cell Surface Glycan Lewis(x) by a Novel Mechanism. J. Biol. Chem. 2007, 282, 17250–17258. [Google Scholar] [CrossRef]
- Awuh, J.A.; Flo, T.H. Molecular Basis of Mycobacterial Survival in Macrophages. Cell. Mol. Life Sci. 2017, 74, 1625–1648. [Google Scholar] [CrossRef]



| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Amplicon Length (bp) |
|---|---|---|---|
| akt2l | CTCGGCTTATCTTTGACCTCTG | CTCCTTCACCAGGATCACTTTAC | 103 |
| casp3a | TGGACGATACAGACGCAAAG | TGGACGATACAGACGCAAAG | 114 |
| CD22 | GGACAGTTGGTGTATAGGAGTTG | CATCTACTCACCTGGCGTTATT | 121 |
| CXCR4 | CGTTTACTCACTAGTCCCACAG | ACCATCCCACACACAAGTT | 100 |
| hsp90aa1 | CTTGTCTGCCACGGTTCTAA | TACTCCGTCTGGTCTTCCTT | 99 |
| ITGB2 | CCAAGCACTGGTGAGAAAGA | CAGCAGTGGATTAGACGACAG | 111 |
| map2k1 | GGGTCTGATTATGGCAAGGAA | GTGCACAAACACTGGAGTAATG | 103 |
| nfkbiab | GACTTCCTCAACAGACAGAATGA | TCTGGAGAGGGACAGGTATTT | 116 |
| tab1 | CTTGACGTCTCCGATATCCTTC | GCCTTCTTATACCCGTCTTTCT | 102 |
| vcam1 | CAGTCTGTGCACTCCCTTTAG | ACCTCCTCACCCGGAATTA | 107 |
| β-actin | GATCTGGCATCACACCTTCTAC | TCTTCTCCCTGTTGGCTTTG | 104 |
| Sample | Total Clean Reads | Total Mapped Reads | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| C0h | 54,708,466 | 43,788,477 (80.04%) | 48.76 | 97.41 | 93.67 |
| B6h | 52,297,476 | 43,565,354 (83.30%) | 48.80 | 97.55 | 93.92 |
| B9h | 36,636,616 | 31,095,881 (84.88%) | 49.68 | 97.62 | 94.00 |
| B15h | 46,424,536 | 38,965,528 (83.93%) | 49.19 | 97.58 | 93.91 |
| B18h | 53,185,078 | 45,678,704 (85.89%) | 48.26 | 97.77 | 94.23 |
| P6h | 47,948,880 | 41,022,046 (85.55%) | 49.37 | 97.85 | 94.40 |
| P9h | 47,849,040 | 41,310,833 (86.34%) | 48.68 | 97.85 | 94.44 |
| P15h | 51,421,080 | 43,273,464 (84.16%) | 49.78 | 97.57 | 94.03 |
| P18h | 54,731,996 | 46,234,883 (84.48%) | 49.50 | 97.43 | 93.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, T.; Fang, Z.; Duarte, D.F.C.; Fernandes, S.A.; Lu, Y.; Guo, J.; Gui, L.; Chen, L. Transcriptome Analysis of Immune Response against Streptococcus agalactiae Infection in the Nile Tilapia GIFT Strain. Fishes 2022, 7, 246. https://doi.org/10.3390/fishes7050246
Zhou T, Fang Z, Duarte DFC, Fernandes SA, Lu Y, Guo J, Gui L, Chen L. Transcriptome Analysis of Immune Response against Streptococcus agalactiae Infection in the Nile Tilapia GIFT Strain. Fishes. 2022; 7(5):246. https://doi.org/10.3390/fishes7050246
Chicago/Turabian StyleZhou, Tao, Zhihua Fang, Daniel F. C. Duarte, Stefan A. Fernandes, Ying Lu, Jing Guo, Lang Gui, and Liangbiao Chen. 2022. "Transcriptome Analysis of Immune Response against Streptococcus agalactiae Infection in the Nile Tilapia GIFT Strain" Fishes 7, no. 5: 246. https://doi.org/10.3390/fishes7050246
APA StyleZhou, T., Fang, Z., Duarte, D. F. C., Fernandes, S. A., Lu, Y., Guo, J., Gui, L., & Chen, L. (2022). Transcriptome Analysis of Immune Response against Streptococcus agalactiae Infection in the Nile Tilapia GIFT Strain. Fishes, 7(5), 246. https://doi.org/10.3390/fishes7050246





